Abstract
In the title molecular salt, [NH2(CH2CH3)2][H2PO4], two unique types of cations and anions, which are configurationally very similar, are present in the asymmetric unit. Both ions form sheets approximately parallel to (-1-1) linked by weak hydrogen bonds. The interconnection within and between the sheets is reinforced by O—H⋯O and N—H⋯O hydrogen bonds involving the tetrahedral H2PO4 anions and the ammonium groups.
Related literature
For preparative details, see: Hanna et al. (1999 ▶). For related structures, see: Averbuch-Pouchot et al. (1987 ▶); Held (2003 ▶). 
Experimental
Crystal data
C4H12N+·H2PO4 −
M r = 171.13
Triclinic,

a = 8.3643 (6) Å
b = 8.8308 (15) Å
c = 11.6446 (12) Å
α = 88.219 (10)°
β = 83.649 (7)°
γ = 79.700 (7)°
V = 841.00 (18) Å3
Z = 4
Mo Kα radiation
μ = 0.29 mm−1
T = 295 K
0.30 × 0.28 × 0.26 mm
Data collection
Nonius MACH3 diffractometer
Absorption correction: ψ scan (North et al., 1968 ▶) T min = 0.858, T max = 0.998
10831 measured reflections
5096 independent reflections
3164 reflections with I > 2σ(I)
R int = 0.037
3 standard reflections every 100 reflections intensity decay: −6.3%
Refinement
R[F 2 > 2σ(F 2)] = 0.040
wR(F 2) = 0.119
S = 0.98
5096 reflections
181 parameters
H-atom parameters constrained
Δρmax = 0.37 e Å−3
Δρmin = −0.39 e Å−3
Data collection: CAD-4 (Enraf–Nonius, 1989 ▶); cell refinement: CAD-4; data reduction: WinGX (Farrugia, 2012 ▶); program(s) used to solve structure: SIR97 (Altomare et al., 1999 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ATOMS (Dowty, 2002 ▶) and ORTEP-3 for Windows (Farrugia, 2012 ▶)’; software used to prepare material for publication: publCIF (Westrip, 2010 ▶).
Supplementary Material
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S1600536814000464/wm2794sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536814000464/wm2794Isup2.hkl
CCDC reference: http://scripts.iucr.org/cgi-bin/cr.cgi?rm=csd&csdid=980665
Additional supporting information: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O13—H13⋯O21i | 0.82 | 1.78 | 2.5851 (19) | 166 |
| O14—H14⋯O12ii | 0.82 | 1.83 | 2.6058 (19) | 158 |
| O24—H24⋯O22iii | 0.82 | 1.95 | 2.585 (2) | 133 |
| O23—H23⋯O11i | 0.82 | 1.84 | 2.620 (2) | 158 |
| N1—H1A⋯O22iv | 0.90 | 1.88 | 2.779 (2) | 174 |
| N1—H1B⋯O21v | 0.90 | 1.87 | 2.769 (2) | 177 |
| N2—H2A⋯O11vi | 0.90 | 1.87 | 2.714 (2) | 155 |
| N2—H2B⋯O12 | 0.90 | 1.91 | 2.795 (2) | 168 |
Symmetry codes: (i)  ; (ii)
; (ii)  ; (iii)
; (iii)  ; (iv)
; (iv)  ; (v)
; (v)  ; (vi)
; (vi)  .
.
supplementary crystallographic information
1. Comment
In the course of a systematic search for new 'double salts' of simple secondary amines and monovalent cations of various inorganic acids (Averbuch-Pouchot et al., 1987), the new structure of (C2N2H10)Li2(SO4)2 was described (Held, 2003). In continuation of these studies, sulfuric acid has been replaced with phosphoric acid in order to get an analogous lithium compound with a tetrahedral phosphoric unit. Moreover, ethylenediamine has been replaced with diethylamine. Surprisingly, lithium was not incorporated in the solid product and only the title compound, [NH2(CH2CH3)2]+[H2PO4]-, was finally obtained, the crystal structure of which is reported herein.
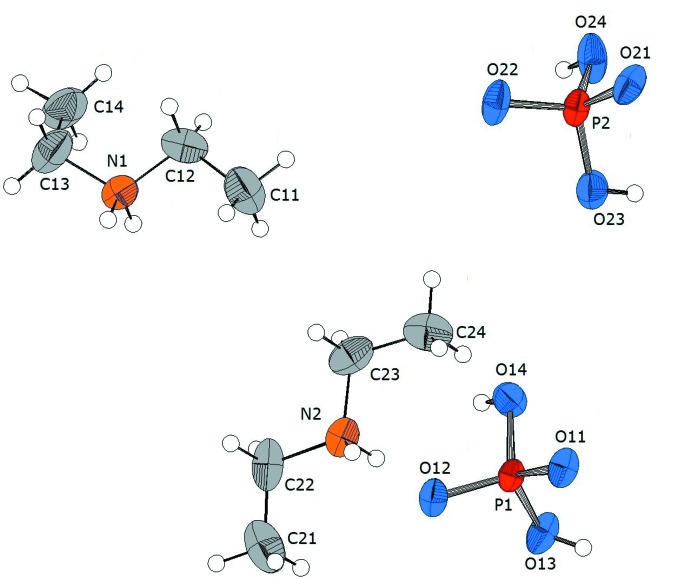
The crystal structure of [NH2(CH2CH3)2]+[H2PO4]- consists of diethylammonium cations, NH2(CH2CH3)2+, and dihydrogen orthophosphate anions, H2PO4- (Fig. 1). The ions form sheets approximately parallel to (112). The interconnection within and between the sheets is reinforced by a hydrogen bonding system between the tetrahedral dihydrogen orthophosphate groups on one hand and between the ammonium function and the H2PO4- units on the other (Figs. 2,3; Table 1).
2. Experimental
The title compound was obtained by reaction of an aqueous solution of lithium dihydrogenposphate with diethylammine in a stoichiometric ratio 1:1 (Hanna et al., 1999). The solution was kept at room temperature by cooling. The title compound crystallized by slow evaporation of the solvent at room temperature in form of colourless crystals with dimensions up to 4 mm within a few days.
Differential scanning calorimetry with a PerkinElmer DSC7 device in the temperature range from 183 K up to 293 K showed no significant feature.
3. Refinement
The H atoms were clearly discernible from difference Fourier maps. However, to all hydrogen atoms riding model contraints were applied in the least squares refinement, with C—H = 0.96 Å for methyl H atoms (Uiso(H) = 1.5Ueq(C)), with C—H = 0.97 (Uiso(H) = 1.2Ueq(C)) for methylene H atoms, with N—H = 0.90 Å (Uiso(H) = 1.2Ueq(N)) and with O—H = 0.82 Å (Uiso(H) = 1.2Ueq(O)).
Figures
Fig. 1.
The molecular entities in the structure of [NH2(CH2CH3)2]+[H2PO4]-, showing the atom-numbering scheme. Displacement ellipsoids are drawn at the 50% probability level. H labels were omitted for clarity.
Fig. 2.
The unit cell of [NH2(CH2CH3)2]+[H2PO4]- with colour scheme: N (orange), O (blue), [H2PO4]-tetrahedra (blue), P (red), C grey) and H (white). Hydrogen bonds (light blue) between H2PO4 tetrahedra and hydrogen bonds (orange) between ammonium groups and H2PO4 tetrahedra are shown.
Fig. 3.
Clinographic projection of eight unit cells of [NH2(CH2CH3)2]+[H2PO4]-. The ionic units form sheets approximately parallel to (112). Hydrogen bonds (grey) interconnect the sheets.
Crystal data
| C4H12N+·H2PO4− | Z = 4 |
| Mr = 171.13 | F(000) = 368 |
| Triclinic, P1 | Dx = 1.352 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation, λ = 0.71073 Å |
| a = 8.3643 (6) Å | Cell parameters from 25 reflections |
| b = 8.8308 (15) Å | θ = 21.0–26.0° |
| c = 11.6446 (12) Å | µ = 0.29 mm−1 |
| α = 88.219 (10)° | T = 295 K |
| β = 83.649 (7)° | Parallelepiped, colourless |
| γ = 79.700 (7)° | 0.30 × 0.28 × 0.26 mm |
| V = 841.00 (18) Å3 |
Data collection
| Nonius MACH3 diffractometer | 3164 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.037 |
| Graphite monochromator | θmax = 30.4°, θmin = 2.5° |
| ω/2θ scans | h = −11→11 |
| Absorption correction: ψ scan (North et al., 1968) | k = −12→12 |
| Tmin = 0.858, Tmax = 0.998 | l = −16→16 |
| 10831 measured reflections | 3 standard reflections every 100 reflections |
| 5096 independent reflections | intensity decay: −6.3% |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.040 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.119 | H-atom parameters constrained |
| S = 0.98 | w = 1/[σ2(Fo2) + (0.0553P)2 + 0.1855P] where P = (Fo2 + 2Fc2)/3 |
| 5096 reflections | (Δ/σ)max < 0.001 |
| 181 parameters | Δρmax = 0.37 e Å−3 |
| 0 restraints | Δρmin = −0.39 e Å−3 |
Special details
| Experimental. A suitable single-crystal was carefully selected under a polarizing microscope and mounted in a glass capillary. |
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| P1 | 0.16567 (5) | 0.37327 (5) | 0.60452 (4) | 0.02736 (12) | |
| O11 | 0.33132 (15) | 0.27994 (16) | 0.61779 (12) | 0.0362 (3) | |
| O12 | 0.16402 (16) | 0.53915 (15) | 0.56977 (12) | 0.0365 (3) | |
| O13 | 0.05039 (16) | 0.36883 (18) | 0.71998 (12) | 0.0426 (4) | |
| H13 | 0.0986 | 0.3119 | 0.7669 | 0.064* | |
| O14 | 0.08747 (18) | 0.29249 (16) | 0.51243 (13) | 0.0418 (3) | |
| H14 | −0.0034 | 0.3416 | 0.5037 | 0.063* | |
| P2 | 0.67897 (6) | −0.09624 (6) | 0.11311 (4) | 0.03332 (13) | |
| O21 | 0.83801 (17) | −0.20760 (18) | 0.10779 (12) | 0.0431 (4) | |
| O22 | 0.68455 (18) | 0.04631 (17) | 0.03943 (13) | 0.0454 (4) | |
| O23 | 0.6256 (2) | −0.04343 (17) | 0.24046 (13) | 0.0502 (4) | |
| H23 | 0.6138 | −0.1185 | 0.2816 | 0.075* | |
| O24 | 0.54703 (19) | −0.18437 (17) | 0.07620 (15) | 0.0540 (4) | |
| H24 | 0.4588 | −0.1259 | 0.0784 | 0.081* | |
| C11 | 0.2499 (4) | 0.5728 (3) | −0.0009 (3) | 0.0663 (7) | |
| H11A | 0.3010 | 0.4677 | −0.0143 | 0.100* | |
| H11B | 0.1790 | 0.5792 | 0.0702 | 0.100* | |
| H11C | 0.3322 | 0.6346 | 0.0040 | 0.100* | |
| C12 | 0.1521 (3) | 0.6303 (3) | −0.0985 (2) | 0.0551 (6) | |
| H12A | 0.2234 | 0.6223 | −0.1705 | 0.066* | |
| H12B | 0.0700 | 0.5668 | −0.1043 | 0.066* | |
| N1 | 0.0712 (2) | 0.7927 (2) | −0.08051 (14) | 0.0389 (4) | |
| H1A | 0.1462 | 0.8472 | −0.0629 | 0.047* | |
| H1B | −0.0049 | 0.7962 | −0.0192 | 0.047* | |
| C13 | −0.0092 (4) | 0.8696 (3) | −0.1815 (2) | 0.0633 (7) | |
| H13A | −0.0518 | 0.9767 | −0.1634 | 0.076* | |
| H13B | 0.0719 | 0.8668 | −0.2481 | 0.076* | |
| C14 | −0.1460 (4) | 0.7938 (4) | −0.2114 (2) | 0.0674 (8) | |
| H14A | −0.1939 | 0.8467 | −0.2759 | 0.101* | |
| H14B | −0.2275 | 0.7978 | −0.1461 | 0.101* | |
| H14C | −0.1040 | 0.6884 | −0.2311 | 0.101* | |
| C21 | 0.2697 (4) | 0.9059 (3) | 0.4832 (3) | 0.0767 (9) | |
| H21A | 0.2125 | 1.0088 | 0.4722 | 0.115* | |
| H21B | 0.3793 | 0.9091 | 0.4988 | 0.115* | |
| H21C | 0.2144 | 0.8581 | 0.5472 | 0.115* | |
| C22 | 0.2739 (3) | 0.8160 (3) | 0.3772 (2) | 0.0530 (6) | |
| H22A | 0.3283 | 0.8650 | 0.3122 | 0.064* | |
| H22B | 0.1632 | 0.8138 | 0.3608 | 0.064* | |
| N2 | 0.36206 (19) | 0.65586 (19) | 0.39194 (14) | 0.0387 (4) | |
| H2A | 0.4643 | 0.6596 | 0.4076 | 0.046* | |
| H2B | 0.3118 | 0.6126 | 0.4535 | 0.046* | |
| C23 | 0.3717 (3) | 0.5556 (3) | 0.2904 (2) | 0.0524 (6) | |
| H23A | 0.4293 | 0.5993 | 0.2242 | 0.063* | |
| H23B | 0.2622 | 0.5520 | 0.2721 | 0.063* | |
| C24 | 0.4584 (3) | 0.3952 (3) | 0.3128 (2) | 0.0599 (7) | |
| H24A | 0.4627 | 0.3337 | 0.2456 | 0.090* | |
| H24B | 0.4005 | 0.3509 | 0.3773 | 0.090* | |
| H24C | 0.5675 | 0.3983 | 0.3297 | 0.090* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| P1 | 0.0192 (2) | 0.0322 (2) | 0.0307 (2) | −0.00441 (17) | −0.00520 (17) | 0.00718 (18) |
| O11 | 0.0191 (6) | 0.0458 (8) | 0.0417 (8) | −0.0025 (5) | −0.0037 (5) | 0.0122 (6) |
| O12 | 0.0325 (7) | 0.0347 (7) | 0.0448 (8) | −0.0102 (6) | −0.0119 (6) | 0.0110 (6) |
| O13 | 0.0254 (7) | 0.0588 (9) | 0.0371 (7) | 0.0040 (6) | 0.0011 (6) | 0.0160 (7) |
| O14 | 0.0397 (8) | 0.0365 (7) | 0.0507 (9) | −0.0017 (6) | −0.0193 (7) | −0.0035 (6) |
| P2 | 0.0281 (2) | 0.0350 (3) | 0.0388 (3) | −0.00980 (19) | −0.0092 (2) | 0.0130 (2) |
| O21 | 0.0312 (7) | 0.0590 (9) | 0.0344 (7) | 0.0011 (6) | −0.0025 (6) | 0.0143 (7) |
| O22 | 0.0428 (8) | 0.0464 (8) | 0.0549 (9) | −0.0229 (7) | −0.0227 (7) | 0.0253 (7) |
| O23 | 0.0586 (10) | 0.0409 (8) | 0.0447 (9) | 0.0048 (7) | −0.0021 (7) | 0.0069 (7) |
| O24 | 0.0470 (9) | 0.0392 (8) | 0.0850 (12) | −0.0202 (7) | −0.0334 (8) | 0.0228 (8) |
| C11 | 0.0626 (17) | 0.0470 (14) | 0.092 (2) | −0.0091 (12) | −0.0194 (16) | −0.0012 (14) |
| C12 | 0.0502 (14) | 0.0593 (15) | 0.0571 (15) | −0.0138 (11) | 0.0012 (11) | −0.0197 (12) |
| N1 | 0.0366 (9) | 0.0481 (10) | 0.0361 (9) | −0.0195 (8) | −0.0036 (7) | 0.0022 (7) |
| C13 | 0.0762 (19) | 0.0782 (18) | 0.0452 (14) | −0.0365 (15) | −0.0179 (13) | 0.0191 (13) |
| C14 | 0.0759 (19) | 0.081 (2) | 0.0537 (15) | −0.0226 (15) | −0.0304 (14) | 0.0031 (14) |
| C21 | 0.074 (2) | 0.0466 (15) | 0.103 (2) | 0.0002 (14) | 0.0004 (18) | 0.0028 (16) |
| C22 | 0.0306 (10) | 0.0525 (13) | 0.0738 (17) | −0.0069 (9) | −0.0045 (10) | 0.0280 (12) |
| N2 | 0.0265 (8) | 0.0485 (10) | 0.0427 (9) | −0.0117 (7) | −0.0050 (7) | 0.0104 (8) |
| C23 | 0.0418 (12) | 0.0802 (18) | 0.0401 (12) | −0.0225 (12) | −0.0078 (10) | 0.0027 (12) |
| C24 | 0.0503 (14) | 0.0718 (18) | 0.0595 (16) | −0.0205 (13) | 0.0058 (12) | −0.0170 (13) |
Geometric parameters (Å, º)
| P1—O11 | 1.5013 (13) | C13—C14 | 1.501 (4) |
| P1—O12 | 1.5056 (14) | C13—H13A | 0.9700 |
| P1—O14 | 1.5673 (14) | C13—H13B | 0.9700 |
| P1—O13 | 1.5691 (14) | C14—H14A | 0.9600 |
| O13—H13 | 0.8200 | C14—H14B | 0.9600 |
| O14—H14 | 0.8200 | C14—H14C | 0.9600 |
| P2—O21 | 1.5027 (14) | C21—C22 | 1.482 (4) |
| P2—O22 | 1.5060 (14) | C21—H21A | 0.9600 |
| P2—O23 | 1.5613 (16) | C21—H21B | 0.9600 |
| P2—O24 | 1.5624 (15) | C21—H21C | 0.9600 |
| O23—H23 | 0.8200 | C22—N2 | 1.487 (3) |
| O24—H24 | 0.8200 | C22—H22A | 0.9700 |
| C11—C12 | 1.497 (4) | C22—H22B | 0.9700 |
| C11—H11A | 0.9600 | N2—C23 | 1.485 (3) |
| C11—H11B | 0.9600 | N2—H2A | 0.9000 |
| C11—H11C | 0.9600 | N2—H2B | 0.9000 |
| C12—N1 | 1.483 (3) | C23—C24 | 1.501 (4) |
| C12—H12A | 0.9700 | C23—H23A | 0.9700 |
| C12—H12B | 0.9700 | C23—H23B | 0.9700 |
| N1—C13 | 1.503 (3) | C24—H24A | 0.9600 |
| N1—H1A | 0.9000 | C24—H24B | 0.9600 |
| N1—H1B | 0.9000 | C24—H24C | 0.9600 |
| O11—P1—O12 | 115.38 (8) | N1—C13—H13B | 109.1 |
| O11—P1—O14 | 107.71 (8) | H13A—C13—H13B | 107.9 |
| O12—P1—O14 | 109.31 (8) | C13—C14—H14A | 109.5 |
| O11—P1—O13 | 110.08 (7) | C13—C14—H14B | 109.5 |
| O12—P1—O13 | 108.20 (8) | H14A—C14—H14B | 109.5 |
| O14—P1—O13 | 105.74 (9) | C13—C14—H14C | 109.5 |
| P1—O13—H13 | 109.5 | H14A—C14—H14C | 109.5 |
| P1—O14—H14 | 109.5 | H14B—C14—H14C | 109.5 |
| O21—P2—O22 | 114.38 (9) | C22—C21—H21A | 109.5 |
| O21—P2—O23 | 109.41 (8) | C22—C21—H21B | 109.5 |
| O22—P2—O23 | 107.50 (9) | H21A—C21—H21B | 109.5 |
| O21—P2—O24 | 107.55 (9) | C22—C21—H21C | 109.5 |
| O22—P2—O24 | 110.16 (8) | H21A—C21—H21C | 109.5 |
| O23—P2—O24 | 107.65 (10) | H21B—C21—H21C | 109.5 |
| P2—O23—H23 | 109.5 | C21—C22—N2 | 110.6 (2) |
| P2—O24—H24 | 109.5 | C21—C22—H22A | 109.5 |
| C12—C11—H11A | 109.5 | N2—C22—H22A | 109.5 |
| C12—C11—H11B | 109.5 | C21—C22—H22B | 109.5 |
| H11A—C11—H11B | 109.5 | N2—C22—H22B | 109.5 |
| C12—C11—H11C | 109.5 | H22A—C22—H22B | 108.1 |
| H11A—C11—H11C | 109.5 | C23—N2—C22 | 114.70 (18) |
| H11B—C11—H11C | 109.5 | C23—N2—H2A | 108.6 |
| N1—C12—C11 | 110.87 (19) | C22—N2—H2A | 108.6 |
| N1—C12—H12A | 109.5 | C23—N2—H2B | 108.6 |
| C11—C12—H12A | 109.5 | C22—N2—H2B | 108.6 |
| N1—C12—H12B | 109.5 | H2A—N2—H2B | 107.6 |
| C11—C12—H12B | 109.5 | N2—C23—C24 | 111.64 (19) |
| H12A—C12—H12B | 108.1 | N2—C23—H23A | 109.3 |
| C12—N1—C13 | 115.31 (19) | C24—C23—H23A | 109.3 |
| C12—N1—H1A | 108.4 | N2—C23—H23B | 109.3 |
| C13—N1—H1A | 108.4 | C24—C23—H23B | 109.3 |
| C12—N1—H1B | 108.4 | H23A—C23—H23B | 108.0 |
| C13—N1—H1B | 108.4 | C23—C24—H24A | 109.5 |
| H1A—N1—H1B | 107.5 | C23—C24—H24B | 109.5 |
| C14—C13—N1 | 112.3 (2) | H24A—C24—H24B | 109.5 |
| C14—C13—H13A | 109.1 | C23—C24—H24C | 109.5 |
| N1—C13—H13A | 109.1 | H24A—C24—H24C | 109.5 |
| C14—C13—H13B | 109.1 | H24B—C24—H24C | 109.5 |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O13—H13···O21i | 0.82 | 1.78 | 2.5851 (19) | 166 |
| O14—H14···O12ii | 0.82 | 1.83 | 2.6058 (19) | 158 |
| O24—H24···O22iii | 0.82 | 1.95 | 2.585 (2) | 133 |
| O23—H23···O11i | 0.82 | 1.84 | 2.620 (2) | 158 |
| N1—H1A···O22iv | 0.90 | 1.88 | 2.779 (2) | 174 |
| N1—H1B···O21v | 0.90 | 1.87 | 2.769 (2) | 177 |
| N2—H2A···O11vi | 0.90 | 1.87 | 2.714 (2) | 155 |
| N2—H2B···O12 | 0.90 | 1.91 | 2.795 (2) | 168 |
Symmetry codes: (i) −x+1, −y, −z+1; (ii) −x, −y+1, −z+1; (iii) −x+1, −y, −z; (iv) −x+1, −y+1, −z; (v) x−1, y+1, z; (vi) −x+1, −y+1, −z+1.
Footnotes
Supporting information for this paper is available from the IUCr electronic archives (Reference: WM2794).
References
- Altomare, A., Burla, M. C., Camalli, M., Cascarano, G. L., Giacovazzo, C., Guagliardi, A., Moliterni, A. G. G., Polidori, G. & Spagna, R. (1999). J. Appl. Cryst. 32, 115–119.
- Averbuch-Pouchot, M. T., Durif, A. & Guitel, J.-C. (1987). Acta Cryst. C43, 1896–1898.
- Dowty, E. (2002). ATOMS Shape Software, Kingsport, Tennessee, USA.
- Enraf–Nonius (1989). CAD-4 Enraf–Nonius, Delft, The Netherlands.
- Farrugia, L. J. (2012). J. Appl. Cryst. 45, 849–854.
- Hanna, A. A., Ali, A. F. & Khalil, M. Sh. (1999). Indian J. Chem. Technol. 6, 43–47.
- Held, P. (2003). Z. Kristallogr. New Cryst. Struct. 218, 13–16.
- North, A. C. T., Phillips, D. C. & Mathews, F. S. (1968). Acta Cryst. A24, 351–359.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Westrip, S. P. (2010). J. Appl. Cryst. 43, 920–925.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S1600536814000464/wm2794sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536814000464/wm2794Isup2.hkl
CCDC reference: http://scripts.iucr.org/cgi-bin/cr.cgi?rm=csd&csdid=980665
Additional supporting information: crystallographic information; 3D view; checkCIF report