Summary
We applied a new Cre-dependent, genetically modified rabies-based tracing system to map direct synaptic connections to CA1 excitatory and inhibitory neuron types in mouse hippocampus. We found common inputs to excitatory and inhibitory CA1 neurons from CA3, CA2, entorhinal cortex and the medial septum (MS), and unexpectedly also from the subiculum. Excitatory CA1 neurons receive inputs from both cholinergic and GABAergic MS neurons while inhibitory CA1 neurons receive a great majority of input from GABAergic MS neurons; both cell types also receive weaker input from glutamatergic MS neurons. Comparisons of inputs to CA1 PV+ interneurons versus SOM+ interneurons showed similar strengths of input from the subiculum, but PV+ interneurons receive much stronger input than SOM+ neurons from CA3, entorhinal cortex and MS. Differential input from CA3 to specific CA1 cell types was also demonstrated functionally using laser scanning photostimulation and whole cell recordings.
Keywords: circuit tracing, inhibitory neurons, genetic, viral, photostimulation
INTRODUCTION
The general anatomy and circuit organization of the hippocampal CA1 area has been particularly well studied, due to its single principal cell layer coupled with a highly organized laminar distribution of its extrinsic inputs (Amaral and Witter, 1989). The CA1 area receives major input connections from several extrinsic sources (Takacs et al., 2012), including CA3 pyramidal cells via their ipsilateral Schaffer collaterals and contralateral commissural fibers (Amaral and Witter, 1989) and layer 3 excitatory cells of the entorhinal cortex through the temporo-ammonic pathway (Steward and Scoville, 1976), as well as the medial septum and diagonal band (MS-DB) area (Freund and Antal, 1988; Gulyas et al., 1990). Functionally, MS-DB inputs are important for hippocampal network oscillations (Buzsaki, 2002), and behavioral evidence indicates functionally separable roles of CA3 and entorhinal inputs to CA1 in hippocampus-dependent learning and memory (Brun et al., 2002; Nakashiba et al., 2008; Remondes and Schuman, 2004; Suh et al., 2011). Like many other cortical areas, CA1 contains diverse types of excitatory and inhibitory neurons that form intricate circuit connections for information processing (Klausberger and Somogyi, 2008). Differential inhibitory control of excitatory cell activity by inhibitory interneurons is largely determined by extrinsic and intrinsic CA1 excitation to different inhibitory cell types (Klausberger and Somogyi, 2008). However, due to technical limitations, there is a limited understanding of whether or how the different sources of input to CA1 are distributed in different strengths onto each of its constituent cell types, which is essential for a mechanistic understanding of hippocampal functional circuit operations.
Until recently, there has been no efficient means for performing cell-type specific circuit analyses in the intact brain over a large scale. New advances in virology and genetic technology are now beginning to complement more traditional approaches and offer powerful tools for mapping cell-type specific circuit connectivity and function (Callaway, 2008). Among them, genetically modified and monosynaptically restricted rabies tracing has proved to be a useful mapping tool for identifying direct circuit inputs to specific cell populations that can be genetically targeted (Marshel et al., 2010; Nakashiba et al., 2012; Wall et al., 2010; Wickersham et al., 2007b).
In the present study, we applied a strategy based upon a Cre-dependent, genetically modified rabies-based tracing system to map local and long-range monosynaptic connections in the intact brain to targeted cell types defined by Cre expression in four different mouse lines including excitatory pyramidal cells (Camk2a-Cre), mixed inhibitory cell types (Dlx5/6-Cre), parvalbumin-expressing (PV-Cre) inhibitory cells and somatostatin-expressing (SOM-Cre) inhibitory cells in CA1 of the mouse hippocampus. Using the new approach, we were able to examine circuit connections of excitatory and inhibitory cell types in the intact brain, and demonstrate quantitative differences in presynaptic local and distant circuit connections to these cell types. These data provide insights into differential circuit mechanisms for hippocampal functional operations.
RESULTS
Cre-dependent rabies tracing approach
Naturally occurring rabies virus has been known for its extremely high efficiency in trans-synaptic labeling, as it propagates exclusively between connected neurons by strictly unidirectional (retrograde) transneuronal transfer (Ugolini, 2008, 2011). We take advantage of the ability to target rabies infection to specific cell types using EnvA pseudotyping and limit trans-synaptic spread to direct inputs by using glycoprotein gene-deleted (ΔG) rabies virus and transcomplementation (Wall et al., 2010; Wickersham et al., 2007b). Specifically, ΔG rabies virus (deletion mutant, SAD-B19 strain) is pseudotyped with the avian sarcoma leucosis virus glycoprotein EnvA (EnvA-SADΔG rabies virus), which can only infect neurons that express avian tumor virus receptor A (TVA), an avian receptor protein that is absent in mammalian cells unless provided through exogenous gene delivery. The deletion-mutant rabies virus then can be transcomplemented with the expression of rabies glycoprotein (RG) in the same TVA-expressing cells to enable its retrograde spread restricted to direct presynaptic neurons.
Our current approach was developed based on a Cre-dependent rabies tracing system reported previously, in which a Cre-dependent helper virus (adeno-associated virus, AAV) targets gene expression of both RG and TVA to Cre-expressing cells to assist with subsequent rabies infection and monosynaptic retrograde tracing (Wall et al., 2010). However, there were two issues with the previous system. With the helper virus used in the previous system, there was leaky TVA expression in non-Cre expressing cells. In addition, GFP that is encoded in the viral construct and intended to mark the starter cells failed to fluoresce. In the current system we have corrected both problems. Rather than using a helper AAV to express TVA, we use a Cre-dependent TVA expressing mouse line (Seidler et al., 2008); there is no leaky TVA expression using the mouse line (see below). A functional helper AAV expressing histone-tagged GFP that robustly labels starter cells as well as RG to complement the SADΔG rabies virus and allow trans-synaptic spread.
As illustrated in Fig. 1A, specific Cre mouse lines are first crossed with a Cre-dependent TVA expressing mouse line, LSL-R26TVA-lacZ (Seidler et al., 2008) so that Cre-expressing neurons express TVA (Fig. S1), thus restricting initial EnvA-ΔG rabies virus infection to Cre + cells. Then a Cre-dependent AAV virus (the helper virus, AAV8-EF1α-FLEX-HB) with a coding sequence of RG required for trans-synaptic rabies virus retrograde spread as well as nuclear localized histone-GFP, is injected into CA1 of the double transgenic mice (Cre:TVA mice). The AAV-targeted subset of Cre+ cells are identified by their nuclear GFP expression from the AAV genome (Fig. 1B, C). Following the AAV injection, a pseudotyped, deletion-mutant rabies virus encoding a red fluorescent protein mCherry (EnvA-SADΔG-mCherry rabies) (Fig. 1B) is injected into the same location of the previous AAV injection. EnvA pseudotyped, ΔG-mCherry rabies enters the Cre+ and TVA+ neurons, and replicates its genome with mCherry expression. Using RGs expressed by the helper vector in the Cre+ cells, ΔG-mCherry rabies undergoes transcomplementation (forming new infectious viral particles), and spreads to the presynaptic partners of the Cre+ starter neurons (Fig. 1C). Because their presynaptic neurons do not express RGs and the ΔG rabies has no RG-coding sequence in its genome, these presynaptic cells cannot produce infectious rabies virus particles, restricting infection to the targeted cell type and its direct monosynaptic inputs. For control experiments, the Cre:TVA animals are only injected with EnVA-SADΔG-mCherry rabies. As expected, without the helper AAV delivering RGs in Cre+ cells, the ΔG-mCherry rabies cannot spread from Cre+ and TVA+ cells so that in control cases rabies labeling is restricted to Cre+, TVA-expressing cells at the rabies injection site (Fig. S2).
Figure 1. Cre-dependent rabies tracing approach.
(A) A mouse transgenic line that expresses Cre in specific type/group of hippocampal neurons is first crossed with the Cre-dependent TVA expressing mouse line, LSL-R26TVA-lacZ (Seidler et al., 2008) to target gene expression and control initial rabies virus infection. (B) The AAV helper virus and EnvA pseudotyped G-deleted rabies virus are used for circuit tracing. Using the AAV8-pEF1α-FLEX-HB (H: nuclear localized histone GFP; B: B19 rabies glycoprotein, RG), the initial AAV injection (0.1 µL, spatially restricted in CA1) allows for expression of RG and GFP transgenes after Cre-recombinase mediated activity in Cre-expressing neurons. The second injection delivers 0.1 µL EnvA-SADΔG-mCherry (ΔG: RG deleted, mCherry: a red fluorescent protein) into the same location of the previous AAV injection. (C) Timeline of viral injections and schematic illustration of rabies-mediated monosynaptic retrograde labeling. Green indicates GFP expression from the helper AAV genome, labeling the rabies receptive target cells while red indicates mCherry expression from SADΔG-mCherry rabies genome in the target cells (starter cells) and their first-order presynaptic neurons. The starter cells are identified as GFP and mCherry double-labeled. At 9 days after the rabies injection, the animal is perfused with 4% paraformaldehyde and the brain extracted for histological processing.
After histological processing, brain sections are imaged, and quantitative examinations across hundreds of sections (every one out of three 30-µm thick sections of the whole brain) are conducted for unbiased analyses of rabies-mediated, direct synaptic connections to targeted cell types in hippocampal CA1 (Fig. S3). In every case, the first analysis step is to visualize the injection site and assure that GFP and mCherry double-labeled starter cells are restricted to CA1 (Fig. 2A, C–D). The starter cells are marked and quantified across sections at and around the targeted injection site. As for the measurement of rabies-labeled presynaptic neurons, every image is examined to identify and mark the locations of mCherry-expressing cell bodies. These labeled cells are assigned to specific anatomical structures, such as different hippocampal subfields, entorhinal cortex and the medial septum for regional input quantification. Thus, through quantitative evaluations of the numbers of targeted postsynaptic (starter) cells and the numbers of their presynaptic cells labeled in various brain structures (Fig. S3), we are able to quantify cell type-specific connections in local and long-range circuits and perform cell-type specific circuit analyses over a large scale. Normalization to the numbers of starter cells also controls for variability between animals and mouse lines.
Figure 2. Rabies labeling of presynaptic neurons shows direct local and more distant circuit connections to CA1 excitatory pyramidal cells in Camk2a-Cre: TVA mouse hippocampus.
(A–B) Ipsilateral and contralateral hippocampal images of the viral injection site. Strong rabies-mediated labeling of putative excitatory neurons is seen in both ipsilateral and contralateral CA3. Local CA1 inhibitory neurons outside stratum pyramidale are also labeled. (C–D) An enlarged view of the white box in A, with GFP and DAPI overlay in C showing restricted AAV-mediated infection and gene expression in stratum pyramidale, and with GFP and mCherry overlay in D showing the GFP-mCherry double labeled starter cells (indicated by the arrows in C and D). (E–J). Rabies-labeled (mCherry-expressing) presynaptic neurons (distant from the injection site) are seen in the subiculum, the medial septum and diagonal band (MS-DB) area, and entorhinal cortex, respectively. F and G show the enlarged view of the two white-boxed regions in E, while J shows the enlarged view of the white-boxed region in I. AP numbers indicate the positions of the coronal sections relative to the bregma landmark.
Comparison of monosynaptic inputs to excitatory versus inhibitory CA1 neurons
Canonical inputs
We first mapped circuit connections to CA1 pyramidal neurons using Camk2a-Cre (T29) mice (Tsien et al., 1996), in which Cre recombinase expression is largely restricted to CA1 pyramidal cells. In the double-transgenic mice (Camk2a-Cre:TVA), we virally traced circuit connections to a small population of starter CA1 pyramidal cells (N = 109.6 ± 16.7 per animal across 5 cases), which are unambiguously identified by their GFP and mCherry expression from the helper AAV and ΔG-mCherry rabies genomes, respectively (Fig. 2 A, C–D). Typically, for each representative case (as shown in Fig. S3), the total number of labeled CA3 cells following rabies infection of CA1 excitatory cells was measured between 3000 and 4500, while the overall number of labeled neurons across different regions was estimated to be ~ 9000, indicating that trans-synaptic rabies labeling is reasonably efficient. Strong labeling of individual neurons presynaptic to CA1 excitatory neurons is seen in ipsilateral and contralateral CA3 (Fig. 2A, B). Essentially all the labeled CA3 cells are located in stratum pyramidale (SP), and morphologically and neurochemically confirmed to be excitatory neurons (Fig. S4A, B). The input connection strength index (CSI) (defined as the ratio of the number of presynaptic neurons versus the number of starter neurons) between CA3 and CA1 excitatory cells is 7.10 ± 0.28 and 3.44 ± 0.22 (mean ± SE, averaged across 5 animals) for ipsilateral and contralateral CA3, respectively (Table S1A). There are putative inhibitory cells labeled outside the pyramidal cell layer of ipsilateral CA3 (Fig. 2A), accounting for a CSI of 0.13 ± 0.03. Almost none of the labeled contralateral CA3 cells are inhibitory.
Local inhibitory interneurons, identified based on their laminar locations and morphology, are also labeled in ipsilateral CA1 with a CSI of 3.26 ± 0.39 for putative inhibitory cells outside CA1 pyramidal cell layer. We did not quantify the labeled inhibitory cells in the pyramidal cell layers due to masking by strong excitatory cell labeling. There are some putative pyramidal cells labeled in ipsilateral CA2 and fewer in contralateral CA2, with their CSIs being 0.47 ± 0.10, and 0.07 ± 0.01, respectively. While contralateral CA1 pyramidal cells provide input with a CSI of 0.54 ± 0.17, few putative inhibitory cells in contralateral CA1 are labeled (CSI = 0.01 ± 0.004). Thus CA1 excitatory cells receive much stronger local inhibition than long-rang inhibition. Robust labeling is also seen in very distant structures such as the medial septum and diagonal band (MS-DB) area (about 3mm anterior to the CA1 injection site) and entorhinal cortex (EC, about 3 mm posterior to the CA1 injection site) (Fig. 2H, I, J). The labeled entorhinal cells are excitatory neurons, mostly located in layer 3. The connection strength of ipsilateral EC is much stronger than contralateral EC, with their CSIs being 0.54 ± 0.08 and 0.01 ± 0.01 (p < 0.01), respectively.
Additionally, following viral tracing in Camk2a-Cre:TVA mice (as well as in other Cre:TVA mice), there is scarce retrograde labeling of neurons in areas known to weakly project to hippocampal CA1 using conventional tracing techniques (Somogyi and Klausberger, 2005). These areas include amygdala, reuniens thalamic nucleus and the raphe nucleus. Note that despite weak inputs, rabies labeling of these neurons is as equally clear as those providing strong synaptic inputs due to self-replication of rabies viral cores in presynaptic neurons and subsequent strong expression of mCherry (Wickersham et al., 2007a).
We then mapped circuit connections to a mixed population of CA1 inhibitory neuron types using Dlx5/6-Cre mice (Monory et al., 2006), in which selective Cre expression is targeted to forebrain GABAergic neurons. As expected, spatially restricted AAV and EnvA rabies viral injections infected a small population of different inhibitory cells located in different CA1 laminae (Fig. 3A, C, D; Fig. S2), with the average numbers of starter cells being 47.75 ± 9.48, 38.35 ± 2.59, 11.25 ± 4.40, and 9.75 ± 4.53 (N = 4 cases, Table S1B) for stratum oriens (SO), stratum pyramidale (SP), stratum radiatum (SR), and stratum lacunosum-moleculare (SLM), respectively. These mixed CA1 inhibitory cell types have strong input connections from putative excitatory cells located in SP of CA1 with a CSI of 9.72 ± 0.49. Overall CA1 inhibitory neurons have a similar connectivity pattern of extrinsic CA1 sources to that of excitatory pyramidal cells, but with differing connection strengths (Fig. S5A; Table S1A, B). The CSIs of inhibitory cell types for ipsilateral CA2, ipsilateral and contralateral CA3 are 0.17 ± 0.01, 1.63 ± 0.37 and 0.60 ± 0.18, respectively, which differ significantly from those of pyramidal cells (p < 0.05 for each comparison). Whereas excitatory connection strength of contralateral CA1 tends to be greater for these inhibitory cells compared with that of pyramidal cells, entorhinal cortex appears to have weaker connections to inhibitory cells (ipsilateral EC CSI: 0.21 ± 0.15).
Figure 3. CA1 inhibitory neurons targeted using the Dlx5/6-Cre: TVA mouse and CA1 excitatory pyramidal cells have similar patterns of input circuit connections.
(A–B) Ipsilateral and contralateral hippocampal images close to the viral injection site. Rabies-mediated labeling of putative excitatory neurons is seen in both ipsilateral and contralateral CA3, as well as in CA2 in these sections. (C–D) An enlarged view of a section at the injection site (AP: −1.94 mm), showing DAPI staining and GFP expression from the helper AAV both in and outside stratum pyramidale in C, and with the GFP and mCherry overlay in D showing the GFP-mCherry double labeled starter cells (indicated by the arrows in C and D). (E–G). Rabies-labeled presynaptic neurons in the subiculum, the medial septum and diagonal band area, and entorhinal cortex, respectively. H shows the enlarged view of the white-boxed region in G.
Non-canonical subicular inputs to both CA1excitatory and inhibitory neurons
Although it is generally believed that there is no direct back-projection from the subiculum to CA1 (however, see (Berger et al., 1980; Kohler, 1985)), our tracing data show that a significant number of subicular cells are retrogradely labeled by rabies tracing from either CA1 pyramidal neurons or inhibitory neurons (Fig. 2E, 3E). We also determined that there are both excitatory and inhibitory subicular cells labeled as determined from their morphology and confirmed using immunochemical staining. Overall, the labeled subicular cells have a connection strength index of 0.81± 0.01 and 1.00 ± 0.20 for these CA1 cell types, respectively.
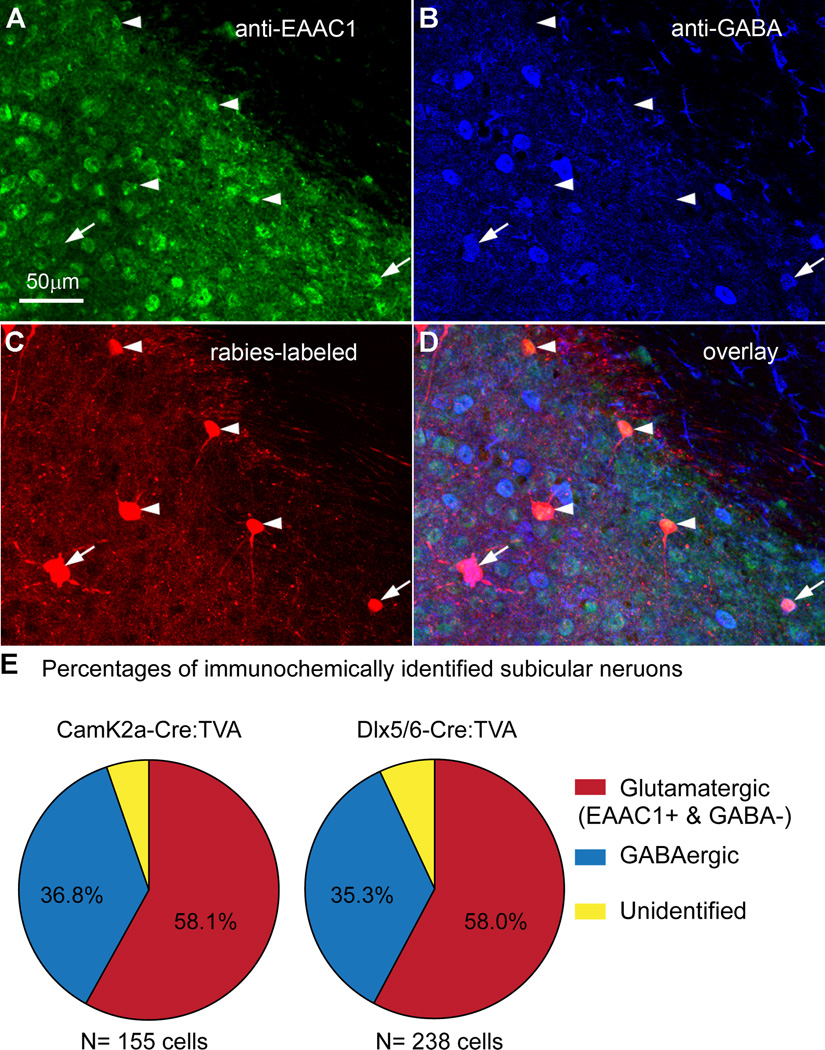
To further understand the nature of this under-described subiculum-CA1 pathway, we performed neurochemical characterization of rabies labeled subicular cells to examine the potential difference of subicular excitatory versus inhibitory cells projecting to different CA1 cell types. Double immunochemical staining against excitatory amino acid transporter type1 (EAAC1) and GABA allowed for identification of glutamatergic or GABAergic CA1-projecting subicular cells in Camk2a-Cre:TVA and Dlx5/6-Cre:TVA sections (Fig. 4A–D). Interestingly, glutamatergic and GABAergic percentages of labeled subicular cells are similar for Camk2a-Cre:TVA and Dlx5/6-Cre:TVA cases with about 58% of CA1-projectinig subicular neurons being glutamatergic neurons, and about 36% of them being GABAergic neurons (Fig. 4E). This indicates no preferential subicular innervation of CA1 excitatory or inhibitory neurons.
Figure 4. Immunochemical characterization and quantification of rabies-labeled CA1-projecting subicular neurons.
(A–D) Immunostaining of excitatory amino acid transporter type1 (EAAC1) and GABA in brain slices with rabies-labeled subicular neurons from a Camk2-Cre: TVA case, in which the immunoreactivity of EAAC1, GABA and rabies mCherry expression is shown in green, blue (pseudocolor from AF647-conjugated secondary antibody), red, respectively. For the rabies-labeled neurons, the arrows point at subicular GABAergic neurons (GABA+) while glutamatergic neurons (EAAC1+ and GABA−) are pointed at by the arrowheads. Note that many GABAergic neurons also show strong EAAC1 staining (Conti et al., 1998). (E) Quantification of rabies-labeled, immunochemically identified subicular glutamatergic (excitatory) neurons and GABAergic inhibitory neurons. There are a small percentage of rabies labeled cells that were neurochemically unidentified, as they did not show robust staining against EAAC1 or GABA.
Neurochemical specific septohippocampal inputs to CA1
As the MS-DB region contains cholinergic, GABAergic and glutamatergic neurons, we are interested in examining whether all these neurochemical specific MS neurons have cell-type specific preferences in innervating CA1 excitatory versus inhibitory cell types. Septohippocampal connections of cholinergic and GABAergic MS cells have been examined in highly localized hippocampal microcircuits using conventional anatomical tracing combined with electron microscopy (Freund and Antal, 1988; Gulyas et al., 1990). As septohippocampal circuit connections of more recently described glutamatergic MS-DB neurons are much less understood than cholinergic and GABAergic MS cells (Colom et al., 2005; Huh et al., 2010; Manseau et al., 2005), we also would like to examine whether glutamatergic MS neurons have direct, monosynaptic connections to CA1 neurons using rabies tracing.
Consistent with the previous studies, our rabies tracing indicates that MS-DB neurons provide significant inputs to CA1 (Fig. 2 and 3). For Camk2a-Cre:TVA cases, the overall CSI for MS-DB neurons to CA1 excitatory starter cells is 0.95 ± 0.05, while for Dlx5/6-Cre:TVA cases, the overall CSI for MS-DB neurons to CA1 inhibitory starter cells is 1.17 ± 0.18. Our data show that cholinergic and GABAergic septohippocampal cells differentially innervate CA1 excitatory and inhibitory cell types, and MS-DB glutamatergic neurons also directly innervate CA1 neurons (Fig. 5). In conjunction with immunochemical examinations, for Camk2a-Cre:TVA cases we found that 66% of rabies-labeled septohippocampal cells are cholinergic (Fig. 5A–D), and that 27% of the labeled septohippocampal cells are GABAergic (Fig. 5J). Therefore GABAergic neurons comprise a significant portion of the septal input to CA1 excitatory neurons. We also found that septohippocampal glutamatergic neurons only account for 7% of rabies labeled cells in MS-DB in Camk2a:TVA cases (Fig. 5J). In contrast, for Dlx5/6-Cre:TVA cases, a great majority of the labeled septohippocampal cells in the MS region innervating CA1 inhibitory neuron are GABAergic (67%), while cholinergic and glutamatergic MS neurons account for 12% and 21% of the total labeled cells, respectively (Fig. 5E–I, K).
Figure 5. Neurochemical characterization and quantification of rabies-labeled neurons in medial septum and diagonal band of broca complex (MS-DB) presynaptic to CA1 excitatory and inhibitory neurons.
(A) Immunostaining of choline acetyltransferase (ChAT) in brain slices with rabies-labeled medial septum neurons from Camk2a-Cre: TVA case to identify septal cholinergic neurons. (B–D) Enlarged view of the region indicated by the white box in A which immunoreactivity of ChAT shows in green, and rabies mCherry shows in red. Arrows point at medial septum cholinergic neurons labeled by rabies virus. (E) Immunostaining of EAAC1 and GABA in brain slices with rabies-labeled medial septum neurons from Dlx5/6-Cre: TVA case to identify septal GABAergic and glutamatergic neurons. (F–I) Enlarged view of the region indicated by the white box in E. The immunoreactivity of EAAC1, GABA and rabies mCherry expression is shown in green, blue (pseudocolor from AF647-conjugated secondary antibody), red, respectively. For rabies-labeled neurons, the arrows point at medial septum GABAergic neurons (GABA+), while glutamatergic neurons (EAAC1+ and GABA−) are pointed at by the arrowheads. (J–K) Quantification of rabies-labeled, immunochemically identified MS-DB cholinergic, GABAergic, and glutamatergic neurons in both Camk2-Cre: TVA and Dlx5/6-Cre: TVA cases.
Comparison of monosynaptic inputs to PV+ versus SOM+ inhibitory CA1 neurons
Given the diversity of inhibitory cells targeted using Dlx5/6-Cre mice, we further examined whether specific sub-groups of inhibitory cells in CA1, targeted by using PV-Cre and SOM-Cre mice, have differential circuit connections. The PV+ and SOM+ inhibitory cell types were examined because they are most numerous and functionally important in the cerebral cortex including the hippocampus (Freund and Buzsaki, 1996; Xu et al., 2010b). Compared with Camk2a-Cre:TVA and Dlx5/6-Cre:TVA cases, the total starter cells are fewer in PV-Cre:TVA and SOM-TVA cases, as expected due to the restriction of Cre-dependent gene expression to targeted subpopulations (Fig. S1 and S2). The average numbers of PV+ starter cells found in SO, SP, SR and SLM are 11.4 ± 1.21, 17.8 ± 1.59, 1.2 ± 0.58 and 0 (N = 5 cases; Table S1C), respectively, while the average numbers of SOM+ starter cells found in SO, SP, SR and SLM are 52.6 ± 11.73, 3.2 ± 1.2, 1.4 ± 0.75 and 0.2 ± 0.2, respectively (N = 5 cases; Table S1D). Immunostaining of PV and SOM validated chemical identities of starter cells in the PV-Cre:TVA and SOM-Cre:TVA mouse, respectively (Fig. S4C–F). Most of SOM+ cells labeled by rabies display O-LM cell morphology (Fig. S2E, K), as their cell bodies and horizontal dendrites are located in stratum oriens but their axons ascend through SP and SR to branch heavily in SLM, which contains distal dendrites and apical tufts of pyramidal neurons. The axons of O-LM cells with strong mCherry expression clearly form a prominent band in SLM.
Although the input connection pattern of PV+ inhibitory neurons mapped using the PV-Cre:TVA mouse generally resemble that of mixed types of inhibitory cells observed with Dlx5/6-Cre:TVA mice (Fig. 6A–B, E–H; Table S1C; Fig. S5B), SOM+ inhibitory neurons targeted using the SOM-Cre: TVA mouse have a different pattern of circuit connections (Fig. 6 C–D, I–L; Table S1D; Fig. S5B). SOM+ cells have predominant excitatory connections from within ipsilateral CA1 with few CA3 inputs and little entorhinal input. The CSIs of SOM+ cells for ipsilateral and contralateral CA3 are 0.41 ± 0.12 and 0.11 ± 0.05, respectively, which are much less than those of PV+ cells (2.97 ± 0.37 and 0.72 ± 0.10, respectively) (p < 0.001 for either comparison). On average, CA1 excitatory input connections to PV+ and SOM+ cells are comparable to those of Dlx5/6-Cre cells, with their CSIs being 9.46 ± 0.36, 9.13 ± 0.54, respectively. Both PV+ cells and SOM+ cells in CA1 have input connections from the subiculum, with their CSIs being 0.69 ± 0.16 versus 1.24 ± 0.31, respectively. In addition, PV+ cells have a greater input connection strength of MS-DB compared with SOM+ cells (CSIs: 2.12 ± 0.2 versus 1.07 ± 0.16, p < 0.05). PV+ cells also have a greater CSI of MS-DB than excitatory and mixed inhibitory cell types (p < 0.05 for either comparison).
Figure 6. Comparison of circuit input connections between parvalbumin-expressing (PV+) inhibitory neurons and somatostatin-expressing (SOM+) inhibitory neurons.
PV+ cells are targeted using the PV-Cre: TVA mouse, while SOM+ cells are targeted using the SOM-Cre: TVA mouse. (A–B) A PV-Cre: TVA case. Ipsilateral and contralateral hippocampal images close to the viral injection site. Rabies-mediated labeling of putative excitatory neurons is seen in both ipsilateral and contralateral CA3, as well as in contralateral CA1 in the section. (C–D) A SOM-Cre: TVA case. Ipsilateral and contralateral hippocampal images around the viral injection site. Rabies labeling is predominately found throughout ipsilateral CA1, while no or little labeling is found in contralateral hippocampus. Rabies-mediated labeling of putative excitatory neurons is seen in stratum pyramidale of ipsilateral CA1. (E–F) Examples from the PV-Cre: TVA case. Rabies-labeled presynaptic neurons in the medial septum and diagonal band area, and entorhinal cortex, respectively. (G–H) The enlarged view of the two white-boxed regions in F. (I–J). Examples from the SOM-Cre:TVA case. Rabies-labeled presynaptic neurons in the medial septum and diagonal band area, and the subiculum, respectively. (K–L) The enlarged view of the two white-boxed regions in G. Note that there is no or little labeling in entorhinal cortex.
Our results above demonstrate that the Cre-dependent rabies-based system can target selected cell groups in hippocampal CA1 and effectively label their monosynaptic connections in the intact brain. The tracing data allow for determination of the relative weight of the distributed synaptic connections to these cell groups in the large circuit context of the entire brain, which helps to evaluate hypotheses about differential functions of these selected cell groups/types (see below).
LSPS local circuit mapping supports rabies tracing
As rabies tracing is a new method for anatomical examination of circuit connections to small Cre-defined neuronal populations in hippocampal CA1, we followed up with circuit mapping using LSPS which allows functional mapping of local synaptic inputs to identified cell types. The LSPS method has been previously used in the neocortex and hippocampus (Antonio et al., 2013; Brivanlou et al., 2004; Xu and Callaway, 2009). Under our experimental conditions, this method has a sufficient spatial resolution to map synaptic inputs to CA1 cells from specific hippocampal subfields (Fig. S6A–B).
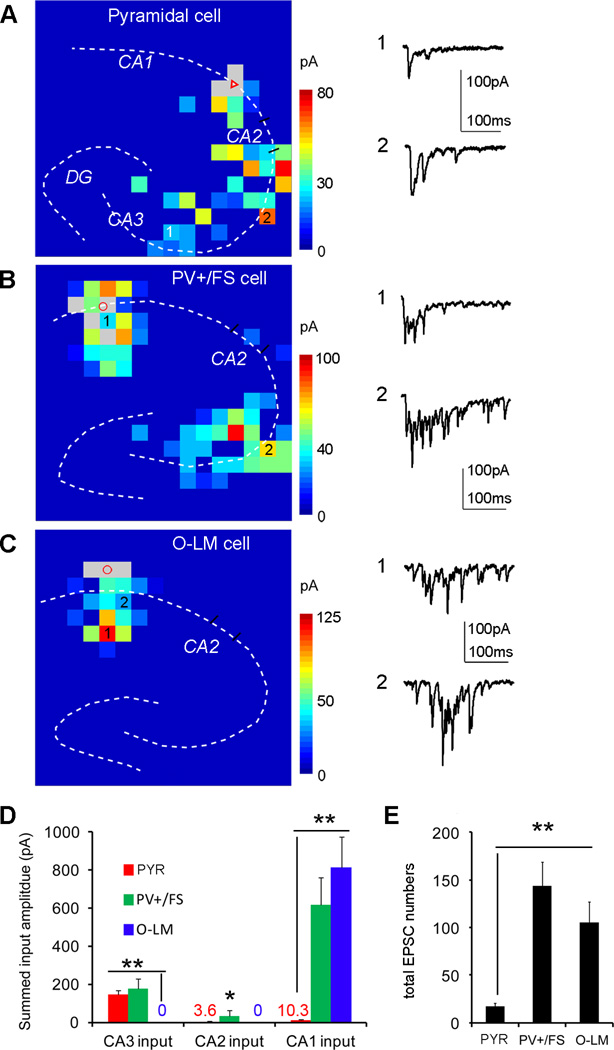
We conducted photostimulation mapping experiments to examine intrahippocampal excitatory circuit connections to excitatory pyramidal cells, PV+/FS basket cells and SOM+ O-LM inhibitory cells. LSPS mapping data confirmed cell-type specific differences of intra-hippocampal circuit connections as identified in rabies tracing-based circuit mapping. Targeted recordings of inhibitory neurons were facilitated by using transgenic mouse lines expressing GFP in inhibitory neurons (Oliva et al., 2000; Tamamaki et al., 2003; Xu and Callaway, 2009) (Fig. S7). As illustrated in Fig. S6C, the LSPS approach involves recording from single neurons, then stimulating at surrounding sites on the LSPS mapping grid in order to generate action potentials from neurons in those sites, thus providing an input map for the recorded neuron based on activation of presynaptic inputs. We found that CA1 pyramidal cells receive a great majority of excitatory input from extensive CA3 locations, while having weak inputs from CA2 and from within CA1 (Fig. 7A, D). PV+/FS basket cells receive strong excitatory input from both CA3 and CA1, as well as some input from CA2 (Fig. 7B, D). In contrast, O-LM cells receive all of their excitatory inputs exclusively from within CA1 (Fig. 7C, D). Generally, the differences in pattern and amplitudes of excitatory inputs to these cell types revealed by LSPS mapping experiments verify cell-type specific intrahippocampal connections to Camk2a-Cre, PV-Cre and SOM-Cre cell groups observed with rabies tracing. However, as the LSPS method allows high resolution mapping of local functional input to identified cell types within the Cre-defined cell groups targeted by the rabies tracing, it is capable of detecting more detailed features in intrahippocampal excitatory inputs at the level of more precise cell types. For example, although strong excitatory connections within CA1 of PV+/FS cells fit well with rabies tracing results, PV+/FS basket cells receive equally strong CA3 excitatory input as pyramidal cells (Fig. 7D), which reflects stronger CA3 excitatory connections than the overall PV+ cell group targeted by rabies tracing in PV-Cre:TVA mice (Table S1). Similarly, as a subtype of SOM+ cells, O-LM cells show more localized excitatory connections than the overall SOM cell group targeted by rabies tracing in SOM-Cre:TVA mice (Table S1).
Figure 7. Laser scanning photostimulation (LSPS) mapping functionally verifies rabies tracing in identifying cell-type specific differences of intra-hippocampal circuit connections.
(A) A color-coded, averaged input map (with each square corresponding to one stimulation site) superimposed with the hippocampal contour, illustrating the pattern and strength of synaptic inputs to a recorded excitatory pyramidal neuron in CA1. Its somatic location is indicated by the red triangle. The grey squares indicate the removal of direct responses from these sites. For the scale of input amplitudes, the warmer color indicates stronger amplitude. The grey squares indicate the removal of direct responses from these sites (see the Supplemental Method). The numbered sites correspond to the illustrated photostimulation response traces plotting from the onset of photostimulation. (B) A color-coded, averaged input map, illustrating the pattern and strength of synaptic inputs to an example PV+/FS inhibitory cell in CA1. (C) A color-coded, averaged input map, illustrating the pattern and strength of synaptic inputs to an example SOM+ O-LM inhibitory cell in CA1. (D) Summary data showing input strength differences across CA3, CA2 and CA1 to targeted pyramidal cells (PYR) (N = 7), PV+/FS inhibitory cells (N = 8) and O-LM inhibitory cells (N = 7). As for CA3 excitatory inputs, the average total input amplitudes of pyramidal cells and PV+/FS inhibitory cells did not differ from each other, but these cell types differed significantly from O-LM cells. PV+/FS inhibitory cells had stronger CA2 inputs than either pyramidal cells or O-LM cells. As for CA1 excitatory inputs, PV+/FS and O-LM inhibitory cells did not differ from each other, but these cell types differed significantly from pyramidal cells. * and ** indicate the statistical significance levels of p < 0.05 and 0.01, respectively, for statistical comparisons. (E) Summary data showing average total EPSC events per cell measured from the records cells. PV+/FS and O-LM inhibitory cells did not differ from each other, but these cell types differed significantly from pyramidal cells.
Furthermore, the LSPS mapping provided functional information such as the amplitudes and numbers of excitatory postsynaptic current (EPSC) events (Fig. 7D, E), which complements anatomical rabies tracing. Excitatory cells, PV+ basket cells and O-LM cells appear to have relatively similar amplitudes of individual EPSCs, but EPSCs of excitatory cells are less frequent with longer durations in response to presynaptic photostimulation compared to those of PV+ basket cells and O-LM cells (Fig. 7A–C, E). This indicates these cell types have differential synaptic contacts, kinetics and connection probabilities of presynaptic neurons.
Discussion
Technical considerations
We have combined transgenic mouse technology and recently developed monosynaptic rabies virus-based tracing techniques (Wall et al., 2010; Wickersham et al., 2007b) for mapping cell type specific circuit connections to hippocampal CA1 in a quantitative manner in the intact brain. This method restricts initial infection and subsequent complementation of EnvA-ΔG rabies viruses to a small population of Cre-defined specific cell types and enables robust monosynaptic retrograde spread of ΔG rabies from these targeted cells to their direct presynaptic cells in local and distant brain structures. We believe that the complemented virus in the starter cell can cross all input synapses with equal efficiency, as previous studies have established that rabies virus receptors are ubiquitously distributed within the CNS and all neuronal populations of the same synaptic order are infected regardless of their neurotransmitters, synaptic strength, and distance (Ugolini, 2011). This is evidenced by consistent labeling of neurons in ipsilateral and contralateral CA3 and no trans-synaptic spread to dentate granule cells, which directly project to CA3 but not CA1. This new approach makes it possible to identify the sources and cell types providing direct monosynaptic input to any Cre-defined population in various mouse lines. However, as Cre mouse lines are crossed to the TVA mouse, all Cre+ cells expressing TVA can be directly infected by local injection of EnvA-ΔG rabies, thus making it difficult to examine local connections between Cre+ starter cells and other Cre+ cells at the injection site. Nevertheless, local circuit connections to specific cell types can be examined more precisely with other methods such as LSPS.
Our data indicate that the current system of rabies labeling works in a non-biased fashion. The new method of rabies labeling is reliable as labeled cells are seen in very distant structures such as the MS-DB area (about 3mm anterior to the CA1 injection site) and other areas (e.g., amygdala, reuniens thalamic nucleus and the raphe nucleus) known to weakly project to hippocampal CA1. However, we do not expect that the method that we use label every input to each neuron. But this limitation does not mean that our method is not useful or not effective. As rabies labels inputs to different cell types in a similar manner, we assess the relative numbers of inputs from each source onto each target cell type, and quantify the numbers of cells that are labeled at various input locations following rabies virus infection of different types of postsynaptic cells in CA1. Different from single cell targeting, this approach benefits from targeting a small population of Cre+ postsynaptic cells and can provide weighted connection strengths for defined cell types. Because the number of postsynaptic starter cells and the numbers of direct presynaptic labeled cells in specified structures across the entire brain can be quantitatively determined, this approach allows for assessment of relative abundance of connected populations. Specifically, we measure the connection strength index (CSI) which is defined as the ratio of the number of presynaptic neurons versus the number of starter neurons and reflects a comparative number of presynaptic cells labeled by rabies in each region. Thus, this approach enables us to address the proposed questions of whether or how the different sources of input to CA1 are distributed in different strengths onto each of its constituent cell types.
Major extrinsic inputs to different CA1 cell types
Our results both confirm previous findings, validating the new methods that we use, and also go on to extend those findings to reveal new details about the sources of input to various CA1 cell types. Our rabies tracing of circuit connections to excitatory and inhibitory cell types show that major extrinsic inputs to CA1 innervate both principal cells and interneurons in differential weights according to the pathways and cell types. Based on our quantification of input connection strengths, we confirm the strongest excitatory input to the CA1 area is from CA3 via the Schaffer collaterals/commissural input (Amaral and Witter, 1989; Takacs et al., 2012; Wittner et al., 2006). We also show its differential innervations of excitatory cells and interneurons, with excitatory cells having about 4–5 fold greater input connection strength over mixed inhibitory cell types. Compared to CA3 Schaffer / commissural inputs, the temporoammonic pathway (the monosynaptic pathway originating from EC layer 3 to CA1), is weak to both excitatory and inhibitory cell types. However, entorhinal innervation of excitatory cells is still significantly greater than inhibitory cell types. Overall, the comparative input strengths of CA3 - CA1 and entorhinal - CA1 monosynaptic pathways to excitatory neurons appear to be related to differential operations of these functionally distinct circuits. The entorhinal - CA circuit is required for long-term spatial memory consolidation and maintenance (Brun et al., 2008; Brun et al., 2002; Remondes and Schuman, 2004) (however see (Suh et al., 2011)) and nonspatial temporal association (Suh et al., 2011), while the trisynaptic pathway including intact CA3 - CA1 connectivity is required for rapid new learning, pattern completion-based memory recall (Nakashiba et al., 2009; Nakashiba et al., 2008).
Interestingly, the rabies tracing shows non-canonical inputs from the subiculum to CA1 excitatory and inhibitory cell types, in contrast to the general belief of unidirectional information flow from CA1 to the subiculum. The data establish the existence of a subicular-CA1 back projection pathway in the mouse as previously indicated in other mammalian species (Berger et al., 1980; Kohler, 1985; Shao and Dudek, 2005). Furthermore, we have found that both subicular excitatory and inhibitory cells project back to CA1, and that the proportions of subicular excitatory versus inhibitory cells projecting to either CA1 excitatory or inhibitory neurons are similar. These findings may have functional implications about how subicular neurons modulate their CA1 input resources.
Our rabies tracing has further provided information on neurochemical specific septo-hippocampal inputs. Consistent with previous reports with more traditional approaches, we find that CA1 excitatory cells receive a great majority (~66%) of MS-DB innervation from cholinergic cells. But CA1 excitatory cells also receive significant GABAergic septohippocampal innervation (~27%) which has not been explicitly examined (Freund and Antal, 1988; Gulyas et al., 1990). In comparison, CA1 inhibitory cell types receive much less cholinergic MS-DB innervation (~12% of the total labeled MS-DB cells) and much stronger GABAergic septohippocampal innervation (~67%). Although glutamatergic MS-DB cells are known to exist (Colom et al., 2005; Huh et al., 2010; Manseau et al., 2005), rabies tracing firmly establishes direct glutamatergic septohippocampal connections to CA1. Compared to CA1 excitatory neurons, CA1 inhibitory neurons receive much stronger glutamatergic septohippocampal projections (~21% vs. 7%). These new findings can have functional implications in understanding septo-hippocampal circuit operations which are likely to be dictated by differential neurochemical interactions between medial septum and CA1 excitatory and inhibitory cell types (Huh et al., 2010).
Functional implications of inhibitory cell type specific connectivity
Different types or groups of inhibitory neurons coordinate or interact with excitatory neurons in space and time; they are believed to differentially contribute to regulation of circuit dynamics and network oscillations (Freund and Katona, 2007; Glickfeld and Scanziani, 2006; Klausberger and Somogyi, 2008). Although functional differences have been measured (Klausberger et al., 2003; Klausberger and Somogyi, 2008), the mechanisms that underlie different activation patterns of distinct inhibitory cell types have not been understood. In the present study, mapping local and more distant circuit connections to specific inhibitory cell types should help us to further understand the contribution of circuit connection differences of specific inhibitory neurons to their functional differences. In particular, we have examined all the potential input sources to specific CA1 neuron types and presented new information on circuit connections to excitatory neurons, mixed inhibitory cells and PV+ and SOM+ cells in CA1, which could not be examined to this scale in the past.
Within the context of previous literature, our new data provide insights into differential circuit mechanisms of inhibitory neuronal control of hippocampal excitatory neurons in terms of their local and long-range input sources. It has been known that inhibitory cells can regulate excitatory cell activity via feedforward and feedback inhibitory mechanisms (Freund and Buzsaki, 1996; Freund and Katona, 2007). Although past studies performed degeneration or stimulation of the known afferents to examine synaptic inputs to identified CA1 inhibitory neuron types (Ali and Thomson, 1998; Blasco-Ibanez and Freund, 1995; Glickfeld and Scanziani, 2006), specific input sources to inhibitory interneurons remain to be further investigated. Now we are able to identify global input sources to mixed inhibitory cell types, PV+ inhibitory cells and SOM+ inhibitory cells with the rabies method, which are confirmed by the LSPS method for intrahippocampal connections. CA1 PV+ and SOM+ interneurons show similar strengths of input from the subiculum, but PV+ cells have a greater strength of MS input than SOM+ cells. PV+ cells have strong excitatory connections from other input source such as CA3 and entorhinal cortex, while having strong CA1 excitatory connections as well. In contrast, the tracing data indicate that SOM+ cells receive predominant excitatory connections within CA1, and do not receive much input from CA3 or entorhinal cortex. Thus, compared to SOM+ cells, PV+ inhibitory cells are apparently a primary mediator of feedforward inhibition from the longer distance input sources. In fact, the differential circuit connections of PV+ and SOM+ inhibitory cells in CA1 are analogous to those cell types in layer 4 of primary sensory cortex, in which PV+/FS cells, but not SOM+ cells receive strong thalamocortical inputs (Cruikshank et al., 2007; Gibson et al., 1999). Similarly PV+/FS cells in cortical layer 2/3 receive strong feedforward and recurrent excitation from layers 4 and 2/3 respectively, while SOM cells receive stronger input from layer 2/3 than layer 4 (Xu and Callaway, 2009). These observations suggest that the differential roles of PV+ versus SOM+ cells in mediating feedforward versus feedback inhibition are conserved between hippocampal cortex and neocortex. Taken together with previous studies, these salient features of specific inhibitory circuit connections are important for inhibitory neuronal functions.
EXPERIMENTAL PROCEDURES
All experiment procedures were approved by the Institutional Animal Care and Use Committee of the University of California, Irvine, and rabies experiments conducted under a protocol approved by the institutional biosafety committee. The mice were anaesthetized under isoflurane for stereotaxic viral injections. The coordinates targeting dorsal hippocampal CA1 were anteroposterior −1.94 mm, lateromedial −1.40 mm; dorsoventral −1.35 mm. A pulled glass pipette (tip diameter, ≈30 µm) was loaded with virus and then lowered into the brain, and a Picospritzer was used to pulse virus into the brain. A total of 0.1 ul of the helper virus (AAV8-EF1α-FLEX-HB, ~2 × 1011 genome units per ml) was injected into each brain. After 3 weeks of the AAV injection, the pseudotyped, RG-deleted rabies virus (EnvA-SADΔG-mCherry, 0.1 ul, ~2 × 109 infectious units per ml) was injected into the same location of the previous injection. The animals were perfused for tissue processing after 9–10 days following the rabies injection. The brain was sectioned coronally in 30 µm thickness on a freezing microtome. Every one out of 3 sections was mounted for examination and quantification of starter cells and their presynaptic cells in different brain structures. These sections were imaged for all subsequent computer-based analyses. Some of the remaining sections were selected for immunostained with various antibodies for neurochemical characterization of starter cells and rabies labeled cells in different regions. To confirm rabies tracing results, we conducted photostimulation mapping experiments to examine intrahippocampal excitatory circuit connections to specific CA1 cell types. The procedure of electrophysiological recording, photostimulation have been previously described (San Antonio et al., 2013; Xu et al., 2010a). For statistical comparisons between groups, a t-test or Mann–Whitney U-test was performed to compare two groups. For statistical tests across more than two groups, we used the Kruskal–Wallis test (non-parametric One-Way ANOVA) and the Mann–Whitney U-test for group comparisons. In all experiments, the level of statistical significance was defined as p < 0.05. All data were presented as mean ± SE. For more detailed information, please refer to the Supplemental Method.
Supplementary Material
Highlights.
Connections to specific CA1 neuron types were identified using monosynaptic rabies
Excitatory and inhibitory neurons receive similar inputs that differ in strength
Non-canonical direct subicular inputs to both neuron types
Septohippocampal connections to CA1 include glutamatergic components.
Acknowledgments
This work was funded by the US National Institutes of Health grants NS078434 to X.X., and MH063912 to E.M.C.. Additional funding was provided by the Gatsby Charitable Foundation to E.M.C. and a NARSAD Young Investigator Award to X.X. and a DFG grant SA 1374/1-2 to D.S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions: Y.S., X.X., A.N., J.N., L.L. conducted experiments, collected data and performed analysis. D.S. provided the TVA mouse. J.C. developed the helper AAV. X.X. designed the experiments and supervised the project. X.X., Y.S. and E.M.C. interpreted data and wrote the manuscript.
References
- Ali AB, Thomson AM. Facilitating pyramid to horizontal oriens-alveus interneurone inputs: dual intracellular recordings in slices of rat hippocampus. J Physiol. 1998;507(Pt 1):185–199. doi: 10.1111/j.1469-7793.1998.185bu.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral DG, Witter MP. The three-dimensional organization of the hippocampal formation: a review of anatomical data. Neuroscience. 1989;31:571–591. doi: 10.1016/0306-4522(89)90424-7. [DOI] [PubMed] [Google Scholar]
- Berger TW, Swanson GW, Milner TA, Lynch GS, Thompson RF. Reciprocal anatomical connections between hippocampus and subiculum in the rabbit evidence for subicular innervation of regio superior. Brain Res. 1980;183:265–276. doi: 10.1016/0006-8993(80)90463-1. [DOI] [PubMed] [Google Scholar]
- Blasco-Ibanez JM, Freund TF. Synaptic input of horizontal interneurons in stratum oriens of the hippocampal CA1 subfield: structural basis of feed-back activation. Eur J Neurosci. 1995;7:2170–2180. doi: 10.1111/j.1460-9568.1995.tb00638.x. [DOI] [PubMed] [Google Scholar]
- Brivanlou IH, Dantzker JL, Stevens CF, Callaway EM. Topographic specificity of functional connections from hippocampal CA3 to CA1. Proc Natl Acad Sci U S A. 2004;101:2560–2565. doi: 10.1073/pnas.0308577100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brun VH, Leutgeb S, Wu HQ, Schwarcz R, Witter MP, Moser EI, Moser MB. Impaired spatial representation in CA1 after lesion of direct input from entorhinal cortex. Neuron. 2008;57:290–302. doi: 10.1016/j.neuron.2007.11.034. [DOI] [PubMed] [Google Scholar]
- Brun VH, Otnass MK, Molden S, Steffenach HA, Witter MP, Moser MB, Moser EI. Place cells and place recognition maintained by direct entorhinal-hippocampal circuitry. Science. 2002;296:2243–2246. doi: 10.1126/science.1071089. [DOI] [PubMed] [Google Scholar]
- Buzsaki G. Theta oscillations in the hippocampus. Neuron. 2002;33:325–340. doi: 10.1016/s0896-6273(02)00586-x. [DOI] [PubMed] [Google Scholar]
- Callaway EM. Transneuronal circuit tracing with neurotropic viruses. Curr Opin Neurobiol. 2008;18:617–623. doi: 10.1016/j.conb.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colom LV, Castaneda MT, Reyna T, Hernandez S, Garrido-Sanabria E. Characterization of medial septal glutamatergic neurons and their projection to the hippocampus. Synapse. 2005;58:151–164. doi: 10.1002/syn.20184. [DOI] [PubMed] [Google Scholar]
- Conti F, DeBiasi S, Minelli A, Rothstein JD, Melone M. EAAC1, a high-affinity glutamate tranporter, is localized to astrocytes and gabaergic neurons besides pyramidal cells in the rat cerebral cortex. Cereb Cortex. 1998;8:108–116. doi: 10.1093/cercor/8.2.108. [DOI] [PubMed] [Google Scholar]
- Cruikshank SJ, Lewis TJ, Connors BW. Synaptic basis for intense thalamocortical activation of feedforward inhibitory cells in neocortex. Nat Neurosci. 2007;10:462–468. doi: 10.1038/nn1861. [DOI] [PubMed] [Google Scholar]
- Freund TF, Antal M. GABA-containing neurons in the septum control inhibitory interneurons in the hippocampus. Nature. 1988;336:170–173. doi: 10.1038/336170a0. [DOI] [PubMed] [Google Scholar]
- Freund TF, Buzsaki G. Interneurons of the hippocampus. Hippocampus. 1996;6:347–470. doi: 10.1002/(SICI)1098-1063(1996)6:4<347::AID-HIPO1>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Freund TF, Katona I. Perisomatic inhibition. Neuron. 2007;56:33–42. doi: 10.1016/j.neuron.2007.09.012. [DOI] [PubMed] [Google Scholar]
- Gibson JR, Beierlein M, Connors BW. Two networks of electrically coupled inhibitory neurons in neocortex. Nature. 1999;402:75–79. doi: 10.1038/47035. [DOI] [PubMed] [Google Scholar]
- Glickfeld LL, Scanziani M. Distinct timing in the activity of cannabinoid-sensitive and cannabinoid-insensitive basket cells. Nat Neurosci. 2006;9:807–815. doi: 10.1038/nn1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulyas AI, Gorcs TJ, Freund TF. Innervation of different peptide-containing neurons in the hippocampus by GABAergic septal afferents. Neuroscience. 1990;37:31–44. doi: 10.1016/0306-4522(90)90189-b. [DOI] [PubMed] [Google Scholar]
- Huh CY, Goutagny R, Williams S. Glutamatergic neurons of the mouse medial septum and diagonal band of Broca synaptically drive hippocampal pyramidal cells: relevance for hippocampal theta rhythm. J Neurosci. 2010;30:15951–15961. doi: 10.1523/JNEUROSCI.3663-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klausberger T, Somogyi P. Neuronal diversity and temporal dynamics: the unity of hippocampal circuit operations. Science. 2008;321:53–57. doi: 10.1126/science.1149381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler C. Intrinsic projections of the retrohippocampal region in the rat brain. I. The subicular complex. J Comp Neurol. 1985;236:504–522. doi: 10.1002/cne.902360407. [DOI] [PubMed] [Google Scholar]
- Manseau F, Danik M, Williams S. A functional glutamatergic neurone network in the medial septum and diagonal band area. J Physiol. 2005;566:865–884. doi: 10.1113/jphysiol.2005.089664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshel JH, Mori T, Nielsen KJ, Callaway EM. Targeting single neuronal networks for gene expression and cell labeling in vivo. Neuron. 2010;67:562–574. doi: 10.1016/j.neuron.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monory K, Massa F, Egertova M, Eder M, Blaudzun H, Westenbroek R, Kelsch W, Jacob W, Marsch R, Ekker M, et al. The endocannabinoid system controls key epileptogenic circuits in the hippocampus. Neuron. 2006;51:455–466. doi: 10.1016/j.neuron.2006.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashiba T, Buhl DL, McHugh TJ, Tonegawa S. Hippocampal CA3 output is crucial for ripple-associated reactivation and consolidation of memory. Neuron. 2009;62:781–787. doi: 10.1016/j.neuron.2009.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashiba T, Cushman JD, Pelkey KA, Renaudineau S, Buhl DL, McHugh TJ, Rodriguez Barrera V, Chittajallu R, Iwamoto KS, McBain CJ, et al. Young dentate granule cells mediate pattern separation, whereas old granule cells facilitate pattern completion. Cell. 2012;149:188–201. doi: 10.1016/j.cell.2012.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashiba T, Young JZ, McHugh TJ, Buhl DL, Tonegawa S. Transgenic inhibition of synaptic transmission reveals role of CA3 output in hippocampal learning. Science. 2008;319:1260–1264. doi: 10.1126/science.1151120. [DOI] [PubMed] [Google Scholar]
- Oliva AA, Jr, Jiang M, Lam T, Smith KL, Swann JW. Novel hippocampal interneuronal subtypes identified using transgenic mice that express green fluorescent protein in GABAergic interneurons. J Neurosci. 2000;20:3354–3368. doi: 10.1523/JNEUROSCI.20-09-03354.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remondes M, Schuman EM. Role for a cortical input to hippocampal area CA1 in the consolidation of a long-term memory. Nature. 2004;431:699–703. doi: 10.1038/nature02965. [DOI] [PubMed] [Google Scholar]
- San Antonio A, Liban K, Ikrar T, Tsyganovskiy E, Xu X. Distinct physiological and developmental properties of hippocampal CA2 subfield revealed using anti-Purkinje cell protein 4 (PCP4) immunostaining. J Comp Neurol. 2013 doi: 10.1002/cne.23486. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidler B, Schmidt A, Mayr U, Nakhai H, Schmid RM, Schneider G, Saur D. A Cre-loxP-based mouse model for conditional somatic gene expression and knockdown in vivo by using avian retroviral vectors. Proc Natl Acad Sci U S A. 2008;105:10137–10142. doi: 10.1073/pnas.0800487105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao LR, Dudek FE. Electrophysiological evidence using focal flash photolysis of caged glutamate that CA1 pyramidal cells receive excitatory synaptic input from the subiculum. J Neurophysiol. 2005;93:3007–3011. doi: 10.1152/jn.00877.2004. [DOI] [PubMed] [Google Scholar]
- Somogyi P, Klausberger T. Defined types of cortical interneurone structure space and spike timing in the hippocampus. J Physiol. 2005;562:9–26. doi: 10.1113/jphysiol.2004.078915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steward O, Scoville SA. Cells of origin of entorhinal cortical afferents to the hippocampus and fascia dentata of the rat. J Comp Neurol. 1976;169:347–370. doi: 10.1002/cne.901690306. [DOI] [PubMed] [Google Scholar]
- Suh J, Rivest AJ, Nakashiba T, Tominaga T, Tonegawa S. Entorhinal cortex layer III input to the hippocampus is crucial for temporal association memory. Science. 2011;334:1415–1420. doi: 10.1126/science.1210125. [DOI] [PubMed] [Google Scholar]
- Takacs VT, Klausberger T, Somogyi P, Freund TF, Gulyas AI. Extrinsic and local glutamatergic inputs of the rat hippocampal CA1 area differentially innervate pyramidal cells and interneurons. Hippocampus. 2012;22:1379–1391. doi: 10.1002/hipo.20974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamamaki N, Yanagawa Y, Tomioka R, Miyazaki J, Obata K, Kaneko T. Green fluorescent protein expression and colocalization with calretinin, parvalbumin, and somatostatin in the GAD67-GFP knock-in mouse. J Comp Neurol. 2003;467:60–79. doi: 10.1002/cne.10905. [DOI] [PubMed] [Google Scholar]
- Tsien JZ, Chen DF, Gerber D, Tom C, Mercer EH, Anderson DJ, Mayford M, Kandel ER, Tonegawa S. Subregion- and cell type-restricted gene knockout in mouse brain. Cell. 1996;87:1317–1326. doi: 10.1016/s0092-8674(00)81826-7. [DOI] [PubMed] [Google Scholar]
- Ugolini G. Use of rabies virus as a transneuronal tracer of neuronal connections: implications for the understanding of rabies pathogenesis. Dev Biol (Basel) 2008;131:493–506. [PubMed] [Google Scholar]
- Ugolini G. Rabies virus as a transneuronal tracer of neuronal connections. Adv Virus Res. 2011;79:165–202. doi: 10.1016/B978-0-12-387040-7.00010-X. [DOI] [PubMed] [Google Scholar]
- Wall NR, Wickersham IR, Cetin A, De La Parra M, Callaway EM. Monosynaptic circuit tracing in vivo through Cre-dependent targeting and complementation of modified rabies virus. Proc Natl Acad Sci U S A. 2010;107:21848–21853. doi: 10.1073/pnas.1011756107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickersham IR, Finke S, Conzelmann KK, Callaway EM. Retrograde neuronal tracing with a deletion-mutant rabies virus. Nat Methods. 2007a;4:47–49. doi: 10.1038/NMETH999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickersham IR, Lyon DC, Barnard RJ, Mori T, Finke S, Conzelmann KK, Young JA, Callaway EM. Monosynaptic restriction of transsynaptic tracing from single, genetically targeted neurons. Neuron. 2007b;53:639–647. doi: 10.1016/j.neuron.2007.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittner L, Henze DA, Zaborszky L, Buzsaki G. Hippocampal CA3 pyramidal cells selectively innervate aspiny interneurons. Eur J Neurosci. 2006;24:1286–1298. doi: 10.1111/j.1460-9568.2006.04992.x. [DOI] [PubMed] [Google Scholar]
- Xu X, Callaway EM. Laminar specificity of functional input to distinct types of inhibitory cortical neurons. J Neurosci. 2009;29:70–85. doi: 10.1523/JNEUROSCI.4104-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Olivas ND, Levi R, Ikrar T, Nenadic Z. High precision and fast functional mapping of cortical circuitry through a combination of voltage sensitive dye imaging and laser scanning photostimulation. J Neurophysiol. 2010a;103:2301–2312. doi: 10.1152/jn.00992.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Roby KD, Callaway EM. Immunochemical characterization of inhibitory mouse cortical neurons: three chemically distinct classes of inhibitory cells. J Comp Neurol. 2010b;518:389–404. doi: 10.1002/cne.22229. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.