Abstract
The role of climate in driving selection of mtDNA as Homo sapiens migrated out of Africa into Eurasia remains controversial. We evaluated the role of mtDNA variation in resting metabolic rate (RMR) and total energy expenditure (TEE) among 294 older, community-dwelling African and European American adults from the Health, Aging and Body Composition Study. Common African haplogroups L0, L2 and L3 had significantly lower RMRs than European haplogroups H, JT and UK with haplogroup L1 RMR being intermediate to these groups. This study links mitochondrial haplogroups with ancestry-associated differences in metabolic rate and energy expenditure.
Keywords: Metabolic rate, Energetics, Mitochondria, Mitochondrial haplogroups, mtDNA, Oxidative phosphorylation
1. Introduction
The vast majority of the energy needs of the human body are met by mitochondrial oxidative phosphorylation (OXPHOS). OXPHOS takes place entirely in mitochondria and is a highly efficient system dependent upon the coordinated expression and interaction of genes encoded in both the nuclear and mitochondrial genomes. The mtDNA is a circular double-stranded DNA molecule of 16.6 kb in humans that encodes 13 essential polypeptides of the OXPHOS system and the necessary RNA machinery for their translation within the mitochondria. MtDNA does not recombine, is maternally inherited (Giles et al., 1980), and has a unique organization in that its structural genes lack introns, intergenic spaces, and 5′ and 3′ noncoding sequences. Each human cell contains hundreds of mitochondria and thousands of copies of mitochondrial DNA (mtDNA) with the number of copies being dependent upon the cell type. Human mtDNA is highly variable and approximately one-third of sequence variants found in the general population may be functionally important (Wallace et al., 2010). The genes of the mtDNA are central to energy production, both to generate ATP and to generate heat to maintain body temperature. Impaired mitochondrial function resulting from mitochondrial DNA (mtDNA) and/or nuclear DNA variation is likely to contribute to an imbalance in cellular energy homeostasis, increase vulnerability to oxidative stress, and increase the rate of cellular senescence and aging.
The evolution of human mtDNA is characterized by the emergence of distinct lineages associated with the major global ethnic groups. Recent analyses support all modern mtDNA haplogroups being ultimately traceable to matrilineal ancestors that lived in Africa approximately 150–230 thousand years ago (kya) (Soares et al., 2009). MtDNA sequence variation evolved as a result of the sequential accumulation of mutations along maternally inherited lineages (Quintana-Murci et al., 1999 and Salas et al., 2002). Macrohaplogroup L is specific to sub-Saharan Africa with the most ancient mtDNA haplogroups L0 (112–188 kya) and L1 (108–174 kya) forming the basal lineages of the human mtDNA tree followed by L2 (68–111 kya) and L3 (57–87 kya) (Soares et al., 2009). Haplogroups L0–L3 are most common to sub-Saharan Africa and largely of East African origin (Soares et al., 2009), however the widespread distribution and diversity of the African haplogroups makes their exact origin points within Africa difficult to trace with confidence (Salas et al., 2002). Dispersal of haplogroup L3 to the Near East ~ 70 kya, possibly associated with an improvement of the climatic conditions after a long period of drought (Soares et al., 2009), gave rise to the major Eurasian haplogroups M and N from which the vast majority of non-Africans are descended (Salas, Richards, De la Fe, Lareu, Sobrino, Sanchez-Diz, Macaulay and Carracedo, 2002). Haplogroup L3 is more related to Eurasian haplogroups than to the most divergent African haplogroups L1 and L2 (Maca-Meyer, Gonzalez, Larruga, Flores and Cabrera, 2001). Individuals bearing M and N mtDNAs colonized Europe and Asia with the major European haplogroups (H, V, J, T, U, K, I, W, and X) derived from macrohaplogroup N and the major Asian, Arctic and Native American haplogroups (A, B, C, D, G, X, Y, and Z) from both macrohaplogroups M and N. Europe was first settled by modern humans from the near east during the Early Upper Paleolithic ~ 45 kya (Soares et al., 2009), possibly with the establishment of haplogroup U (43–65 kya) (Richards et al., 2000). This first colonization contributed to ~ 10% of extant mtDNA lineages (Richards et al., 2000). Modern humans arriving during the Middle Upper Paleolithic (~ 26–45 kya) and the Late Upper Paleolithic (~ 9–15 kya) contributed ~ 45–60% of extant mtDNA lineages (Richards et al., 2000), including the arrival of aplogroups H (19–21 kya), J (22–27 kya), and T (33–40 kya) from the Near East.
It has been proposed that the geographic distribution of mtDNA lineages resulted from selection mainly driven by adaptation to climate and nutrition (Mishmar et al., 2003, Ruiz-Pesini et al., 2004, Ruiz-Pesini and Wallace, 2006 and Wallace et al., 2003). According to this hypothesis, certain ancient mtDNA variants permitted humans to adapt to colder climates resulting in the regional enrichment of specific lineages. Underlying this selection were functional mtDNA variants that altered OXPHOS coupling efficiency, shifting the energetic balance from ATP generation to heat production consequently allowing Homo sapiens to adapt to colder environments after leaving Africa (Mishmar et al., 2003 and Ruiz-Pesini et al., 2004).
While there is strong evidence supporting selection as an important factor in the evolution of human mtDNA (Balloux et al., 2009, Elson et al., 2004, Kivisild et al., 2006, Marcuello et al., 2009, Martinez-Redondo et al., 2010, Mishmar et al., 2003, Moilanen et al., 2003, Moilanen and Majamaa, 2003, Montiel-Sosa et al., 2006,Ruiz-Pesini et al., 1998, Ruiz-Pesini et al., 2000, Ruiz-Pesini et al., 2004 and Ruiz-Pesini and Wallace, 2006), not all studies support climate as the driving force for human mtDNA evolution (Amo and Brand, 2007,Amo et al., 2008, Elson et al., 2004, Kivisild et al., 2006 and Moilanen et al., 2003). Evidence that climatic adaptation has influenced the geographic distribution of mtDNA diversity was obtained by examining patterns of genetic variation across the mtDNA coding region, including the 13 mtDNA OXPHOS genes (Balloux et al., 2009, Mishmar et al., 2003 and Ruiz-Pesini et al., 2004). An examination of regional (tropical, temperate and arctic) gene-specific variation in mitochondrial OXPHOS genes provided support for adaptive selection influencing mtDNA diversity (Mishmar et al., 2003). ATP6 was highly variable in the mtDNAs from the arctic, cytb was more variable in temperate Europe, and COI was highly variable in tropical Africa (Mishmar et al., 2003). These genes were largely invariant in the regions outside of their high adaptation zones (e.g. ATP6 was strongly conserved in the temperate and tropical zones). These results were interpreted as evidence for regional gene-specific selection since this pattern of variation would not be expected if all mtDNA mutations were random and neutral. The frequency of conserved, non-synonymous (missense) mutations across the mtDNA coding region was also found to increase from tropical Africa to temperate Europe and arctic northeastern Siberia (Ruiz-Pesini et al., 2004). This excess of non-synonymous mutations in the colder latitudes was interpreted as evidence for adaptive selection playing an important role as people migrated out of Africa into temperate and arctic Eurasia.
Other analyses do not support a simple model in which climatic adaptation has been a major force during human mtDNA evolution (Elson et al., 2004, Kivisild et al., 2006 and Moilanen et al., 2003). For example, the excess non-synonymous substitutions observed in some OXPHOS genes may not reflect positive selection but the relaxation of negative selection in specific populations (Elson et al., 2004) or may be a feature of the terminal branches of the phylogenetic tree, independent of geographical region (Kivisild et al., 2006). Others have observed significant differences in the frequency of non-synonymous mutations among the European haplogroups (Moilanen et al., 2003), suggesting some mutations may be non-neutral within specific phylogenetic lineages but neutral within others.
Functional evidence supporting metabolic differences between haplogroups is equally inconsistent (Amo and Brand, 2007, Amo et al., 2008, Gomez-Duran et al., 2010, Marcuello et al., 2009, Martinez-Redondo et al., 2010, Montiel-Sosa et al., 2006, Ruiz-Pesini et al., 1998 and Ruiz-Pesini et al., 2000). Human spermatozoa motility is fully dependent on the functionality of the OXPHOS system and the haplogroup T samples showed 23% and 29% reductions, respectively, in complexes I and IV activity compared with haplogroup H samples (Ruiz-Pesini et al., 2000). Comparisons of spermatozoa motility among several European haplogroups revealed that sperm from haplogroups H subjects swam significantly faster than those from haplogroups T subjects (Ruiz-Pesini et al., 2000). Interestingly, no differences in complex II activity were observed between haplogroups H and T (complex II is exclusively encoded by the nuclear genome). Spermatozoa motility is directly correlated with activities of OXPHOS complexes I–IV (Ruiz-Pesini et al., 1998). Within the broadly distributed European haplogroup U, sublineages of the group exhibited differences in sperm motility (Montiel-Sosa et al., 2006). Results from a comparison of cybrids containing haplogroups H and Uk (Gomez-Duran et al., 2010) suggest that there are differences in mtDNA and mtRNA levels, mitochondrial protein synthesis, cytochrome oxidase activity and amount, normalized oxygen consumption, mitochondrial inner membrane potential and growth capacity. Differences in endogenous, leaking and uncoupled respiration, however, were not observed in comparisons of haplogroups H and T (Amo et al., 2008) or haplogroups H and Uk (Gomez-Duran et al., 2010).
Conserved cytb mutations were enriched in northern Europe and less prevalent in southern Europe, which is suggestive of selection allowing adaptation to a colder northern climate (Montiel-Sosa et al., 2006). Maximal oxygen uptake (VO2 max) and mitochondrial oxidative damage have been shown to be higher in human subjects from European haplogroup H compared with haplogroup J (Marcuello et al., 2009 and Martinez-Redondo et al., 2010). By contrast, studies of cytoplasmic hybrids (cybrids) harboring mitochondria with either haplogroup H or haplogroup T in cultured cells with identical nuclear backgrounds show no functionally important differences in bioenergetic capacities and coupling efficiencies (Amo et al., 2008) and results were similar for both isolated mitochondria and mitochondria within cells. Furthermore, cybrid studies comparing arctic (A, C and D) or tropical (L1, L2 and L3) haplogroups yielded no overall differences between arctic and tropical mtDNA haplogroups with regard to the overall kinetics of substrate oxidation (Amo and Brand, 2007). Intriguingly, mitochondria from Arctic haplogroups had similar or greater coupling efficiency than mitochondria from tropical haplogroups, which is contrary to the hypothesis that mitochondrial haplogroups with lower coupling efficiency were positively selected during radiations of modern humans (Amo and Brand, 2007).
To date, the relationship between mitochondrial genetic variation and metabolic rate has not been directly evaluated in human subjects. In this study we examined data gathered in the Health, Aging and Body Composition (Health ABC) Study, a longitudinal study of community-dwelling African and European American adults. We compare resting metabolic rate (RMR) measured by indirect calorimetry and total energy expenditure (TEE) measured by the doubly-labeled water method across the major African and European haplogroups and identify individual mtDNA variants that influence RMR and TEE. Both activity energy expenditure and physical activity level decrease with age (Black et al., 1996 and Elia et al., 2000) and are considered to underlie a range of age-associated pathological conditions, including CVD, neuromuscular and neurological impairments and other life-style changes generally associated with senescence (Linnane, 1992). Higher levels of physical activity are also associated with reductions in coronary heart disease (Wannamethee et al., 1998), cancer incidence (Gregg et al., 2003), falls (Gregg et al., 2000), and physical disability (Ferrucci et al., 1999). Identifying genetic variants that influence metabolic rate and energy expenditure may have broad implications for investigations of functional decline and disease risk.
2. Materials and methods
2.1. Participants
Participants were part of the Health, Aging and Body Composition (Health ABC) Study, a prospective cohort study of 3075 community-dwelling black and white men and women living in Memphis, TN, or Pittsburgh, PA, and aged 70–79 years at recruitment in 1997. To identify potential participants, a random sample of white and all black Medicare-eligible elders, within designated zip code areas, were contacted. To be eligible, participants had to report no difficulty with activities of daily living, walking a quarter of a mile, or climbing 10 steps without resting. They also had to be free of life-threatening cancer diagnoses and have no plans to move out of the study area for at least 3 years. The sample was approximately balanced for sex (51% women) and 42% of participants were black. Participants self-designated race/ethnicity from a fixed set of options (Asian/Pacific Islander, black/African American, white/Caucasian, Latino/Hispanic, do not know, other). The study was designed to have sufficient numbers of Blacks to allow separate estimates of the relationship of body composition to functional decline. All eligible participants signed a written informed consent, approved by the institutional review boards at the clinical sites. This study was approved by the institutional review boards of the clinical sites and the coordinating center (University of California, San Francisco).
2.2. Resting metabolic rate
Resting metabolic rate was measured via indirect calorimetry on a Deltatrac II respiratory gas analyzer (Datex Ohmeda Inc, Helsinki), detailed procedures have been described elsewhere (Blanc et al., 2004). While in a fasting state and after 30 min of rest, a respiratory gas exchange hood was placed over the participant’s head and RMR was measured minute-by-minute for 40 min. To avoid gas exchange created by the initial placement of the hood, only the final 30 min were used in subsequent calculations. Movement or sleeping during the test was noted and those time periods were excluded from the RMR calculation. Methanol burn tests were performed in duplicate once or twice per month. Carbon dioxide recovery averaged 100.1 ± 1.4% at the Pittsburgh site and 100.5 ± 1.5% at the Memphis site. The gas exchange ratios for methanol differed by 2.5% between sites (Memphis: 0.66 ± 0.01, Pittsburgh: 0.68 ± 0.01, p < 0.001) and this difference did not demonstrate a trend over time. Therefore, a correction factor was employed to equate the two study sites by dividing the respiratory ratios for participants enrolled at Pittsburgh by 1.025.
2.3. Total energy expenditure
Total energy expenditure was measured using the 2-point doubly-labeled water (DLW) technique that has been previously described in detail (Blanc et al., 2002). Briefly, on the first visit, participants ingested 2 g/kg estimated total body water (TBW) dose of DLW, composed of 1.9 g/kg estimated TBW of 10% H 182O and 0.12 g/kg estimated TBW of 99.9% 2H2O. After dosing, urine samples were obtained at approximately 2, 3, and 4 h. Two consecutive urine voids were taken during a second visit to the laboratory, approximately fourteen days after the first visit. Plasma from a 5 mL blood sample was obtained from everyone but only used for those who had evidence of delayed isotopic equilibration likely caused from urine retention in the bladder (n = 28) (Blanc et al., 2002). Urine and plasma samples were stored at − 20 °C until analysis by isotope ratio mass spectrometry.
Dilution spaces for 2H and 18O were calculated according to Coward (1990). Total body water was calculated as the average of the dilutions spaces of 2H and 18O after correction for isotopic exchange (1.041 for 2H and 1.007 for 18O). Carbon dioxide production was calculated using the two-point DLW method outlined by Schoeller and colleagues (Schoeller et al., 1986 and Schoeller and van Santen, 1982) and TEE was derived using Weir’s equation (Weir, 1949). A food quotient of 0.86 was used from the third National Health and Nutrition Examination Survey (Kant, 2002) and from Black et al. (1986). All values of energy expenditure were converted to kilocalories per day (kcal/day) and the thermic effect of meals was assumed to be 10% of TEE (Bloesch et al., 1988). For measurement of total body water, the intra-subject repeatability calculated as the average percentage difference between the two analyses, was − 0.1 ± 1.2%. The intra-tester repeatability of TEE based on blinded, repeat, urine isotopic analysis was excellent (mean difference = 1.2 ± 5.4%, n = 16) and compared well with a recent review article (Elia et al., 2000).
2.4. Body composition
Lean mass (FFM) was measured annually using a Hologic 4500A Scanner (Hologic Inc, Watham, MA). Body composition analysis was performed using HOLOGIC software (version 8.21; Hologic Inc). Calibration was performed 3 times per week using whole body quality control phantoms outlined in the Hologic manual. Absolute variation between the clinic sites was monitored by cross-calibrating the two scanners using separate phantoms. Validation analyses on the scanners detected a systematic overestimation of FFM that was subsequently corrected by multiplying by a factor of 0.964 (Tylavsky et al., 2003). Values of lean mass were calculated after removing mass due to bone mineral content (BMC).
2.5. Genotyping
Genomic DNA was extracted from buffy coat collected using PUREGENE® DNA Purification Kit during the baseline exam. Genotyping was performed by the Center for Inherited Disease Research (CIDR) using the Illumina Human1M-Duo BeadChip system. This platform includes 138 common mtDNA SNPs with ≥ 1% frequency including the majority of haplogroup-defining variants (Saxena et al., 2006). Samples were excluded from the dataset for the reasons of sample failure, genotypic sex mismatch, and first-degree relative of an included individual based on genotype data. Genotyping of 138 mtDNA SNPs (including 137 variant sites) was successful for 2797 unrelated individuals including 294 participants with RMR and TEE measurements who this analysis is based upon. The major African (L: L0, L1, L2, L3) and European (N: H, JT, UK) haplogroups were defined using PhyloTree (van Oven and Kayser, 2009) (Table S1).
2.6. Data analysis
Baseline comparisons were performed between African (L) and European (N) haplogroups using analysis of variance (ANOVA) for continuous variables and the χ2 statistic for categorical variables. Pairwise ANOVA comparisons of RMR and TEE were performed for the major African (L: L0, L1, L2, L3) and European (N: H, JT, UK) haplogroups using PROC GLM in SAS adjusting for age, sex and lean mass. Individual mtDNA variants were also examined for associations with RMR and TEE. To account for confounding by ancestry we derived eigenvectors using principal components analysis (PCA) for all Health ABC participants with mtDNA genotype data (Biffi et al., 2010). Since missing genotypes can eliminate individual samples from PCA, we based PCA on coding region variants (mt750 to mt15930) and variants missing in < 1% of the sample. A total of 107 mtDNA variants generated eigenvectors for 2471 Health ABC samples. The first 6 eigenvectors account for 71% of the variance in the dataset. Analyses of individual mtDNA variants were adjusted for age, study site, sex, lean mass, and the first 6 eigenvectors of mitochondrial genetic ancestry derived from principal component analysis. All analyses were carried out using SAS version 9.1 (SAS Institute Inc, Cary, NC).
3. Results
A total of 294 participants with RMR and TEE measurements were genotyped for 137 variant mtDNA SNPs. We identified 132 participants from African haplogroup L and 162 participants from European haplogroup N. Descriptive characteristics of haplogroups L and N are listed in Table 1. Participants from haplogroups L and N had similar age, sex composition, and lean mass. Haplogroup N had an elevated number of participants from Pittsburgh than haplogroup L (p = 0.03). RMR did not differ between participants from the two clinic sites (p = 0.48) but TEE was significantly higher (p = 0.008) among participants from Pittsburgh (2217 ± 27 kcal/day) when compared with those from Memphis (2120 ± 25 kcal/day) after adjustment for baseline age, sex, haplogroup, and lean mass. Both RMR and TEE were significantly elevated in haplogroup N participants compared to haplogroup L participants after adjustment for baseline age, sex, clinic site, and lean mass (Table 2).
Table 1.
Baseline characteristics of participants stratified by the major African and European haplogroups.
| African | European | ||
|---|---|---|---|
| Haplogroup | L | N | |
| N | 132 | 162 | p-value |
| Age, years, mean (SD) | 73 (2.9) | 74 (2.8) | 0.12 |
| Sex | |||
| Female, n (%) | 64 (44) | 81 (56) | 0.29 |
| Male, n (%) | 68 (46) | 64 (48) | 0.16 |
| Study site | |||
| Memphis, n (%) | 78 (49) | 82 (51) | 0.75 |
| Pittsburgh, n (%) | 54 (40) | 80 (60) | 0.03 |
| Lean mass, kg, mean (SD) | 48.7 (9.6) | 46.6 (10) | 0.07 |
Table 2.
Comparison of resting metabolic rate and total energy expenditure between the major African and European haplogroups. Values are Least Squares means and Standard Errors adjusted for age, sex, clinic site and lean mass.
| African | European | ||
|---|---|---|---|
| Haplogroup | L | N | |
| N | 132 | 162 | P-value |
| RMR kcal/day, Mean (SE)* | 1221 (11) | 1326 (9) | <0.0001 |
| TEE kcal/day, Mean (SE)* | 2079 (27) | 2237 (25) | <0.0001 |
RMR = Resting metabolic rate determined by indirect calorimetry.
TEE = Total energy expenditure determined using doubly-labeled water.
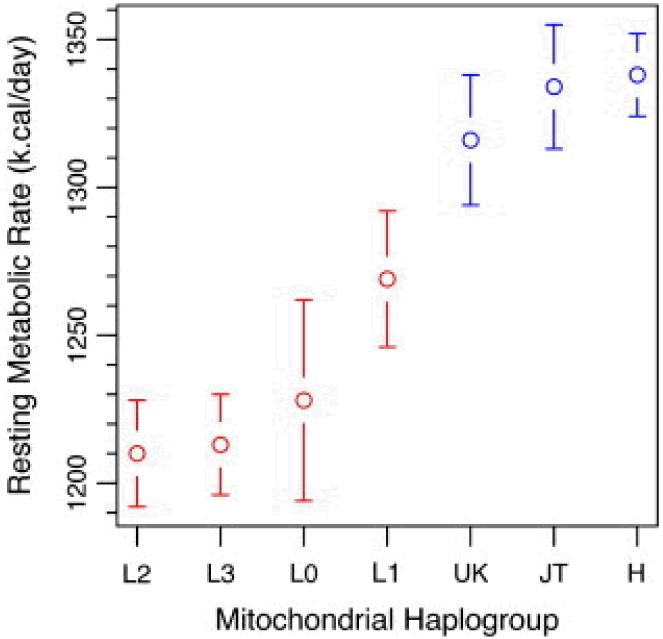
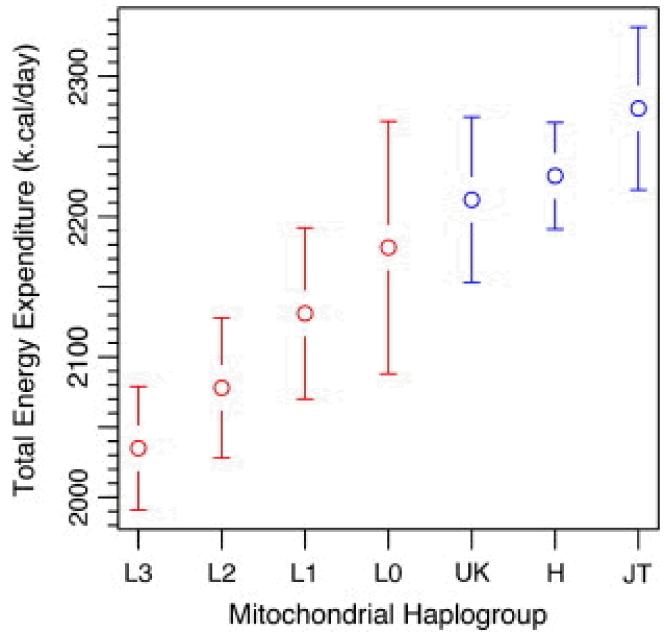
Fig. 1 and Fig. 2 illustrate the differences in RMR and TEE for the common African and European in haplogroups after adjustment for age, sex, clinic site, and lean mass. Haplogroups were ranked in order of increasing RMR (Fig. 1) and TEE (Fig. 2) to allow visual comparisons. African haplogroups L0 (n = 12), L2 (n = 42), and L3 (n = 49) had significantly lower RMRs than the three European haplogroups, UK (n = 31), JT (n = 30), and H (n = 70). African haplogroup L1 (n = 27) had an RMR intermediate to the European and remaining African haplogroups. For African haplogroup L1, RMR was significantly lower (p = 0.01) than the highest RMR from European haplogroup H and significantly higher (p = 0.03) than the lowest RMR from African haplogroup L3 (Table 3). African haplogroup L3 had a significantly lower TEE than the three European haplogroups and haplogroup L2 had a significantly lower TEE than haplogroups H and JT (Table 4).
Fig. 1.
Summary of resting metabolic rate (kcal/day) for major mitochondrial haplogroups. Values are LS means and SEs adjusted for age, sex, and lean mass.
Fig. 2.
Summary of total energy expenditure (kcal/day) for major mitochondrial haplogroups. Values are LS means and SEs adjusted for age, sex, and lean mass.
Table 3.
Summary of pairwise ANOVA comparisons (p-values) for differences in resting metabolic rate (kcal/day). Values are Least Squares means and Standard Errors adjusted for age, sex, and lean mass.
| L1 | JT | UK | H | |
|---|---|---|---|---|
| L3 | 0.03 | 0.0003 | <0.0001 | <0.0001 |
| L2 | ns | 0.001 | 0.0001 | <0.0001 |
| L0 | ns | 0.04 | 0.01 | 0.002 |
| L1 | ns | ns | ns | 0.01 |
Table 4.
Summary of pairwise ANOVA comparisons (p-values) for differences in total energy expenditure (kcal/day). Values are Least Squares means and Standard Errors adjusted for age, sex, and lean mass.
| UK | H | JT | |
|---|---|---|---|
| L3 | 0.01 | 0.0004 | 0.004 |
| L2 | ns | 0.01 | 0.02 |
Individual mtDNA variants for associations were examined for associations with RMR and TEE (Table S2 and Table S3). After adjustment for age, sex, lean mass, and 6 eigenvectors of mitochondrial genetic ancestry, variants in mt-RNR2, and ND5 were associated with RMR (Table S2). In addition, one participant carrying mtDNA variants Mito9093G (ATP6, complex V) and Mito11377A (ND4, complex I) had a RMR of 1647 kcal/day compared with Mito9093A and Mito11377G carriers (n = 293) having a RMR of 1284 ± 8 kcal/day. Variants in mt-RNR2, COI, ND5 and CYTB were associated with TEE after multivariate adjustment (Table S3). We examined individual mtDNA variants controlling for mtDNA population stratification using mitochondrial PCA (Biffi et al., 2010). This method has been demonstrated to outperform haplogroup-stratified or adjusted association analyses with no loss in power for detection of true associations (Biffi et al., 2010). Additionally, correlation between nuclear and mitochondrial principal components was limited and adjustment for nuclear PCs had no effect on mitochondrial analysis (Biffi et al., 2010).
4. Discussion
There has been a long-standing debate over the role of climate in driving adaptive selection of mtDNA asHomo sapiens migrated out of Africa into temperate and arctic Eurasia. In this study we observed significant bioenergetic differences between the major African and European mitochondrial haplogroups and among the common African haplogroups. A specific mechanism remains to be identified but the differences could be due to effects on respiratory rates, specific dietary make-up, mitochondrial coupling efficiencies, or ATP supply. Some previous cybrid studies do not support a role for selection of lower OXPHOS coupling efficiencies during radiations of modern humans (Amo and Brand, 2007 and Amo et al., 2008), suggesting that more subtle mechanisms may ultimately be responsible for the differences observed in this study. The greatest differences in RMR and TEE were observed when African haplogroup L participants were compared to European haplogroup N participants. Both RMR and TEE were significantly elevated in haplogroup N participants compared to those from haplogroup L after adjustment for baseline age, sex, clinic site, and lean mass. Determining which variants, genes or OXPHOS complexes contribute to the metabolic differences observed between haplogroups L and N will be complicated by the fact that variants in complexes I (ND3,ND4), III (CytB), IV (COIII) and V (ATP6) all define this split (van Oven and Kayser, 2009).
Within African haplogroup L, we observed that haplogroup L1 had an RMR intermediate to the European and remaining African haplogroups. African haplogroups L2 and L3 exhibited significantly lower RMR values than all others examined. Determining which variants or genes contribute to the metabolic differences observed for haplogroups L1, L2, and L3 may be less difficult since variants in OXPHOS complexes I (ND1, ND3, ND5, and ND6) and IV(COI and COIII) define these haplogroups (van Oven and Kayser, 2009). Generally, haplogroups L0–L3 are most common to sub-Saharan Africa and largely of East African origin (Soares et al., 2009) but the widespread distribution and diversity of these haplogroups makes their exact points of origin within Africa difficult to trace (Salas et al., 2002). Approximately 90% of the African–American mtDNA pool originates from West and Southwest Africa (Salas et al., 2005). An examination of the African mtDNA sub-haplogroups that are prominent in African Americans reveals that haplogroup L1 possibly has a different point of origin than haplogroups L2 and L3 (Salas et al., 2005), which may in part contribute to the RMR differences observed between these groups. The two L1 sub-haplogroups found in African–Americans, L1b and L1c, have a likely origin in the Congo River region of Central Africa which today includes the equatorial forests (Salas et al., 2005). The L2 and L3 sub-haplogroups common in African–Americans, L2a, L2b, L2c, L3b, L3d, and L3e, likely originated in the Niger River region of Western African near the Sahara Desert (Salas et al., 2005). It is not clear whether adaptation to different regions played a role in the development of West African sub-haplogroups of L1, L2, and L3 leading to differences in RMR observed in this study. Of interest, though, is that we have documented RMR differences among West African sub-haplogroups (L1 vs. L2–L3) that had independent origins from isolated geographic regions.
The observed haplogroup differences in TEE were not as apparent as those for RMR. The discrepancy between TEE and RMR is largely attributable to the addition of physical activity as part of TEE. TEE is comprised of three major components including: RMR; activity energy expenditure; and the thermic effect of food metabolism Manini (2010). RMR accounts for 60–80% of TEE, thermic effect of food metabolism uses 10% of TEE, and activity energy expenditure can comprise 20–40% of TEE Manini (2010). RMR is an innate feature of the tissue and the cells that make up that tissue. It is far less determined by environmental factors leading to a clearer distinction among haplogroups. The assessment TEE, however, includes 20–40% of energy that is due to environmental factors outside the control of haplogroup categorization. By adding a large environmentally determined factor, the haplogroup differences generally become less distinct. Differences in TEE among European haplogroup N members emerged for TEE, however, with haplogroup JT exhibiting a nominally higher TEE than all other haplogroups examined in this study. Haplogroup JT is of Levantine origins (42–54 kya) including present-day Israel, Jordan, Lebanon, Palestine, Syria and the Sinai Peninsula (Richards et al., 2000). The original peoples associated with haplogroup JT brought farming and herding to Europe beginning about 10 kya. This contrasts with all other West Eurasian-origin groups (H, V, U, K, I, W, X) which were largely hunter–gatherers. As with the West African sub-haplogroups, it is not clear whether local adaptation played a role in the development of haplogroup JT. Haplogroup J is over-represented in long-lived people and centenarians from several populations (De Benedictis et al., 1999,Niemi et al., 2003 and Ross et al., 2001). The mtDNA control region variant that defines Caucasian haplogroup J (mtDNA 295C/T) has been found to change mitochondrial transcription and copy number (Suissa et al., 2009). Cybrids containing haplogroup J mtDNA had a greater than 2-fold increase in mtDNA copy number compared with cybrids containing haplogroup H mtDNA. This is one of the few examples demonstrating functional consequences for a polymorphism underlying a specific haplogroup. In this case, the impact of the haplogroup J regulatory region mutation on mtDNA replication or stability may partially account for population-based observations that haplogroup J is associated with human longevity.
Although functional analyses of the polymorphisms underlying haplogroup definitions are needed, there are few reports that identify functional effects of mtDNA polymorphisms — mainly due to methodological difficulties (Kazuno et al., 2006). MtDNAs harboring the two founder macrohaplogroup N missense mutations, ND3 (complex I) nucleotide 10398A/G and ATP6 (complex V) nucleotide 8701A/G, exhibited alterations in mitochondrial matrix pH and intracellular calcium dynamics (Kazuno et al., 2006). A second study of common Asian haplogroups identified distinctive intracellular calcium dynamics among subhaplogroups G3/G4 and D4a (Kazuno et al., 2008). Subhaplogroup G3/G4 (characterized by mtDNA polymorphisms at sites 1413, 2109, 3434, 5460, 7521, 9011, 9670 and 15940) showed lower cytosolic and mitochondrial calcium levels (Kazuno et al., 2008). By contrast, subhaplogroup D4a (characterized by mtDNA variant 13651A/G) showed higher mitochondrial and cytosolic calcium levels (Kazuno et al., 2008). Haplogroup studies can mask the effects of individual nucleotide changes because most mitochondrial haplogroups can be defined by several control-region and/or coding-region variants. For example, the large number of variants that are closely associated with one another and that define macrohaplogroup N (characterized by mtDNA variants 8701, 9540, 10398, 10873, and 15301 (van Oven and Kayser, 2009)) complicate interpretation of the functional and association data. This implies that functional studies should account for all mtDNA variants within the mitochondrial genome, especially for haplogroups that can be defined by several variants. The analysis of mtDNA is further complicated by recurrent mutation at the same mtDNA site across divergent haplogroups, especially in the control region, which can sometimes erase diagnostic specificity of a particular variant. That the same mutations have been observed repeatedly on different mtDNA backgrounds (e.g. haplogroups) has been cited as evidence of convergent adaptive evolution of particular mtDNA mutations (Wallace, 2010).
Determining how specific mitochondrial variants influence physiology is not only complicated by the complexities of mtDNA evolution as outlined above, but also requires the coordinated expression of hundreds of nuclear genes and a few dozen mitochondrial genes. This complex genetic architecture suggests that there might be a complex set of gene interactions (epistases) involving genetic variation in the nuclear and mitochondrial genomes (Mishmar et al., 2006, Pravenec et al., 2007, Rand et al., 2001, Rand et al., 2006, Rand et al., 2004, Rand and Kann, 1998 and Willett and Burton, 2003). This is especially important for admixed populations (e.g. African–Americans and Latinos) where it is not uncommon to find individuals with mtDNA and nuclear DNA of different ancestries (e.g. individuals with African mtDNA in a European genetic background or Native American mtDNA in an African genetic background). It is not known how breaking up potentially coadapted mitochondrial and nuclear OXPHOS gene complexes affects human health, but increasing our understanding of mitochondrial–nuclear epistasis in admixed populations may be important for examining the genetic basis of chronic disease.
Because mitochondria play a central role in energy metabolism it is likely that mitochondrial–nuclear epistasis has important fitness effects (Dowling et al., 2007, Hutter and Rand, 1995, Rand et al., 2001, Rawson and Burton, 2002, Rhode and Cruzan, 2005,Schmidt et al., 2001, Tranah, 2011 and Willett and Burton, 2003) and is evolutionarily important (Rand et al., 2004). Both activity energy expenditure and physical activity level decrease with age (Black et al., 1996 and Elia et al., 2000) and are considered to underlie a range of age-associated pathological conditions (Gregg et al., 2003, Gregg et al., 2000, Linnane, 1992 and Wannamethee et al., 1998). TEE demonstrates an inverted U pattern across the lifespan. In the first two decades of life, TEE increases 2-fold and declines dramatically after the age of 40 years (Manini, 2010). Aging is also associated with a progressive decline in whole-body RMR at a rate of 1–2% per decade after 20 years of age (Elia et al., 2000). Conditions such as Alzheimer’s, chronic obstructive pulmonary disease, congestive heart failure, Parkinson’s, diabetes, malnourishment and cancer are associated with a change in RMR (Toth and Poehlman, 2000). It is not clear, however, why bioenergetic decline occurs and how increased energetic expenditure protects older adults from physical disability and premature mortality. Future studies seeking to link mitochondrial genetic variation to bioenergetics, physiological traits or disease risk will need to address the analytic challenges posed both by the unique mode of mtDNA evolution and the complexities of examining mitochondrial – nuclear epistasis. Identifying mitochondrial genetic variants that influence energy expenditure and metabolic rate could lead to interventions that have a very significant impact on prolonging the productive and healthy years of human life.
Supplementary Material
Acknowledgments
This research is supported in part by the Intramural Research Program of the NIH, National Institute on Aging. This research was supported by NIA contracts N01AG62101, N01AG62103, N01AG62106 and NIA grant 1R03AG032498-01. The genome-wide association study was funded by NIA grant 1R01AG032098-01A1 to Wake Forest University Health Sciences and genotyping services were provided by the Center for Inherited Disease Research (CIDR). CIDR is fully funded through a federal contract from the National Institutes of Health to The Johns Hopkins University, contract number HHSN2682007820962C.
Footnotes
Conflict of interest statement
The authors declare no conflict of interest.
References
- Amo T, Brand MD. Were inefficient mitochondrial haplogroups selected during migrations of modern humans? A test using modular kinetic analysis of coupling in mitochondria from cybrid cell lines. Biochem. J. 2007;404:345–351. doi: 10.1042/BJ20061609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amo T, Yadava N, Oh R, Nicholls DG, Brand MD. Experimental assessment of bioenergetic differences caused by the common European mitochondrial DNA haplogroups H and T. Gene. 2008;411:69–76. doi: 10.1016/j.gene.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balloux F, Handley LJ, Jombart T, Liu H, Manica A. Climate shaped the worldwide distribution of human mitochondrial DNA sequence variation. Proc Biol Sci. 2009;276:3447–3455. doi: 10.1098/rspb.2009.0752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biffi A, Anderson CD, Nalls MA, Rahman R, Sonni A, Cortellini L, Rost NS, Matarin M, Hernandez DG, Plourde A, de Bakker PI, Ross OA, Greenberg SM, Furie KL, Meschia JF, Singleton AB, Saxena R, Rosand J. Principal-component analysis for assessment of population stratification in mitochondrial medical genetics. Am. J. Hum. Genet. 2010;86:904–917. doi: 10.1016/j.ajhg.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black AE, Prentice AM, Coward WA. Use of food quotients to predict respiratory quotients for the doubly-labelled water method of measuring energy expenditure. Human nutrition. 1986;40:381–391. [PubMed] [Google Scholar]
- Black AE, Coward WA, Cole TJ, Prentice AM. Human energy expenditure in affluent societies: an analysis of 574 doubly-labelled water measurements. Eur. J. Clin. Nutr. 1996;50:72–92. [PubMed] [Google Scholar]
- Blanc S, Colligan AS, Trabulsi J, Harris T, Everhart JE, Bauer D, Schoeller DA. Influence of delayed isotopic equilibration in urine on the accuracy of the (2)H(2) method in the elderly. J. Appl. Physiol. 2002;92:1036–1044. doi: 10.1152/japplphysiol.00743.2001. [DOI] [PubMed] [Google Scholar]
- Blanc S, Schoeller DA, Bauer D, Danielson ME, Tylavsky F, Simonsick EM, Harris TB, Kritchevsky SB, Everhart JE. Energy requirements in the eighth decade of life. Am. J. Clin. Nutr. 2004;79:303–310. doi: 10.1093/ajcn/79.2.303. [DOI] [PubMed] [Google Scholar]
- Bloesch D, Schutz Y, Breitenstein E, Jequier E, Felber JP. Thermogenic response to an oral glucose load in man: comparison between young and elderly subjects. J. Am. Coll. Nutr. 1988;7:471–483. doi: 10.1080/07315724.1988.10720263. [DOI] [PubMed] [Google Scholar]
- Coward WA, Report of an IDECG Expert Working Group . Calculation of pool sizes and flux rates. In: Prentice AM, editor. the doubly labeled water method: technical recommendations for use in humans. Vienna, Austria, AERA: 1990. pp. 48–68. [Google Scholar]
- De Benedictis G, Rose G, Carrieri G, De Luca M, Falcone E, Passarino G, Bonafe M, Monti D, Baggio G, Bertolini S, Mari D, Mattace R, Franceschi C. Mitochondrial DNA inherited variants are associated with successful aging and longevity in humans. Faseb J. 1999;13:1532–1536. doi: 10.1096/fasebj.13.12.1532. [DOI] [PubMed] [Google Scholar]
- Dowling DK, Friberg U, Hailer F, Arnqvist G. Intergenomic epistasis for fitness: within-population interactions between cytoplasmic and nuclear genes in Drosophila melanogaster. Genetics. 2007;175:235–244. doi: 10.1534/genetics.105.052050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia M, Ritz P, Stubbs RJ. Total energy expenditure in the elderly. Eur. J. Clin. Nutr. 2000;54(Suppl 3):S92–S103. doi: 10.1038/sj.ejcn.1601030. [DOI] [PubMed] [Google Scholar]
- Elson JL, Turnball DM, Howell N. Comparative genomics and the evolution of human mitochondrial DNA: assessing the effects of selection. Am. J. Hum. Genet. 2004;74:229–238. doi: 10.1086/381505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrucci L, Izmirlian G, Leveille S, Phillips CL, Corti MC, Brock DB, Guralnik JM. Smoking, physical activity, and active life expectancy. Am. J. Epidemiol. 1999;149:645–653. doi: 10.1093/oxfordjournals.aje.a009865. [DOI] [PubMed] [Google Scholar]
- Giles RE, Blanc H, Cann HM, Wallace DC. Maternal inheritance of human mitochondrial DNA. Proc Natl Acad Sci USA. 1980;77:6715–6719. doi: 10.1073/pnas.77.11.6715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Duran A, Pacheu-Perez D, Lopez-Gallardo E, Diez-Sanchez C, Montoya J, Lopez-Perez MJ, Ruiz-Pesini E. Unmasking the causes of multifactorial disorders: OXPHOS differences between mitochondrial haplogroups. Hum. Mol. Genet. 2010;19:3343–3353. doi: 10.1093/hmg/ddq246. [DOI] [PubMed] [Google Scholar]
- Gregg EW, Pereira MA, Caspersen CJ. Physical activity, falls, and fractures among older adults: a review of the epidemiologic evidence. J. Am. Geriatr. Soc. 2000;48:883–893. doi: 10.1111/j.1532-5415.2000.tb06884.x. [DOI] [PubMed] [Google Scholar]
- Gregg EW, Cauley JA, Stone K, Thompson TJ, Bauer DC, Cummings SR, Ensrud KE. Relationship of changes in physical activity and mortality among older women. Jama. 2003;289:2379–2386. doi: 10.1001/jama.289.18.2379. [DOI] [PubMed] [Google Scholar]
- Hutter CM, Rand DM. Competition between mitochondrial haplotypes in distinct nuclear genetic environments: Drosophila pseudoobscura vs D. persimilis. Genetics. 1995;140:537–548. doi: 10.1093/genetics/140.2.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kant AK. Nature of dietary reporting by adults in the third National Health and Nutrition Examination Survey, 1988-1994. J. Am. Coll. Nutr. 2002;21:315–327. doi: 10.1080/07315724.2002.10719229. [DOI] [PubMed] [Google Scholar]
- Kazuno AA, Munakata K, Nagai T, Shimozono S, Tanaka M, Yoneda M, Kato N, Miyawaki A, Kato T. Identification of mitochondrial DNA polymorphisms that alter mitochondrial matrix pH and intracellular calcium dynamics. PLoS Genet. 2006;2:e128. doi: 10.1371/journal.pgen.0020128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazuno AA, Munakata K, Tanaka M, Kato N, Kato T. Relationships between mitochondrial DNA subhaplogroups and intracellular calcium dynamics. Mitochondrion. 2008;8:164–169. doi: 10.1016/j.mito.2007.12.002. [DOI] [PubMed] [Google Scholar]
- Kivisild T, Shen P, Wall DP, Do B, Sung R, Davis K, Passarino G, Underhill PA, Scharfe C, Torroni A, Scozzari R, Modiano D, Coppa A, de Knijff P, Feldman M, Cavalli-Sforza LL, Oefner PJ. The role of selection in the evolution of human mitochondrial genomes. Genetics. 2006;172:373–387. doi: 10.1534/genetics.105.043901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnane AW. Mitochondria and aging: the universality of bioenergetic disease. Aging Milano. 1992;4:267–271. doi: 10.1007/BF03324106. [DOI] [PubMed] [Google Scholar]
- Maca-Meyer N, Gonzalez AM, Larruga JM, Flores C, Cabrera VM. Major genomic mitochondrial lineages delineate early human expansions. BMC Genet. 2001;2:13. doi: 10.1186/1471-2156-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manini TM. Energy expenditure and aging. Ageing Res. Rev. 2010;9:1–11. doi: 10.1016/j.arr.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcuello A, Martinez-Redondo D, Dahmani Y, Casajus JA, Ruiz-Pesini E, Montoya J, Lopez-Perez MJ, Diez-Sanchez C. Human mitochondrial variants influence on oxygen consumption. Mitochondrion. 2009;9:27–30. doi: 10.1016/j.mito.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Martinez-Redondo D, Marcuello A, Casajus JA, Ara I, Dahmani Y, Montoya J, Ruiz-Pesini E, Lopez-Perez MJ, Diez-Sanchez C. Human mitochondrial haplogroup H: the highest VO2max consumer — is it a paradox? Mitochondrion. 2010;10:102–107. doi: 10.1016/j.mito.2009.11.005. [DOI] [PubMed] [Google Scholar]
- Mishmar D, Ruiz-Pesini E, Golik P, Macaulay V, Clark AG, Hosseini S, Brandon M, Easley K, Chen E, Brown MD, Sukernik RI, Olckers A, Wallace DC. Natural selection shaped regional mtDNA variation in humans. Proc Natl Acad Sci USA. 2003;100:171–176. doi: 10.1073/pnas.0136972100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishmar D, Ruiz-Pesini E, Mondragon-Palomino M, Procaccio V, Gaut B, Wallace DC. Adaptive selection of mitochondrial complex I subunits during primate radiation. Gene. 2006;378:11–18. doi: 10.1016/j.gene.2006.03.015. [DOI] [PubMed] [Google Scholar]
- Moilanen JS, Majamaa K. Phylogenetic network and physicochemical properties of nonsynonymous mutations in the protein-coding genes of human mitochondrial DNA. Mol. Biol. Evol. 2003;20:1195–1210. doi: 10.1093/molbev/msg121. [DOI] [PubMed] [Google Scholar]
- Moilanen JS, Finnila S, Majamaa K. Lineage-specific selection in human mtDNA: lack of polymorphisms in a segment of MTND5 gene in haplogroup J. Mol. Biol. Evol. 2003;20:2132–2142. doi: 10.1093/molbev/msg230. [DOI] [PubMed] [Google Scholar]
- Montiel-Sosa F, Ruiz-Pesini E, Enriquez JA, Marcuello A, Diez-Sanchez C, Montoya J, Wallace DC, Lopez-Perez MJ. Differences of sperm motility in mitochondrial DNA haplogroup U sublineages. Gene. 2006;368:21–27. doi: 10.1016/j.gene.2005.09.015. [DOI] [PubMed] [Google Scholar]
- Niemi AK, Hervonen A, Hurme M, Karhunen PJ, Jylha M, Majamaa K. Mitochondrial DNA polymorphisms associated with longevity in a Finnish population. Hum. Genet. 2003;112:29–33. doi: 10.1007/s00439-002-0843-y. [DOI] [PubMed] [Google Scholar]
- Pravenec M, Hyakukoku M, Houstek J, Zidek V, Landa V, Mlejnek P, Miksik I, Dudova-Mothejzikova K, Pecina P, Vrbacky M, Drahota Z, Vojtiskova A, Mracek T, Kazdova L, Oliyarnyk O, Wang J, Ho C, Qi N, Sugimoto K, Kurtz T. Direct linkage of mitochondrial genome variation to risk factors for type 2 diabetes in conplastic strains. Genome Res. 2007;17:1319–1326. doi: 10.1101/gr.6548207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana-Murci L, Semino O, Bandelt HJ, Passarino G, McElreavey K, Santachiara-Benerecetti AS. Genetic evidence of an early exit of Homo sapiens sapiens from Africa through eastern Africa. Nat. Genet. 1999;23:437–441. doi: 10.1038/70550. [DOI] [PubMed] [Google Scholar]
- Rand DM, Kann LM. Mutation and selection at silent and replacement sites in the evolution of animal mitochondrial DNA. Genetica. 1998;102-103:393–407. [PubMed] [Google Scholar]
- Rand DM, Clark AG, Kann LM. Sexually antagonistic cytonuclear fitness interactions in Drosophila melanogaster. Genetics. 2001;159:173–187. doi: 10.1093/genetics/159.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand DM, Haney RA, Fry AJ. Cytonuclear coevolution: the genomics of cooperation. Trends Ecol Evol. 2004;19:645–653. doi: 10.1016/j.tree.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Rand DM, Fry A, Sheldahl L. Nuclear-mitochondrial epistasis and drosophila aging: introgression of Drosophila simulans mtDNA modifies longevity in D. melanogaster nuclear backgrounds. Genetics. 2006;172:329–341. doi: 10.1534/genetics.105.046698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawson PD, Burton RS. Functional coadaptation between cytochrome C and cytochrome C oxidase within allopatric populations of a marine copepod. Proc Natl Acad Sci USA. 2002;99:12955–12958. doi: 10.1073/pnas.202335899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhode JM, Cruzan MB. Contributions of heterosis and epistasis to hybrid fitness. Am. Nat. 2005;166:E124–E139. doi: 10.1086/491798. [DOI] [PubMed] [Google Scholar]
- Richards M, Macaulay V, Hickey E, Vega E, Sykes B, Guida V, Rengo C, Sellitto D, Cruciani F, Kivisild T, Villems R, Thomas M, Rychkov S, Rychkov O, Rychkov Y, Golge M, Dimitrov D, Hill E, Bradley D, Romano V, Cali F, Vona G, Demaine A, Papiha S, Triantaphyllidis C, Stefanescu G, Hatina J, Belledi M, Di Rienzo A, Novelletto A, Oppenheim A, Norby S, Al-Zaheri N, Santachiara-Benerecetti S, Scozari R, Torroni A, Bandelt HJ. Tracing European founder lineages in the Near Eastern mtDNA pool. Am. J. Hum. Genet. 2000;67:1251–1276. [PMC free article] [PubMed] [Google Scholar]
- Ross OA, McCormack R, Curran MD, Duguid RA, Barnett YA, Rea IM, Middleton D. Mitochondrial DNA polymorphism: its role in longevity of the Irish population. Exp. Gerontol. 2001;36:1161–1178. doi: 10.1016/s0531-5565(01)00094-8. [DOI] [PubMed] [Google Scholar]
- Ruiz-Pesini E, Wallace DC. Evidence for adaptive selection acting on the tRNA and rRNA genes of human mitochondrial DNA. Hum. Mutat. 2006;27:1072–1081. doi: 10.1002/humu.20378. [DOI] [PubMed] [Google Scholar]
- Ruiz-Pesini E, Diez C, Lapena AC, Perez-Martos A, Montoya J, Alvarez E, Arenas J, Lopez-Perez MJ. Correlation of sperm motility with mitochondrial enzymatic activities. Clin. Chem. 1998;44:1616–1620. [PubMed] [Google Scholar]
- Ruiz-Pesini E, Lapena AC, Diez-Sanchez C, Perez-Martos A, Montoya J, Alvarez E, Diaz M, Urries A, Montoro L, Lopez-Perez MJ. Human mtDNA haplogroups associated with high or reduced spermatozoa motility. Am. J. Hum. Genet. 2000;67:682–696. doi: 10.1086/303040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Pesini E, Mishmar D, Brandon M, Procaccio V, Wallace DC. Effects of purifying and adaptive selection on regional variation in human mtDNA. Science. 2004;303:223–226. doi: 10.1126/science.1088434. [DOI] [PubMed] [Google Scholar]
- Salas A, Richards M, De la Fe T, Lareu MV, Sobrino B, Sanchez-Diz P, Macaulay V, Carracedo A. The making of the African mtDNA landscape. Am. J. Hum. Genet. 2002;71:1082–1111. doi: 10.1086/344348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas A, Carracedo A, Richards M, Macaulay V. Charting the ancestry of African Americans. Am. J. Hum. Genet. 2005;77:676–680. doi: 10.1086/491675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena R, de Bakker PI, Singer K, Mootha V, Burtt N, Hirschhorn JN, Gaudet D, Isomaa B, Daly MJ, Groop L, Ardlie KG, Altshuler D. Comprehensive association testing of common mitochondrial DNA variation in metabolic disease. Am. J. Hum. Genet. 2006;79:54–61. doi: 10.1086/504926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt TR, Wu W, Goodman M, Grossman LI. Evolution of nuclear- and mitochondrial-encoded subunit interaction in cytochrome c oxidase. Mol. Biol. Evol. 2001;18:563–569. doi: 10.1093/oxfordjournals.molbev.a003836. [DOI] [PubMed] [Google Scholar]
- Schoeller DA, van Santen E. Measurement of energy expenditure in humans by doubly labeled water method. J. Appl. Physiol. 1982;53:955–959. doi: 10.1152/jappl.1982.53.4.955. [DOI] [PubMed] [Google Scholar]
- Schoeller DA, Ravussin E, Schutz Y, Acheson KJ, Baertschi P, Jequier E. Energy expenditure by doubly labeled water: validation in humans and proposed calculation. Am. J. Physiol. 1986;250:R823–R830. doi: 10.1152/ajpregu.1986.250.5.R823. [DOI] [PubMed] [Google Scholar]
- Soares P, Ermini L, Thomson N, Mormina M, Rito T, Rohl A, Salas A, Oppenheimer S, Macaulay V, Richards MB. Correcting for purifying selection: an improved human mitochondrial molecular clock. Am. J. Hum. Genet. 2009;84:740–759. doi: 10.1016/j.ajhg.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suissa S, Wang Z, Poole J, Wittkopp S, Feder J, Shutt TE, Wallace DC, Shadel GS, Mishmar D. Ancient mtDNA genetic variants modulate mtDNA transcription and replication. PLoS Genet. 2009;5:e1000474. doi: 10.1371/journal.pgen.1000474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth MJ, Poehlman ET. Energetic adaptation to chronic disease in the elderly. Nutr. Rev. 2000;58:61–66. doi: 10.1111/j.1753-4887.2000.tb01840.x. [DOI] [PubMed] [Google Scholar]
- Tranah GJ. Mitochondrial-nuclear epistasis: implications for human aging and longevity. Ageing Res. Rev. 2011;10:238–252. doi: 10.1016/j.arr.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tylavsky F, Lohman T, Blunt BA, Schoeller DA, Fuerst T, Cauley JA, Nevitt MC, Visser M, Harris TB. QDR 4500A DXA overestimates fat-free mass compared with criterion methods. J. Appl. Physiol. 2003;94:959–965. doi: 10.1152/japplphysiol.00732.2002. [DOI] [PubMed] [Google Scholar]
- van Oven M, Kayser M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum. Mutat. 2009;30:E386–E394. doi: 10.1002/humu.20921. [DOI] [PubMed] [Google Scholar]
- Wallace DC. Colloquium paper: bioenergetics, the origins of complexity, and the ascent of man. Proc Natl Acad Sci USA. 2010;107(Suppl 2):8947–8953. doi: 10.1073/pnas.0914635107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC, Ruiz-Pesini E, Mishmar D. mtDNA variation, climatic adaptation, degenerative diseases, and longevity. Cold Spring Harb. Symp. Quant. Biol. 2003;68:479–486. doi: 10.1101/sqb.2003.68.471. [DOI] [PubMed] [Google Scholar]
- Wallace DC, Fan W, Procaccio V. Mitochondrial energetics and therapeutics. Annu. Rev. Pathol. 2010;5:297–348. doi: 10.1146/annurev.pathol.4.110807.092314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wannamethee SG, Shaper AG, Walker M. Changes in physical activity, mortality, and incidence of coronary heart disease in older men. Lancet. 1998;351:1603–1608. doi: 10.1016/S0140-6736(97)12355-8. [DOI] [PubMed] [Google Scholar]
- Weir JB. New methods for calculating metabolic rate with special reference to protein metabolism. J. Physiol. 1949;109:1–9. doi: 10.1113/jphysiol.1949.sp004363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willett CS, Burton RS. Environmental influences on epistatic interactions: viabilities of cytochrome C genotypes in interpopulation crosses. Evolution. 2003;57:2286–2292. doi: 10.1111/j.0014-3820.2003.tb00240.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.