Abstract

Discovery of protein biomarkers in clinical samples necessitates significant prefractionation prior to liquid chromatography–mass spectrometry (LC–MS) analysis. Integrating traveling wave ion mobility spectrometry (TWIMS) enables in-line gas phase separation which when coupled with nanoflow liquid chromatography and data independent acquisition tandem mass spectrometry, confers significant advantages to the discovery of protein biomarkers by improving separation and inherent sensitivity. Incorporation of TWIMS leads to a packet of concentrated ions which ultimately provides a significant improvement in sensitivity. As a consequence of ion packeting, when present at high concentrations, accurate quantitation of proteins can be affected due to detector saturation effects. Human plasma was analyzed in triplicate using liquid-chromatography data independent acquisition mass spectrometry (LC-DIA-MS) and using liquid-chromatography ion-mobility data independent acquisition mass spectrometry (LC-IM-DIA-MS). The inclusion of TWIMS was assessed for the effect on sample throughput, data integrity, confidence of protein and peptide identification, and dynamic range. The number of identified proteins is significantly increased by an average of 84% while both the precursor and product mass accuracies are maintained between the modalities. Sample dynamic range is also maintained while quantitation is achieved for all but the most abundant proteins by incorporating a novel data interpretation method that allows accurate quantitation to occur. This additional separation is all achieved within a workflow with no discernible deleterious effect on throughput. Consequently, TWIMS greatly enhances proteome coverage and can be reliably used for quantification when using an alternative product ion quantification strategy. Using TWIMS in biomarker discovery in human plasma is thus recommended.
Biomarkers are molecules which alter as a consequence of disease etiology and progression and reflect the status of the disease.1 They are also required for the assessment of treatment efficacy, particularly in preventative regimes.2 Discovering protein biomarkers is not trivial and requires significant analytical methodology to enable sufficient separation of highly complex biological samples. The complexity of samples such as blood or urine is amplified by digesting the thousand-plus proteins to peptides such that there can be tens of thousands of peptides in each sample.3 This level of complexity can be partially tackled by ultrahigh pressure liquid chromatography combined with modern chromatography chemistries which through improving column capacity enable a higher degree of analytical separation to occur.4,5 However, because of the intense complexity of a tryptically treated plasma sample, the analytical space is still crowded and additional strategies are required to enable sufficient separation of analytes.
In order to investigate a greater proportion of the plasma proteome, prefractionation strategies including 2D-reversed phase (RP)-RP,6 ion exchange (IEX)-RP,7,8 strong cation exchange (SCX)-RP,8,9 liquid electrophoresis,10 and 1D-gel LC–MS11 are often utilized, and these lead to successful levels of separation but with significant decreases in throughput, making large clinical studies unviable.12 Incorporating traveling wave ion mobility orthogonally to the time-of-flight (TOF) analyzer (oa-TOF)13 enables a gas phase separation which, because of its orthogonality, provides additional separation at no additional cost to throughput.14 Consequently, the incorporation of traveling wave ion mobility spectrometry (TWIMS) can be used to replace prefractionation strategies and maintain a reasonable throughput or used within a prefractionation pipeline to significantly improve the overall coverage. Previous studies have shown that quantitation of proteins can be reliably achieved in complex mixtures using a data independent approach.15 However, up until now there has been no assessment as to the ability to reliably quantitate proteins in human plasma using TWIMS augmented LC–MS.
Traveling wave ion mobility enables gas phase electrophoretic separation by creating a dynamic pulse of alternating voltages that result in a “travelling wave” of ions. Ions are separated by their size, shape, or charge state.13 Ion mobility (IM) separation is on the millisecond time scale, nesting within the time scale of nano-LC separation (seconds) and TOF-MS acquisition (∼100 μs), allowing multiple mass spectra to be taken of each ion mobility separation16 and providing an additional degree of peak capacity.17
In order to achieve quantitation, liquid chromatography-data independent acquisition mass spectrometry (LC-DIA-MS) takes advantage of the observation that the quantity of a protein can be stoichiometrically related to the mean average of the intensities of the three most intense peptides following the addition of an internal standard.18 The method, also described as LC–MSE, relies on carrying out an iterative analysis of low collision induced dissociation (CID) energy followed by an analysis at elevated CID energy. This iterative process occurs throughout an entire run, and software is used to bring together the precursor ion with the dissociated product ions by virtue of their retention time alignment.19 An LC-IM-DIA-MS experiment uses the quantitative capacity of LC-DIA-MS coupled with the resolving capabilities of traveling wave ion mobility spectrometry. In a Synapt G2 HDMS instrument, the region prior to the TOF analyzer contains a TriWave device comprised of three conjoined stacked ring-electrode ion guides designated trap, (IMS) separator, and transfer. The separator has a helium cell at the entrance, which enables higher pressures to be used without encroaching on the gas phase stability of the molecules.17 The ion mobility experiment is carried out within the TriWave device. Ions are accumulated within the trap ion guide before a packet of ions are released into the separator, where mobility separation occurs. Finally, the ions are transferred into the oaTOF mass analyzer via the transfer ion guide which maintains the mobility separation of the ions to the oaTOF. CID occurs in the transfer ion guide after ions are mobility separated. This means that precursor ions, undergoing dissociation, exhibit the same mobility drift time as their product ions. Product ions are thus associated with their precursors by (a) mobility drift time and (b) chromatographic retention time thus affording greater specificity with such associations.
Previous work has demonstrated that LC-IM-DIA-MS presents no compromise in quantitative precision across sample replicates; however, some attenuation in quantitative accuracy is observed in proteins of higher abundance. Shliaha et al.20 identifies the reduction of signal linearity at the upper end of the dynamic range as the primary challenge of label-free quantitation on the q-TWIMS-TOF Synapt G2 design. Additionally, the authors reveal a clear loss in transmission which is reproducible and consequently not deleterious to quantitation. Moreover, it is suggested by the authors that the ion transmission loss could be partially ameliorated by careful use of specific parameters. Thus, in order to exploit the clear benefits of incorporating IM into a workflow, steps to overcome the effects of ion saturation are required. Bond et al. has attempted to improve quantitation by combining an ion mobility experiment and non-ion mobility experiment within a single run and then combining the improved separation qualities of IMS enhanced DIA with the less saturated signals associated with non-ion mobility runs.21
MS2 or product ion based quantitation strategies have been previously utilized for relative quantitation in isobaric tagging experiments22 and label-free techniques.23−25 Relative quantitation aims to detect differential protein expression between samples of interest; however, amount estimation of proteins offers the greatest relevance to biomarker studies as absolute amounts of proteins can be related to cellular processes or pharmacological interventions. Immunological techniques for absolute quantitation involve targeting the protein of interest with antibodies, an expensive and low-throughput process. Using MS2 quantitation allows for the saturation effects to be reduced by virtue of the fact that the signal intensity of the product ions are likely to be significantly less intense than the precursor ion.
Mass spectrometry with absolute quantitation allows a global survey of a sample’s proteome with identification and quantitation in a single step, a feature of particular utility in the analysis of scarce clinical samples. For this reason, label free quantitation methods are of particular interest in the search for biomarkers.26 Sample quality is not compromised by additional preparation procedures as all data required for quantitation are collected as part of the standard analytical workflow.27
In this paper, the impact of incorporating TWIMS into the identification and quantification of proteins is explored by looking at multiple characteristics that define standard proteomic indices. More specifically, the impact of using LC-IM-DIA-MS in the discovery of biomarkers in human plasma is assessed. Data shown in this paper demonstrate that the challenges in quantitation when incorporating TWIMS, which are associated with signal saturation, can be overcome using an alternative, MS2-based, label-free, absolute quantitation method.
Methods
Reagents
HPLC grade water, HPLC grade acetonitrile, 98% molecular biology dithiothreitol, purified grade ethylenediaminetetraacetic acid (EDTA), and reagent grade iodoacetamide were purchased from Sigma-Aldrich (Poole, Dorset, U.K.). Ultra grade (≥99.5%) ammonium bicarbonate was purchased from Fluka (Buchs, Switzerland). Mass spectrometry grade Trypsin Gold was purchased from Promega (Madison WI). Rapigest, enolase digestion standard (Saccharomyces cerevisiae) and alcohol dehydrogenase digestion standard (S. cerevisiae) were purchased from Waters Corp. (Milford, MA). Lo-Bind sample tubes were purchased from Eppendorf (Hamburg, Germany). LCMS grade formic acid was purchased from Fisher Scientific (Loughborough, U.K.). Trasylol was purchased from Bayer (Newbury, U.K.). The 5 kDa molecular weight cut off spin concentrator tubes were purchased from Agilent Technologies Inc. (Cheadle, U.K.).
Blood Sample Collection and Plasma Extraction
Human blood sample was collected from a healthy donor following informed consent. A volume of 20 mL of blood was collected in a Sterilin tube containing 330 μL of Trasylol (= 3000 KIU (Kallikrein Inhibitor Units)) and 80 μL of 1 M EDTA per 20 mL of blood. The blood was mixed before centrifugation at 15 000g at 4 °C for 30 min. The plasma layer was separated from the buffy layer and red blood cells, and stored at −80 °C.
Experimental Design
To reduce biological variation and allow complete assessment of the effects of traveling wave ion mobility and alternative quantitation methods on proteome analysis, a single human plasma sample was prepared with immunodepletion, reduction, and alkylation. To test the reproducibility of the analysis method, this sample was divided between six vials and analyzed in triplicate as a randomized sample list.
Immunodepletion
The top 14 most abundant proteins were depleted from human plasma using a Seppro IGY14 LC2 immunodepletion column (Sigma Aldrich) using the manufacturer’s recommended protocols and buffers.
Digestion
In order to render all proteins in the sample soluble for reduction, alkylation, and digestion, Rapigest was used as a denaturant. A 1% solution was produced by adding 100 μL of water to the 1 mg vial of Rapigest. The 1% Rapigest solution was then added to the sample vial to give a 0.1% final concentration. Sample was vortexed before incubation at 80 °C for 45 min. After incubation, sample was centrifuged for 10 s to resettle. Sample was then reduced with 100 mM aqueous dithiothreitol (DTT) solution added to give a final concentration of DTT of 5 mM prior to incubation at 60 °C for 30 min. A 200 mM iodoacetamide (IAA) solution was added to the sample to give a final concentration of 10 mM before incubation in the dark at room temperature for 30 min. A trypsin solution of 1 μg/μL was added to the sample in a 1:50 w/w ratio. Sample was vortexed and incubated at 37 °C overnight. Digestion was halted, and Rapigest cleaved with the addition of 100% formic acid to the sample to give a final concentration of 0.5%. Sample was centrifuged at 13 000 rpm for 10 min to remove insoluble material. Supernatant was transferred as aliquots to six clean sample vials. Sample was spiked with alcohol dehydrogenase (S. cerevisiae) for quantitation. Each analysis injection contained 100 fmol of alcohol dehydrogenase.
Nanoscale Liquid Chromatography
Each sample was analyzed on a Waters NanoAcquity system (Waters Corporation, Milford, MA). The peptides were initially loaded onto a Symmetry C18 180 μm × 20 mm 5 μm trap column to desalt and chromatographically focus the peptides prior to elution onto a HSS T3 C18 75 μm × 150 mm, 1.7 μm analytical column. Solvent A, HPLC grade water with 0.1% formic acid; solvent B, acetonitrile with 0.1% formic acid was used. The flow rate was set at 0.3 μL/min. The gradient began following a 3 min (5 μL/min) trapping stage on the trap column. At time zero, solvent A was 99% while solvent B was 1%. Solvent B increased linearly to 40% at 90 min and to 85% at 92 min. The gradient was held at 85% solvent B at 93 min and returned to starting conditions at 95 min to equilibrate.
Mass Spectrometry
The LC system was coupled to a Waters Synapt G2 HDMS (Waters Corporation, Milford, MA). The instrument was run in positive ion nanoelectrospray ionization mode. The capillary voltage was set at 3.4 kV and cone voltage at 30 V. Picotip emitters (10 μm internal diameter, Presearch, Basingstoke, U.K.) were used for the nanostage to direct flow from the analytical column through to the source. A helium gas flow of 180 mL/min and ion mobility separator gas flow (N2) of 90 mL/min with a pressure of 2.5 mbar was used. An IM wave velocity of 600 m/s and wave height of 40 V was used throughout each run. During LC-IM-DIA-MS low CID energy, 2 V was applied across the transfer ion guide. During high CID energy, a ramp of 27–50 V was applied. Argon was used as the CID gas. Lockspray of [Glu1]-Fibrinopeptide (GFP) m/z at 785.84265 was used to maintain mass accuracy throughout the chromatographic run. Data were acquired using MassLynx 4.1.
Data Processing
The raw data was processed with ProteinLynx Global Server (PLGS) 2.5.2 Data were extracted, aligned, and searched against the Uniprot human proteomic database, version 2012-07, appended with the alcohol dehydrogenase (S. cerevisiae) sequence. The ion accounting algorithm used has been described previously.28 PLGS 2.5.2 utilizes the drift time of mobility separated peptides to increase the specificity of alignment/association for precursor and product ions. Data were further processed with Microsoft Excel 2010, Graphpad Prism 5.03, Stata/IC 12.1 and Origin 8.6.
Product Ion Quantitation
MS2-High3 quantitation takes advantage of the observation that MS2 signals compared to their corresponding MS1 precursors are less likely to lead to signal saturation. MS2-High3 quantitation was performed manually using the search output files from ProteinLynx Global Server 2.5.2 (PLGS). PLGS assigns peptide identifications to proteins through an iterative matching process.28 Peptides assigned to proteins in the first matching pass were selected for use in product ion quantitation.
MS2-High3 was performed for each identified protein by summing the product ion (MS2) intensity of all associated peptides. The top three most abundant peptides were selected for onward processing. The MS2 intensities of these three peptides (MS2 pep) were summed to give each protein a product ion intensity value (P), eq 1.
| 1 |
This value was compared with the product ion intensity value of the internal standard protein (IS), alcohol dehydrogenase (S. cerevisiae) calculated by the same means, eq 2.
| 2 |
The ratio of the product ion intensity value of the identified protein to the internal standard product ion intensity value is multiplied by the absolute quantity in fmol of internal standard (ABSIS) in the sample run, in this case, 100 fmol, to give the absolute fmol quantity of the identified protein in the run (ABSP), eq 3.
| 3 |
Proteins with fewer than three peptide matches were quantitated with the corresponding number of alcohol dehydrogenase peptides. For accuracy, proteins with 100% sequence homology were excluded from quantitation.
Results
Effects of the Incorporation of Traveling Wave Ion Mobility on the Characterization of the Proteome
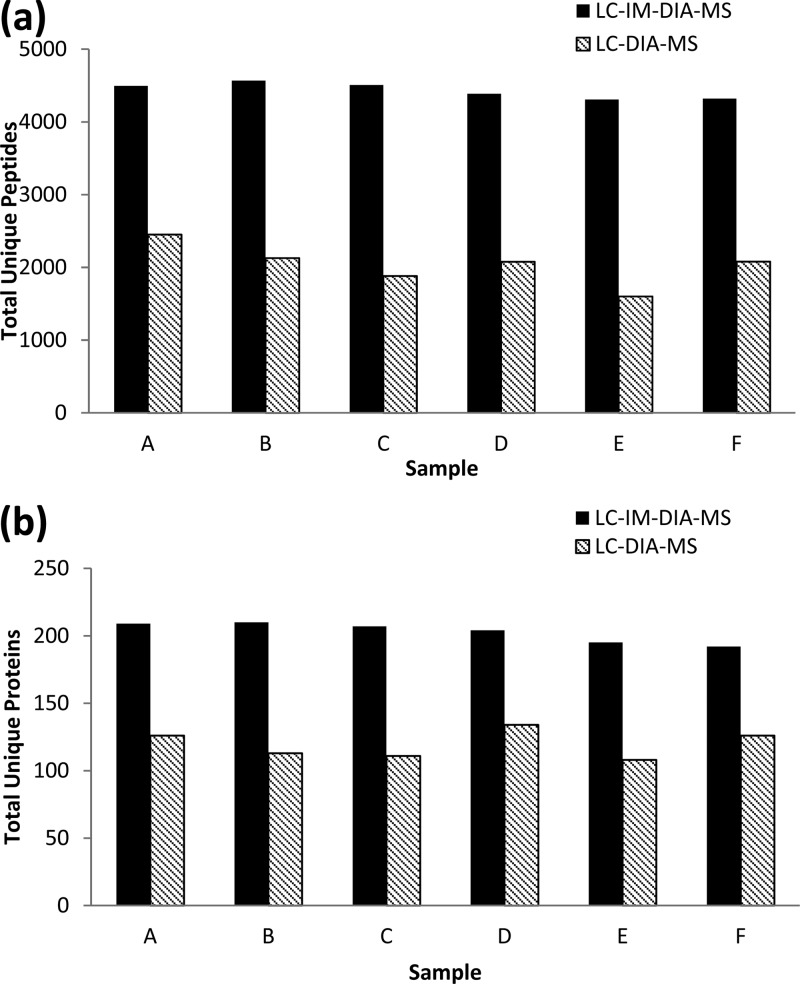
Initial comparisons center upon the effect of characterization of the plasma proteome. More specifically, the sample coverage in terms of the number of proteins identified was investigated. The results shown in Figure 1 demonstrate that the incorporation of ion mobility into the workflow results in a more thorough analysis of the sample content. More cogently, for all samples, more peptides were identified with LC-IM-DIA-MS reporting an average of 4430 unique peptides per sample than with LC-DIA-MS, reporting an average of 2036 unique peptides per sample (Figure 1a). This marked increase in peptide observation translated to the same trend in protein identification between the two methods, with an average of 70% more protein identifications reported for LC-IM-DIA-MS experiments (Figure 1b).
Figure 1.
Results plasma analyzed with (black bars) and without (hatched bars) ion mobility enhanced DIA for different characteristics: (a) the total number of unique peptides identified and the (b) total number of unique proteins identified.
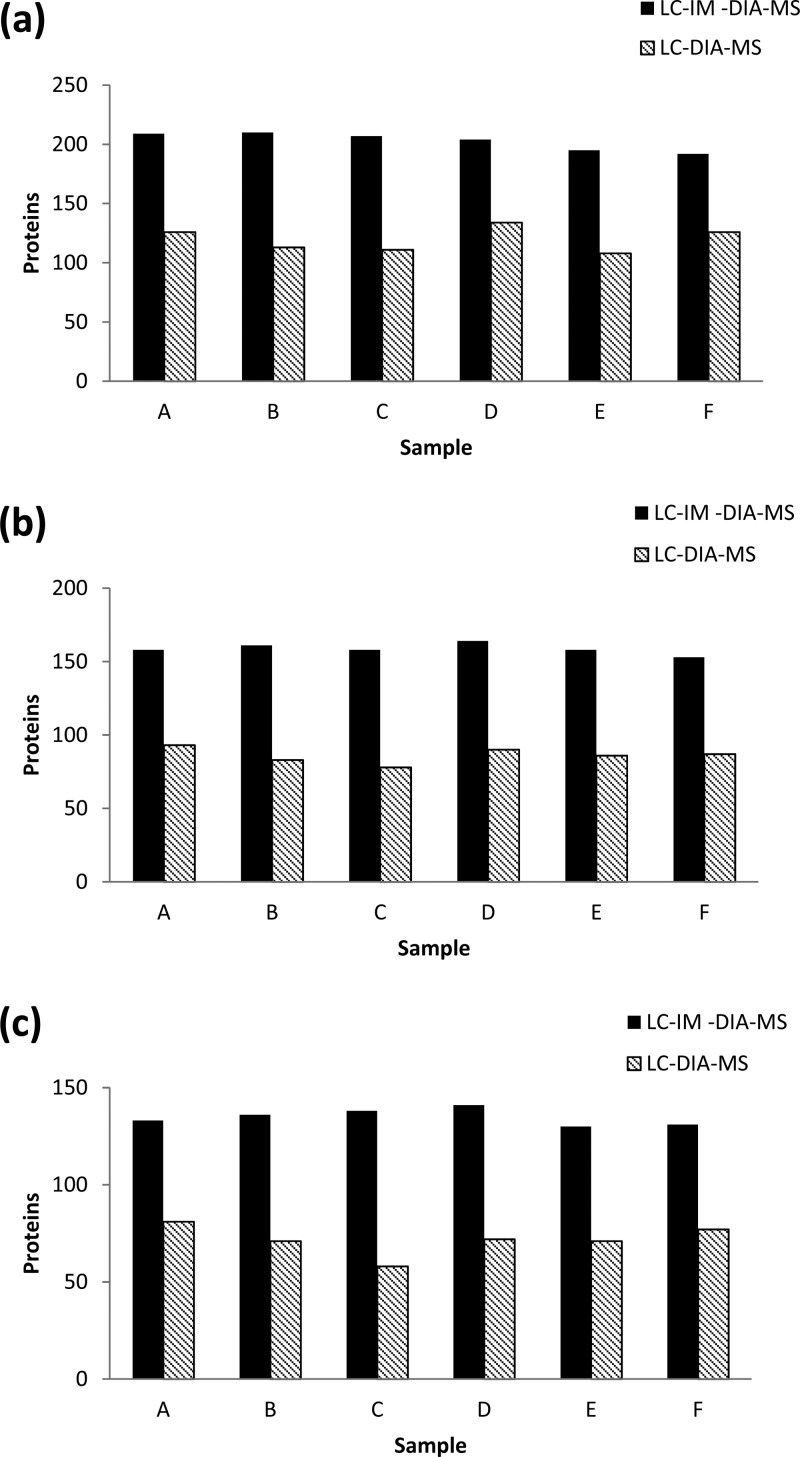
Samples were analyzed in triplicate; Figure 2 examines the reproducibility of the two analysis methods within the triplicate sets for each sample. When all proteins identified in at least one run of a sample triplicate run are examined, LC-IM-DIA-MS yields an average of 70% more protein identifications than LC-DIA-MS (Figure 2a). The difference between the two methods rises to 84% when proteins identified in at least two runs of a triplicate are counted, excluding single-run identifications (Figure 2b). When proteins appearing in all three runs of a triplicate for each sample are counted, LC-IM-DIA-MS yields an average of 88% more protein identifications than LC-DIA-MS (Figure 2c). These statistics imply that the inclusion of ion mobility separation into the proteomic workflow confers a further degree of reproducibility to the sample analysis.
Figure 2.
Analysis methods reproducibility: (a) Proteins identified in at least one run of a sample triplicate. (b) Proteins identified in at least two runs of a sample triplicate. (c) Proteins identified in all three runs of a triplicate suggesting that LC-IM-DIA-MS is a more reproducible analysis method. LC-IM-DIA-MS produces on average, 70% more protein identifications for replicating identification. Considering also the proteins found in all runs of the triplicate analysis this value rises to LC-IM-DIA-MS producing 88% more protein identifications compared to LC-DIA-MS.
Effects of Ion Mobility on the Accuracy of the Analysis
The statistical assignment of proteins relies on the combination of a number of parameters to increase confident assignment of protein identifications. Key proteomic indices of the two analysis methods were compared, shown in Table 1. In addition a greater proportion of sequence coverage with LC-IM-DIA-MS was evident and the precursor and product mass errors are slightly better, displaying improved precision.
Table 1. Key Mass Spectrometry Indicesa.
| parameter | LC-IM-DIA-MS | LC-DIA-MS |
|---|---|---|
| coverage (%) | 53.9 (4.1) | 45.0 (5.4) |
| precursor mass error (ppm) | 3.0 (0.8) | 3.0 (1.8) |
| product mass error (ppm) | 9.8 (0.6) | 10.5 (1.0) |
Coverage and product and precursor mass error values indicate average values by the method of 48 universally replicated proteins, each value representing 18 experimental runs, with the standard deviation in brackets.
Effect on Quantitation
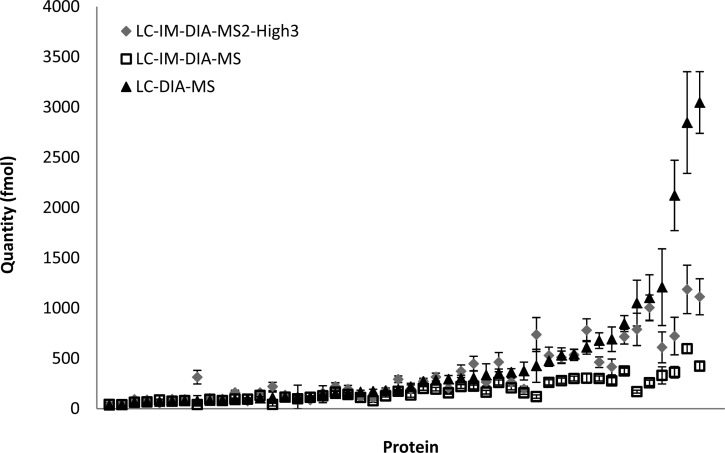
Quantitation is achieved by using the intensities of the top three peptides (MS1). However, for highly abundant ions, the number of detectable ions reaching the detector results in signal saturation. As intensities of the product ions are a composite of the precursor ion, the intensities of the individual product ions will be less intense and consequently are far less likely to result in saturation of the detector. Consequently, we investigated whether using the composite product ions for each of three most intense peptides for each protein might more accurately depict the quantity of the protein measured. This approach improves the accuracy of the LC-IM-DIA-MS approach by reducing the impact of detector saturation on quantitation of highly abundant proteins. This effect is seen in Figure 3 where proteins are quantified using LC-DIA-MS (non ion mobility with MS1 label free quantitation), LC-IM-DIA-MS1 (ion mobility with MS1 label free quantitation), and LC-IM-DIA-MS2-High3 (ion mobility with MS2 label free quantitation). In Figure 3, the triangles represent LC-DIA-MS. Here, a series of measurements fulfills an almost exponential relationship as highly dominant proteins are measured with the expected abundance in relation to the majority of the proteins. However, when LC-IM-DIA-MS1 is carried out, the measurements for the majority of ions is broadly in agreement with LC-DIA-MS but diverge as the abundant proteins predominate. The difference in measurements between these two experiments demonstrates the effect of signal saturation. Using the same data, but instead using LC-IM-DIA-MS2-High3 for the quantitation method (gray diamonds), we can see that the divergence between ion mobility and nonion mobility is reduced considerably for many proteins except the very highly abundant proteins.
Figure 3.
Standard LC-DIA-MS precursor ion quantitation with LC-IMS-DIA-MS1(diamonds) and LC-IM-DIA-MS2-High3 (squares) quantitation methods. Comparison drawn between proteins replicated in all sample runs.
Clustering Analysis
In order to evaluate the relationship between these three modalities (LC-DIA-MS, LC-IM-DIA-MS, and LC-IM-DIA-MS-MS2-High3), a statistical assessment of the closeness of fit was executed. Normalized quantitation values for three exemplar proteins were plotted on 3D axes. Absolute quantitation data for three example proteins were taken and compared for all three analysis methods. These proteins were selected as representative examples of high (Ig γ-4 chain C region), medium (β-2-glycoprotein 1), and low (zinc-α-2-glycoprotein) abundance proteins consistently observed across all analysis runs.
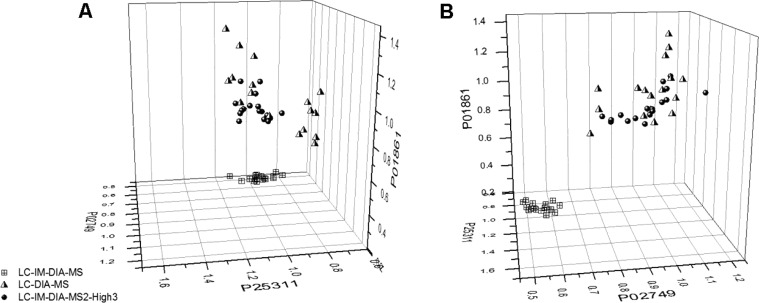
These data were normalized to the average absolute quantity of each protein produced by LC-DIA-MS. The normalized protein quantities were then plotted on 3D axes to demonstrate quantitation across the full range of protein abundance by the three analysis methods (Figure 4). This figure demonstrates that although all three analysis methods are in quantitative agreement for zinc-α-2-glycoprotein, the protein of low abundance (Figure 4A), LC-IM-DIA-MS is unable to match the LC-DIA-MS quantitation for the medium and high abundance proteins (Figure 4B). These data also illustrate that LC-IM-DIA-MS2-High3 quantitation is in agreement with LC-DIA-MS at all levels of protein abundance. This is also demonstrated by Table 2.
Figure 4.
Two views of a 3D scatter plot of normalized protein quantities showing differential quantitation between analysis methods. Normalized values for LC-IM-DIA-MS (squares), LC-DIA-MS (triangles), and LC-IM-DIA-MS2 (circles) were plotted on axes of high (Ig γ-4 chain C region, P01861), medium (β-2-glycoprotein 1, P02749), and low (zinc-α-2-glycoprotein, P25311) abundance proteins.
Table 2. Centroid Distance from Origin and RMS Spread Values of Protein Analysis and Quantitation Methodsa.
| method | centroid distance from origin | RMS spread |
|---|---|---|
| LC-IM-DIA-MS | 1.12 | 0.10 |
| LC-DIA-MS | 1.73 | 0.30 |
| LC-IM-DIA-MS2-High3 | 1.74 | 0.16 |
Similar centroid distance values indicate quantitative agreement between methods across the range of protein abundance. Lower RMS spread values indicate a greater signal to noise ratio.
Table 2 shows the distance of the centroid of each cluster from the origin and the root mean squares (RMS) spread of the data points around the centroid. The almost identical centroid distances between LC-DIA-MS and LC-IM-DIA-MS2-High3 demonstrate that the product ion quantitation method is capable of matching absolute protein quantitation with LC-DIA-MS across the range of protein abundance. The lower RMS values for the ion mobility enabled methods indicate a better signal-to-noise ratio, borne out of the increased sensitivity associated with ion mobility separation. This data further demonstrates that MS2-High3 quantitation provides the increased sensitivity and protein identifications of ion mobility separation combined with the quantitative accuracy of LC-DIA-MS.
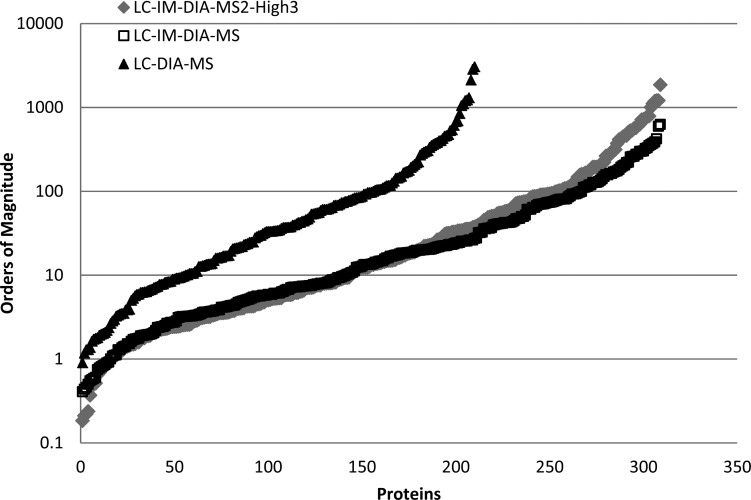
Figure 5 demonstrates the dynamic range of LC-DIA-MS, LC-IM-DIA-MS, and LC-IM-DIA-MS2-High3 analysis. The identified protein quantities span 4 orders of magnitude. A key observation is the increased sensitivity of LC-IM-DIA-MS to detect lower abundance proteins, as observed by a number of low-abundance proteins previously unobserved using LC-DIA-MS. Figure 5 shows the attenuation in quantitation of highly abundant proteins observed by LC-IM-DIA-MS. This signal saturation effect is overcome by LC-IM-DIA-MS2-High3 quantitation, demonstrated by the quantitation of abundant proteins in line with LC-DIA-MS quantitation. These data demonstrate that the experimental dynamic range is increased with the use of the MS2-High3 quantitation method.
Figure 5.
Experimental dynamic range for LC-IMS-DIA-MS and LC-DIA-MS, illustrating that effective experimental dynamic range is maintained.
If individual proteins are more closely examined (Figure 6), then a further interesting observation is revealed. Six individual proteins that represent putative biomarkers found in plasma29−34 are found to be within the acceptable analytical range when measured using LC-IM-DIA-MS2-High3 (as compared to LC-DIA-MS1). Moreover, it appears that reproducibility is more consistent for these proteins. This final observation is directly as a consequence of the increased peak capacity that IM provides. This additional peak capacity provides greater resolution and, particularly following the CID that takes place in the transfer region, product ions are more confidently matched with their precursors by virtue of their chromatographic retention time and mobility drift time alignment.
Figure 6.

Box and whisker plots showing variance between the two quantitation methods of LC-IM-DIA-MS1 data and LC-DIA-MS2-High3 data, illustrating that when LC-IM-DIA-MS sample analysis is utilized, more accurate quantitation of proteins of potential interest as biomarkers is achieved with MS2 quantitation. Proteins shown: P02749, β-2-glycoprotein 1; P02787, serotransferrin; P00747, plasminogen; Q14520, hyaluronan-binding protein 2; P06681, Complement C2; P05155, plasma protease C1 inhibitor. Boxes represent the bounds of the first and third quartiles for each protein quantity, whiskers extend to the lowest data point within 1.5 times the interquartile range of the lower quartile and the highest data point within 1.5 times the interquartile range of the upper quartile.
Discussion
Proteomic analysis of human plasma has undoubtedly been compromised by the inability to deconvolute the complexity of the plasma proteome in terms of both protein numbers and protein concentration dynamic range. Two of the potentially 12 orders of magnitude35 are plausibly overcome by the judicious use of immunoaffinity columns although there still remains a significant dynamic range to overcome.36 In addition to this, there is a need to improve the overall resolution of the analytical platform. Significant improvements in chromatography of peptides have come about as a consequence of new column chemistries, increasing both reproducibility and peak capacity.37 However, in a complex sample such as plasma there are still thousands of analytes which require separation.
Investigating biology through proteomics is a long way off in terms of throughput from genomics. Long LC runs are employed to overcome the complexity of human plasma. Furthermore for many LC runs, the actual period in which analytes are eluting represents less than 50% of the analysis time. Thus, the requirement for an increased role of gas phase separation is compelling. This separation can be achieved by incorporating traveling wave ion mobility spectrometry (TWIMS) into the analytical platform. This dispersive technology has the key advantage over other ion mobility techniques that the ions are maintained and not lost through ion selection or filtering of ions.38 It has been investigated whether TWIMS can be used for the reliable identification and quantification of proteins. Characteristics associated with protein identification and quantification to see the effect of using TWIMS within a LC-IM-DIA-MS (HDMSE) experiment has also been assessed.
The use of LC-IM-DIA-MS results in a greater number of proteins assigned. Moreover, because the number of sequenced peptides is greater, not only are more identified proteins observed but the proteins exhibit higher amino acid sequence coverage. In particular, TWIMS confers greater advantage because of increased specificity for product and precursor ion alignment/association which results in a marked reduction in “chimeric” spectra (which can lead to ambiguity for peptide/protein ID algorithms). This ultimately results in higher confidence identifications of low abundance peptides/proteins. Another characteristic improved by the specificity is the technical reproducibility (median average of RSD decreased from 13% to 9.5%).
In this experiment, only a one-dimensional chromatographic separation was executed. The result is a sample that is rich in highly abundant proteins which constitute a large proportion of observed proteins. In a sample that has undergone considerable fractionation, a greater proportion of observed proteins would be in the range classified as low-abundance markers and consequently less affected by the underreporting of the proteins amounts. However, as mentioned above, the measurements are reproducible and cross-sample comparisons can be made in the context of highly reproducible analysis.
The inclusion of traveling wave ion mobility into the workflow provides greater resolution of the complex plasma samples than standard LC-DIA-MS. The increase in peptide and protein identifications is comparable to that observed with extended LC gradients or prefractionation of samples;12 however, there is no time penalty or additional preparation steps required for the incorporation of TWIMS. The inclusion of TWIMS into the proteomic workflow will confer advantages beyond plasma proteomics into the wider search for biomarkers as the observed enhancement in the analysis of plasma may well be replicated in other complex biological mixtures such as cell lysates, urine, and sputum.
However, in many cases, for proteomics to be purposeful, it must be quantitative especially in clinical proteomics. The observation that proteins of higher abundance are quantitatively under reported implies a reduction of the in-spectra dynamic range of the Synapt G2 HDMS instrument. There appears to be a trade-off between the additional resolution of the plasma proteome, resulting in a greater number of protein identifications and the quantitation of more highly abundant proteins. We have shown in this study that the effect of signal saturation can be reduced when quantitation uses a MS2 strategy which is particularly beneficial for highly abundant proteins. This novel strategy is robust and leads to tangible improvements in reproducibility of quantitation. We chose six proteins, all of which have been implicated as putative biomarkers in a range of different human pathological conditions. Revealingly, the quantitation using MS2 mirrored more closely to literature values and the quantitation found in LC-DIA-MS, a methodology having widespread acceptance for reliable quantitation. Additionally, in the specific application of plasma biomarker discovery, any loss in accuracy is not affected by this quantitative effect as the analysis of proteins germane to biomarker research remains well within the tolerance limits of quantitation.
Summary
This work examines the effect of incorporating traveling wave ion mobility spectrometry into the LC-DIA-MS workflow for examining the human plasma proteome. The results demonstrate that the additional resolution afforded by the method allows a significant deconvolution of the complex sample. The effects of the observed peptide signal saturation of abundant proteins are overcome through the use of the presented MS2-High3 quantitation workflow.
Acknowledgments
The authors wish to thank the following organizations for their financial support of this work: The Biotechnology and Biological Sciences Research Council (BBSRC) and Waters Corporation for joint funding of a BBSRC CASE studentship, The John and Lucille van Geest Foundation for funding of laboratories, and the National Institute for Health Research Leicester Cardiovascular Biomedical Research Unit in which the work was carried out. The authors also thank Matthew Blades (BBASH Bioinformatics, University of Leicester, U.K.) for helpful discussions and encouragement of this work.
The authors declare no competing financial interest.
References
- Clin. Pharmacol. Ther. 2001, 69, 89.11240971 [Google Scholar]
- Scott E. N.; Gescher A. J.; Steward W. P.; Brown K. Cancer Prev. Res. 2009, 2, 525–530. [DOI] [PubMed] [Google Scholar]
- McDonald W. Dis. Markers 2002, 18, 99–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu N.; Clausen A. M. J. Sep. Sci. 2007, 30, 1167–1182. [DOI] [PubMed] [Google Scholar]
- Kay R. G.; Gregory B.; Grace P. B.; Pleasance S. Rapid Commun. Mass Spectrom. 2007, 21, 2585–2593. [DOI] [PubMed] [Google Scholar]
- Chen Y.; Wall D.; Lubman D. Rapid Commun. Mass Spectrom. 1998, 12, 1994–2003. [DOI] [PubMed] [Google Scholar]
- Yates J. A. J. Nat. Biotechnol. 1999, 17, 676–682. [DOI] [PubMed] [Google Scholar]
- Peng J.; Elias J. E.; Thoreen C. C.; Licklider L. J.; Gygi S. P. J. Proteome Res. 2003, 2, 43–50. [DOI] [PubMed] [Google Scholar]
- Washburn M.; Wolters D.; Yates J. Nat. Biotechnol. 2001, 19, 242–247. [DOI] [PubMed] [Google Scholar]
- Zue X.; Speicher D. Electrophoresis 2000, 21, 3035–3047. [DOI] [PubMed] [Google Scholar]
- Wilm M.; Shevchenko A.; Houthaeve T.; Breit S.; Schweigerer L.; Fotsis T.; Mann M. Nature 1996, 379, 466–469. [DOI] [PubMed] [Google Scholar]
- Cao Z.; Tang H.; Wang H.; Liu Q.; Speicher D. W. J. Proteome Res. 2012, 11, 3090–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles K.; Pringle S. D.; Worthington K. R.; Little D.; Wildgoose J. L.; Bateman R. H. Rapid Commun. Mass Spectrom. 2004, 18, 2401–2414. [DOI] [PubMed] [Google Scholar]
- Pringle S.; Wildgoose J.; Williams J.; Slade S.; Thalassinos K. Int. J. Mass Spectrom. 2007, 261, 1–12. [Google Scholar]
- Levin Y.; Hradetzky E.; Bahn S. Proteomics 2011, 11, 3273–3287. [DOI] [PubMed] [Google Scholar]
- Angel T. E.; Aryal U. K.; Hengel S. M.; Baker E. S.; Kelly R. T.; Robinson E. W.; Smith R. D. Chem. Soc. Rev. 2012, 41, 3912–3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles K.; Williams J. P.; Campuzano I. Rapid Commun. Mass Spectrom. 2011, 25, 1559–1566. [DOI] [PubMed] [Google Scholar]
- Silva J.; Li G.; Vissers J.; Geromanos S. Mol. Cell. Proteomics 2006, 5, 144–156. [DOI] [PubMed] [Google Scholar]
- Silva J. C.; Denny R.; Dorschel C. A.; Gorenstein M.; Kass I. J.; Li G.; McKenna T.; Nold M. J.; Richardson K.; Young P.; Geromanos S. Anal. Chem. 2005, 77, 2187–2200. [DOI] [PubMed] [Google Scholar]
- Shliaha P. V.; Bond N. J.; Gatto L.; Lilley K. S. J. Proteome Res. 2013, 12, 2323–2339. [DOI] [PubMed] [Google Scholar]
- Bond N. J.; Shliaha P. V.; Lilley K. S.; Gatto L. J. Proteome Res. 2013, 12, 2340–2353. [DOI] [PubMed] [Google Scholar]
- Thompson A.; Schafer J.; Kuhn K.; Kienle S.; Schwarz J.; Schmidt G.; Neumann T.; Hamon C. Anal. Chem. 2003, 75, 4942–4942. [DOI] [PubMed] [Google Scholar]
- Freund D. M.; Prenni J. E. J. Proteome Res. 2013, 12, 1996–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin N. M.; Yu J.; Long F.; Oh P.; Shore S.; Li Y.; Koziol J. A.; Schnitzer J. E. Nat. Biotechnol. 2010, 28, 83–U116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q.; Zhao Q.; Liang Z.; Qu Y.; Zhang L.; Zhang Y. Analyst 2012, 137, 3146–3153. [DOI] [PubMed] [Google Scholar]
- Ahrné E.; Molzahn L.; Glatter T.; Schmidt A. Proteomics 2013, 13, 2567–2578. [DOI] [PubMed] [Google Scholar]
- Asara J. M.; Christofk H. R.; Freimark L. M.; Cantley L. C. Proteomics 2008, 8, 994–999. [DOI] [PubMed] [Google Scholar]
- Li G.; Vissers J. P. C.; Silva J. C.; Golick D.; Gorenstein M. V.; Geromanos S. J. Proteomics 2009, 9, 1696–1719. [DOI] [PubMed] [Google Scholar]
- Aiyaz M.; Lupton M. K.; Proitsi P.; Powell J. F.; Lovestone S. Immunobiology 2012, 217, 204–215. [DOI] [PubMed] [Google Scholar]
- Giannakopoulos B.; Krilis S. A. N. Engl. J. Med. 2013, 368, 1033–1044. [DOI] [PubMed] [Google Scholar]
- Heywood W.; Mills K.; Wang D.; Hogg J.; Madgett T. E.; Avent N. D.; Chitty L. S. J. Proteomics 2012, 75, 2621–2628. [DOI] [PubMed] [Google Scholar]
- Kumara H. M. C. S.; Tohme S. T.; Yan X.; Nasar A.; Senagore A. J.; Kalady M. F.; Hyman N.; Kim I. Y.; Whelan R. L. Surg. Endosc. 2011, 25, 1939–1944. [DOI] [PubMed] [Google Scholar]
- Parahuleva M. S.; Hoelschermann H.; Zandt D.; Pons-Kuehnemann J.; Parviz B.; Weiskirchen R.; Staubitz A.; Tillmanns H.; Erdogan A.; Kanse S. M. Circ. J. 2012, 76, 2653–2661. [DOI] [PubMed] [Google Scholar]
- Vidotto A.; Henrique T.; Raposo L. S.; Maniglia J. V.; Tajara E. H. Cancer Biomark. 2010, 8, 95–107. [DOI] [PubMed] [Google Scholar]
- Anderson N.; Anderson N. Mol. Cell. Proteomics 2002, 1, 845–867. [DOI] [PubMed] [Google Scholar]
- Smith M. P. W.; Wood S. L.; Zougman A.; Ho J. T. C.; Peng J.; Jackson D.; Cairns D. A.; Lewington A. J. P.; Selby P. J.; Banks R. E. Proteomics 2011, 11, 2222–2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie F.; Smith R. D.; Shen Y. J. Chromatogr., A 2012, 1261, 78–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X.; Plasencia M.; Ragg S.; Valentine S. J.; Clemmer D. E. Brief. Funct. Genomics Proteomics 2004, 3, 177–186. [DOI] [PubMed] [Google Scholar]