Abstract
In the title molecule, C14H17NO, the 5,5-dimethylcyclohex-2-enone moiety is attached to an aniline group, the dihedral angle subtended [54.43 (3)°] indicating a significant twist. The hexaneone ring has a half-chair conformation with the C atom bearing two methyl groups lying 0.6384 (8) Å above the plane of the five remaining atoms (r.m.s. deviation = 0.0107 Å). The crystal packing can be described as alternating layers parallel to (-101), which are consolidated by N—H⋯O hydrogen bonds and C—H⋯π interactions.
Related literature
For the synthesis of the title compound, see: Amini et al. (2013 ▶); Machacek et al. (2002 ▶). For its reactivity, see: Wang et al. (2007 ▶); Mohammadizadeh et al. (2009 ▶); Gao et al. (2008 ▶). For our previous work [inspired by Assy (1996 ▶)] on the preparation and the reactivity of imidazole derivatives, see: Zama et al. (2013a
▶,b
▶); Chelghoum et al. (2011 ▶); Bahnous et al. (2012 ▶) For enamine derivatives as precursors in the synthesis ofcompounds of pharmaceutical interest, see: Palko et al. (2008 ▶) Park & Jahng (1998 ▶); Tadesse et al. (1999 ▶); Thummel & Jahng (1985 ▶); When enamines are treated with alkyl halides, an alkylation occurs to give an iminium salt, see: Adams (2000 ▶); Kempf et al. (2003 ▶).
Experimental
Crystal data
C14H17NO
M r = 215.29
Monoclinic,

a = 10.1766 (19) Å
b = 13.159 (2) Å
c = 9.2877 (17) Å
β = 104.062 (7)°
V = 1206.5 (4) Å3
Z = 4
Mo Kα radiation
μ = 0.07 mm−1
T = 150 K
0.15 × 0.12 × 0.09 mm
Data collection
Bruker APEXII diffractometer
Absorption correction: multi-scan (SADABS; Sheldrick, 2002 ▶) T min = 0.667, T max = 0.747
12403 measured reflections
5021 independent reflections
3873 reflections with I > 2σ(I)
R int = 0.030
Refinement
R[F 2 > 2σ(F 2)] = 0.047
wR(F 2) = 0.138
S = 1.04
5021 reflections
147 parameters
H-atom parameters constrained
Δρmax = 0.45 e Å−3
Δρmin = −0.20 e Å−3
Data collection: APEX2 (Bruker, 2001 ▶); cell refinement: SAINT (Bruker, 2001 ▶); data reduction: SAINT; program(s) used to solve structure: SIR2002 (Burla et al., 2005 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 for Windows (Farrugia, 2012 ▶) and DIAMOND (Brandenburg & Berndt, 2001 ▶); software used to prepare material for publication: WinGX (Farrugia, 2012 ▶) and CRYSCAL (T. Roisnel, local program).
Supplementary Material
Crystal structure: contains datablock(s) I. DOI: 10.1107/S160053681400186X/bq2392sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S160053681400186X/bq2392Isup2.hkl
Supporting information file. DOI: 10.1107/S160053681400186X/bq2392Isup3.cml
CCDC reference: http://scripts.iucr.org/cgi-bin/cr.cgi?rm=csd&csdid=983637
Additional supporting information: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
Cg1 is the centroid of C10–C15 ring.
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1⋯O1i | 0.86 | 2.02 | 2.8587 (11) | 165 |
| C8—H8A⋯Cg1ii | 0.96 | 2.61 | 3.5547 (12) | 157 |
Symmetry codes: (i)  ; (ii)
; (ii)  .
.
Acknowledgments
Thanks are due to MESRS (Ministére de l’Enseignement Supérieur et de la Recherche Scientifique - Algeria) for financial support.
supplementary crystallographic information
1. Comment
The reaction of primary amines with ketones leads to imines; however, secondary amines give enamines. The preparation of enamines takes place when an aldehyde or ketone containing an α hydrogen is treated with a secondary amine. When enamines are treated with alkyl halides, an alkylation occurs to give an iminium salt via an electron transfer from the electron pair on nitrogen, through the C=C to the electrophilic carbon of the alkyl halide (Adams, 2000). In fact, an enamine behaves as a "nitrogen enolate" and generally react as carbon nucleophiles (Kempf et al., 2003). The use of enamine derivatives as intermediates in organic synthesis has been extensively investigated and they proved to be versatile precursors in the synthesis of a large variety of compounds of pharmaceutical interest (Tadesse et al., 1999; Park et al., 1998; Thummel & Jahng 1985; Palko et al., 2008). Inspired by the works of Assy 1996 and also as an extension of our ongoing research on the preparation and the reactivity of imidazole derivatives (Zama et al., 2013a; Zama et al., 2013b; Chelghoum et al., 2011; Bahnous et al., 2012), we describe herein the single-crystal X-ray structure of the enamine 5,5-dimethyl-3-(phenylamino)cyclohex-2-enone (I). This latter has been recovered from our attempt to coupling the dihydropyridine entity with imidazole unit using a "One Pot reaction" strategy implicating aniline, 5,5-dimethyl-1,3-cyclohexandione and N-methylimidazolmethylenemalononitrile. The molecular geometry and the atom-numbering scheme of (I) are shown in Fig. 1. The asymmetric unit of (I) consists of the 5,5-dimethyl-cyclohex-2-enone moiety and its attached phenylamino group. The crystal packing can be described as alternating layers parallel to the (-101)(Fig. 2). It is stabilized by N—H···O hydrogen bond (Fig.3) and C—H···π interactions (Table. 1). These interaction bonds link the molecules within the layers and also link the layers together, reinforcing the cohesion of the structure.
2. Experimental
5,5-dimethyl-3-(phenylamino)cyclohex-2-enone (I) has been obtained from the reaction of aniline and 5,5-dimethyl-1,3-cyclohexandione. In a typical reaction, 1 mmol of 5,5-dimethylcyclohex-1,3-dione, and 1 mmol aniline, in ethanol were placed in a 10 ml round-bottomed flask fitted with a condenser and a magnetic stirrer bar. The reaction mixture was stirred at reflux for 24 h, then 1 mmol of N-methylimidazolmethylenemalononitrile was added to this solution and the reaction mixture was heated for an additional 24 h. The solvent was distilled off and flash chromatographic purification furnished the 5,5-dimethyl-3-(phenylamino)cyclohex-2-enone I and the recovered N-methylimidazolmethylenemalononitrile. Suitable crystals for X-ray experiments of I were obtained by slow evaporation from an ethanol/CH2Cl2 solution at room temperature.
3. Refinement
Approximate positions for all the H atoms were first obtained from the difference electron density map. However, the H atoms were situated into idealized positions and the H-atoms have been refined within the riding atom approximation. The applied constraints were as follow: Caryl—Haryl = 0.93 Å; Cmethylene—Hmethylene = 0.97 Å; Cmethyl—Hmethyl = 0.96 Å and N—H = 0.86 Å; The idealized methyl group was allowed to rotate about the C—C bond during the refinement by application of the command AFIX 137 in SHELXL97 (Sheldrick, 2008). Uiso(Hmethyl) = 1.5Ueq(Cmethyl) or Uiso(Haryl, methylene, amine) = 1.2 Ueq(Caryl, methylene or N).
Figures
Fig. 1.
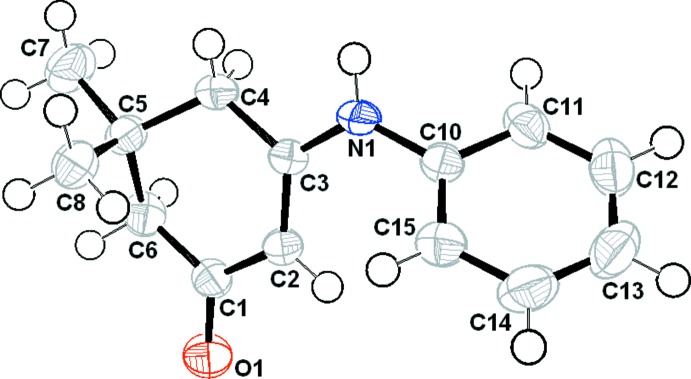
(Farrugia, 2012) The molecular geometry of (I) with the atom-labelling scheme. Displacement ellipsoids are drawn at the 50% probability level. H atoms are represented as small spheres of arbitrary radius.
Fig. 2.

(Brandenburg & Berndt, 2001) Alternating layers parallel to (-101) planes of (I) viewed down the b axis.
Fig. 3.
(Brandenburg & Berndt, 2001) A diagram of the layered crystal packing of (I) viewed down the a axis showing hydrogen bond as dashed line.
Crystal data
| C14H17NO | F(000) = 464 |
| Mr = 215.29 | Dx = 1.185 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 4703 reflections |
| a = 10.1766 (19) Å | θ = 2.6–34.3° |
| b = 13.159 (2) Å | µ = 0.07 mm−1 |
| c = 9.2877 (17) Å | T = 150 K |
| β = 104.062 (7)° | Prism, colorless |
| V = 1206.5 (4) Å3 | 0.15 × 0.12 × 0.09 mm |
| Z = 4 |
Data collection
| Bruker APEXII diffractometer | 3873 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.030 |
| CCD rotation images, thin slices scans | θmax = 34.3°, θmin = 2.6° |
| Absorption correction: multi-scan (SADABS; Sheldrick, 2002) | h = −16→14 |
| Tmin = 0.667, Tmax = 0.747 | k = −13→20 |
| 12403 measured reflections | l = −12→14 |
| 5021 independent reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.047 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.138 | H-atom parameters constrained |
| S = 1.04 | w = 1/[σ2(Fo2) + (0.0725P)2 + 0.1791P] where P = (Fo2 + 2Fc2)/3 |
| 5021 reflections | (Δ/σ)max < 0.001 |
| 147 parameters | Δρmax = 0.45 e Å−3 |
| 0 restraints | Δρmin = −0.20 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | 0.64385 (8) | 0.73838 (6) | 0.82567 (9) | 0.01988 (15) | |
| C2 | 0.53143 (8) | 0.78529 (6) | 0.72449 (9) | 0.01973 (15) | |
| H2 | 0.4742 | 0.7455 | 0.6531 | 0.024* | |
| C3 | 0.50602 (8) | 0.88768 (6) | 0.73022 (9) | 0.01767 (14) | |
| C4 | 0.59803 (8) | 0.95491 (6) | 0.84239 (10) | 0.02194 (16) | |
| H4A | 0.5593 | 0.963 | 0.9274 | 0.026* | |
| H4B | 0.6012 | 1.0216 | 0.7987 | 0.026* | |
| C5 | 0.74309 (8) | 0.91494 (6) | 0.89647 (9) | 0.01943 (15) | |
| C6 | 0.73478 (9) | 0.80376 (6) | 0.94261 (9) | 0.02232 (16) | |
| H6A | 0.8253 | 0.775 | 0.9661 | 0.027* | |
| H6B | 0.7019 | 0.8017 | 1.0322 | 0.027* | |
| C7 | 0.81684 (10) | 0.97776 (8) | 1.03068 (11) | 0.0320 (2) | |
| H7A | 0.7703 | 0.9715 | 1.1086 | 0.048* | |
| H7B | 0.8185 | 1.0478 | 1.0024 | 0.048* | |
| H7C | 0.908 | 0.9533 | 1.0652 | 0.048* | |
| C8 | 0.81959 (9) | 0.92285 (7) | 0.77337 (10) | 0.02490 (17) | |
| H8A | 0.8268 | 0.9929 | 0.7477 | 0.037* | |
| H8B | 0.7712 | 0.886 | 0.6876 | 0.037* | |
| H8C | 0.9086 | 0.8945 | 0.8079 | 0.037* | |
| C10 | 0.29024 (8) | 0.89121 (6) | 0.53767 (9) | 0.01887 (15) | |
| C11 | 0.15721 (8) | 0.90959 (7) | 0.54586 (10) | 0.02316 (16) | |
| H11 | 0.1403 | 0.95 | 0.6215 | 0.028* | |
| C12 | 0.04984 (9) | 0.86746 (7) | 0.44081 (12) | 0.0306 (2) | |
| H12 | −0.0388 | 0.88 | 0.4461 | 0.037* | |
| C13 | 0.07470 (11) | 0.80686 (7) | 0.32830 (13) | 0.0351 (2) | |
| H13 | 0.003 | 0.7781 | 0.2587 | 0.042* | |
| C14 | 0.20729 (11) | 0.78931 (8) | 0.31997 (12) | 0.0343 (2) | |
| H14 | 0.2239 | 0.7488 | 0.2443 | 0.041* | |
| C15 | 0.31524 (9) | 0.83168 (7) | 0.42365 (10) | 0.02586 (18) | |
| H15 | 0.4037 | 0.8203 | 0.4168 | 0.031* | |
| N1 | 0.39813 (7) | 0.93664 (5) | 0.64459 (8) | 0.02126 (14) | |
| H1 | 0.3945 | 1.0014 | 0.6559 | 0.026* | |
| O1 | 0.66676 (7) | 0.64544 (5) | 0.82303 (9) | 0.03086 (17) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.0190 (3) | 0.0142 (3) | 0.0280 (4) | −0.0002 (2) | 0.0086 (3) | 0.0044 (3) |
| C2 | 0.0182 (3) | 0.0134 (3) | 0.0268 (4) | −0.0001 (2) | 0.0040 (3) | −0.0003 (3) |
| C3 | 0.0170 (3) | 0.0144 (3) | 0.0217 (3) | 0.0010 (2) | 0.0048 (3) | −0.0012 (2) |
| C4 | 0.0214 (3) | 0.0170 (3) | 0.0261 (4) | 0.0017 (3) | 0.0032 (3) | −0.0060 (3) |
| C5 | 0.0198 (3) | 0.0172 (3) | 0.0202 (3) | −0.0011 (3) | 0.0027 (3) | −0.0009 (3) |
| C6 | 0.0225 (3) | 0.0210 (4) | 0.0226 (3) | 0.0010 (3) | 0.0036 (3) | 0.0053 (3) |
| C7 | 0.0320 (4) | 0.0310 (5) | 0.0280 (4) | −0.0039 (4) | −0.0024 (4) | −0.0070 (4) |
| C8 | 0.0245 (4) | 0.0214 (4) | 0.0300 (4) | −0.0041 (3) | 0.0092 (3) | 0.0014 (3) |
| C10 | 0.0191 (3) | 0.0144 (3) | 0.0221 (3) | 0.0013 (2) | 0.0031 (3) | 0.0013 (2) |
| C11 | 0.0212 (3) | 0.0206 (3) | 0.0278 (4) | 0.0043 (3) | 0.0061 (3) | 0.0046 (3) |
| C12 | 0.0201 (4) | 0.0254 (4) | 0.0437 (5) | −0.0011 (3) | 0.0025 (3) | 0.0103 (4) |
| C13 | 0.0317 (5) | 0.0228 (4) | 0.0420 (5) | −0.0068 (3) | −0.0080 (4) | 0.0015 (4) |
| C14 | 0.0397 (5) | 0.0256 (4) | 0.0328 (5) | 0.0008 (4) | −0.0004 (4) | −0.0097 (4) |
| C15 | 0.0254 (4) | 0.0237 (4) | 0.0278 (4) | 0.0026 (3) | 0.0052 (3) | −0.0053 (3) |
| N1 | 0.0210 (3) | 0.0135 (3) | 0.0266 (3) | 0.0034 (2) | 0.0007 (3) | −0.0026 (2) |
| O1 | 0.0290 (3) | 0.0135 (3) | 0.0487 (4) | 0.0018 (2) | 0.0067 (3) | 0.0065 (3) |
Geometric parameters (Å, º)
| C1—O1 | 1.2463 (10) | C7—H7C | 0.96 |
| C1—C2 | 1.4320 (11) | C8—H8A | 0.96 |
| C1—C6 | 1.5126 (12) | C8—H8B | 0.96 |
| C2—C3 | 1.3754 (11) | C8—H8C | 0.96 |
| C2—H2 | 0.93 | C10—C15 | 1.3895 (12) |
| C3—N1 | 1.3523 (10) | C10—C11 | 1.3954 (12) |
| C3—C4 | 1.5069 (11) | C10—N1 | 1.4209 (10) |
| C4—C5 | 1.5323 (12) | C11—C12 | 1.3911 (13) |
| C4—H4A | 0.97 | C11—H11 | 0.93 |
| C4—H4B | 0.97 | C12—C13 | 1.3857 (16) |
| C5—C7 | 1.5316 (12) | C12—H12 | 0.93 |
| C5—C6 | 1.5326 (12) | C13—C14 | 1.3895 (16) |
| C5—C8 | 1.5351 (12) | C13—H13 | 0.93 |
| C6—H6A | 0.97 | C14—C15 | 1.3896 (13) |
| C6—H6B | 0.97 | C14—H14 | 0.93 |
| C7—H7A | 0.96 | C15—H15 | 0.93 |
| C7—H7B | 0.96 | N1—H1 | 0.86 |
| O1—C1—C2 | 122.25 (8) | C5—C7—H7C | 109.5 |
| O1—C1—C6 | 119.18 (7) | H7A—C7—H7C | 109.5 |
| C2—C1—C6 | 118.54 (7) | H7B—C7—H7C | 109.5 |
| C3—C2—C1 | 121.65 (7) | C5—C8—H8A | 109.5 |
| C3—C2—H2 | 119.2 | C5—C8—H8B | 109.5 |
| C1—C2—H2 | 119.2 | H8A—C8—H8B | 109.5 |
| N1—C3—C2 | 125.26 (7) | C5—C8—H8C | 109.5 |
| N1—C3—C4 | 113.97 (7) | H8A—C8—H8C | 109.5 |
| C2—C3—C4 | 120.72 (7) | H8B—C8—H8C | 109.5 |
| C3—C4—C5 | 114.36 (7) | C15—C10—C11 | 119.93 (8) |
| C3—C4—H4A | 108.7 | C15—C10—N1 | 121.09 (7) |
| C5—C4—H4A | 108.7 | C11—C10—N1 | 118.95 (8) |
| C3—C4—H4B | 108.7 | C12—C11—C10 | 119.99 (9) |
| C5—C4—H4B | 108.7 | C12—C11—H11 | 120 |
| H4A—C4—H4B | 107.6 | C10—C11—H11 | 120 |
| C7—C5—C4 | 108.91 (7) | C13—C12—C11 | 120.13 (9) |
| C7—C5—C6 | 109.70 (7) | C13—C12—H12 | 119.9 |
| C4—C5—C6 | 107.77 (7) | C11—C12—H12 | 119.9 |
| C7—C5—C8 | 109.43 (7) | C12—C13—C14 | 119.69 (9) |
| C4—C5—C8 | 110.80 (7) | C12—C13—H13 | 120.2 |
| C6—C5—C8 | 110.20 (7) | C14—C13—H13 | 120.2 |
| C1—C6—C5 | 114.07 (7) | C13—C14—C15 | 120.64 (10) |
| C1—C6—H6A | 108.7 | C13—C14—H14 | 119.7 |
| C5—C6—H6A | 108.7 | C15—C14—H14 | 119.7 |
| C1—C6—H6B | 108.7 | C10—C15—C14 | 119.61 (9) |
| C5—C6—H6B | 108.7 | C10—C15—H15 | 120.2 |
| H6A—C6—H6B | 107.6 | C14—C15—H15 | 120.2 |
| C5—C7—H7A | 109.5 | C3—N1—C10 | 126.13 (7) |
| C5—C7—H7B | 109.5 | C3—N1—H1 | 116.9 |
| H7A—C7—H7B | 109.5 | C10—N1—H1 | 116.9 |
| O1—C1—C2—C3 | 178.90 (8) | C8—C5—C6—C1 | −69.18 (9) |
| C6—C1—C2—C3 | 0.93 (12) | C15—C10—C11—C12 | 0.66 (12) |
| C1—C2—C3—N1 | −176.05 (8) | N1—C10—C11—C12 | 178.82 (7) |
| C1—C2—C3—C4 | 1.17 (12) | C10—C11—C12—C13 | 0.27 (13) |
| N1—C3—C4—C5 | −157.43 (7) | C11—C12—C13—C14 | −0.70 (15) |
| C2—C3—C4—C5 | 25.06 (11) | C12—C13—C14—C15 | 0.19 (16) |
| C3—C4—C5—C7 | −168.72 (7) | C11—C10—C15—C14 | −1.17 (13) |
| C3—C4—C5—C6 | −49.78 (9) | N1—C10—C15—C14 | −179.28 (8) |
| C3—C4—C5—C8 | 70.86 (9) | C13—C14—C15—C10 | 0.74 (15) |
| O1—C1—C6—C5 | 152.80 (8) | C2—C3—N1—C10 | 2.19 (14) |
| C2—C1—C6—C5 | −29.16 (11) | C4—C3—N1—C10 | −175.20 (7) |
| C7—C5—C6—C1 | 170.28 (7) | C15—C10—N1—C3 | −54.91 (12) |
| C4—C5—C6—C1 | 51.84 (9) | C11—C10—N1—C3 | 126.96 (9) |
Hydrogen-bond geometry (Å, º)
Cg1 is the centroid of C10–C15 ring.
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1···O1i | 0.86 | 2.02 | 2.8587 (11) | 165 |
| C8—H8A···Cg1ii | 0.96 | 2.61 | 3.5547 (12) | 157 |
Symmetry codes: (i) −x+1, y+1/2, −z+3/2; (ii) −x+1, −y+2, −z+1.
Footnotes
Supporting information for this paper is available from the IUCr electronic archives (Reference: BQ2392).
References
- Adams, J. P. (2000). J. Chem. Soc. Perkin Trans. 1, pp. 125–128.
- Amini, M., Edraki, N., Sarkarzadeh, H., Shafiee, A., Edraki, N., Firuzi, O., Miri, R. & Razzaghi-Asl, N. (2013). Arch. Pharm. Res. 36, 436–447. [DOI] [PubMed]
- Assy, A. M. (1996). Indian J. Chem. Sect. B, 35, 608–610.
- Bahnous, M., Bouraiou, A., Bouacida, S., Roisnel, T. & Belfaitah, A. (2012). Acta Cryst. E68, o1391. [DOI] [PMC free article] [PubMed]
- Brandenburg, K. & Berndt, M. (2001). DIAMOND Crystal Impact, Bonn, Germany.
- Bruker (2001). APEX2 and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Burla, M. C., Caliandro, R., Camalli, M., Carrozzini, B., Cascarano, G. L., De Caro, L., Giacovazzo, C., Polidori, G. & Spagna, R. (2005). J. Appl. Cryst. 38, 381–388.
- Chelghoum, M., Bahnous, M., Bouacida, S., Roisnel, T. & Belfaitah, A. (2011). Acta Cryst. E67, o1890. [DOI] [PMC free article] [PubMed]
- Farrugia, L. J. (2012). J. Appl. Cryst. 45, 849–854.
- Gao, S., Tsai, C. H., Tseng, C. & Yao, C.-F. (2008). Tetrahedron, 64, 9143–9149.
- Kempf, B., Hampel, N., Ofial, A. R. & Mayr, H. (2003). Chem. Eur. J. 9, 2209–2213. [DOI] [PubMed]
- Machacek, V., Simunek, P. & Lycka, A. (2002). Eur. J. Org. Chem. 16, 2764–2769.
- Mohammadizadeh, M. R., Hasaninejad, A., Bahramzadeh, M. & Khanjarlou, Z. S. (2009). Synth. Commun. 39, 1152–1165.
- Palko, R., Egyed, O., Bombicz, P., Riedl, Z. & Hajós, G. (2008). Tetrahedron, 64, 10375–10380.
- Park, J. G. & Jahng, Y. (1998). Bull. Korean Chem. Soc. 19, 436–439.
- Sheldrick, G. M. (2002). SADABS Bruker AXS Inc., Madison, Wisconsin, USA.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Tadesse, S., Bhandari, A. & Gallop, M. A. (1999). J. Comb. Chem. 1, 184–187.
- Thummel, R. P. & Jahng, Y. (1985). J. Org. Chem. 50, 2407–2412.
- Wang, W.-S., Zhang, M.-M., Jiang, H., Chang-Sheng Yao, S.-S. & Tu, S.-J. (2007). Tetrahedron, 63, 4439–4449.
- Zama, S., Bouraiou, A., Bouacida, S., Roisnel, T. & Belfaitah, A. (2013a). Tetrahedron Lett. 54, 5605–5607.
- Zama, S., Bouraiou, A., Bouacida, S., Roisnel, T. & Belfaitah, A. (2013b). Acta Cryst. E69, o837–o838. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I. DOI: 10.1107/S160053681400186X/bq2392sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S160053681400186X/bq2392Isup2.hkl
Supporting information file. DOI: 10.1107/S160053681400186X/bq2392Isup3.cml
CCDC reference: http://scripts.iucr.org/cgi-bin/cr.cgi?rm=csd&csdid=983637
Additional supporting information: crystallographic information; 3D view; checkCIF report



