Abstract
In the title compound, C12H13N3S, the 4,5-dihydro-3H-1,2,4-triazole system is nearly planar [maximum deviation = 0.014 (2) Å], while the cyclopentane ring adopts a half-chair conformation. The dihedral angle between the mean plane of the 4,5-dihydro-3H-1,2,4-triazole-3-thione ring and the phenyl ring is 85.49 (14)°, with the S atom 0.046 (1) Å out of the former plane. The crystal structure is stabilized only by van der Waals interactions. The investigated crystal was found to be a non-merohedral two-component twin by a 180° rotation about c*, with a refined value of the minor twin fraction of 0.12203 (18).
Related literature
For wide-spectrum medicinal applications of spiro compounds incorporating heterocyclic substructures, see: Sar et al. (2006 ▶); Park et al. (2007 ▶); Nakao et al. (2008 ▶); Obniska & Kamiński (2006 ▶); Kamiński et al. (2008 ▶); Obniska et al. (2006 ▶); Chin et al. (2008 ▶); Wang et al. (2007 ▶); Pawar et al. (2009 ▶); Thadhaney et al. (2010 ▶); (Chande et al., 2005 ▶); Shimakawa et al. (2003 ▶); Sarma et al. (2010 ▶). For industrial uses of heterocyclic spiro compounds, see: Rongbao et al. (2009 ▶); Hu et al. (2006 ▶); Méhes et al. (2012 ▶); Billah et al. (2008 ▶). For the crystal structure of a similar compound, see: Akkurt et al. (2013 ▶). For ring-puckering parameters, see: Cremer & Pople (1975 ▶). For the indexing program for twinned crystals, see: Sheldrick (2008a
▶).
Experimental
Crystal data
C12H13N3S
M r = 231.32
Monoclinic,

a = 11.4780 (12) Å
b = 12.0452 (12) Å
c = 17.0439 (17) Å
β = 101.3060 (14)°
V = 2310.7 (4) Å3
Z = 8
Mo Kα radiation
μ = 0.26 mm−1
T = 150 K
0.15 × 0.13 × 0.12 mm
Data collection
Bruker SMART APEX CCD diffractometer
Absorption correction: multi-scan (TWINABS; Sheldrick, 2009 ▶) T min = 0.96, T max = 0.97
29725 measured reflections
29725 independent reflections
21519 reflections with I > 2σ(I)
R int = 0.050
Refinement
R[F 2 > 2σ(F 2)] = 0.059
wR(F 2) = 0.168
S = 1.05
29725 reflections
146 parameters
44 restraints
H-atom parameters constrained
Δρmax = 0.62 e Å−3
Δρmin = −0.65 e Å−3
Data collection: APEX2 (Bruker, 2013 ▶); cell refinement: SAINT (Bruker, 2013 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXTL (Sheldrick, 2008b ▶); program(s) used to refine structure: SHELXL2014 (Sheldrick, 2008b ▶); molecular graphics: DIAMOND (Brandenburg & Putz, 2012 ▶) and ORTEP-3 for Windows (Farrugia, 2012 ▶); software used to prepare material for publication: SHELXTL.
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536814005418/rz5108sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536814005418/rz5108Isup2.hkl
Supporting information file. DOI: 10.1107/S1600536814005418/rz5108Isup3.cml
CCDC reference: 990878
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
Manchester Metropolitan University, Tulane University and Erciyes University are gratefully acknowledged for supporting this study.
supplementary crystallographic information
1. Comment
Heterocyclic spirocompounds are an important class of chemicals due to their great applications in medicinal and industrial fields. Beside the wide spectrum of their biological activities such as antibacterial agents (Sar et al., 2006; Park et al., 2007), anti-dermatitis agents (Nakao et al., 2008), anticonvulsant agents (Obniska et al., 2006; Kamiński et al., 2008; Obniska et al., 2006), anticancer agents (Chin et al., 2008; Wang et al., 2007), antimicrobial agents (Pawar et al., 2009; Thadhaney et al., 2010), anti-tuberculosis agents (Chande et al., 2005), and recently as anti-oxidants (Shimakawa et al., 2003; Sarma et al., 2010), they act also as pesticides (Rongbao et al., 2009), antifungal agents (Hu et al., 2006), electroluminescent devices (Méhes et al., 2012) and laser dyes (Billah et al., 2008). We were inspired by these findings to synthesize the title compound as part of our on-going study on synthesis and biological activity of spirocompound-based triazole derivatives.
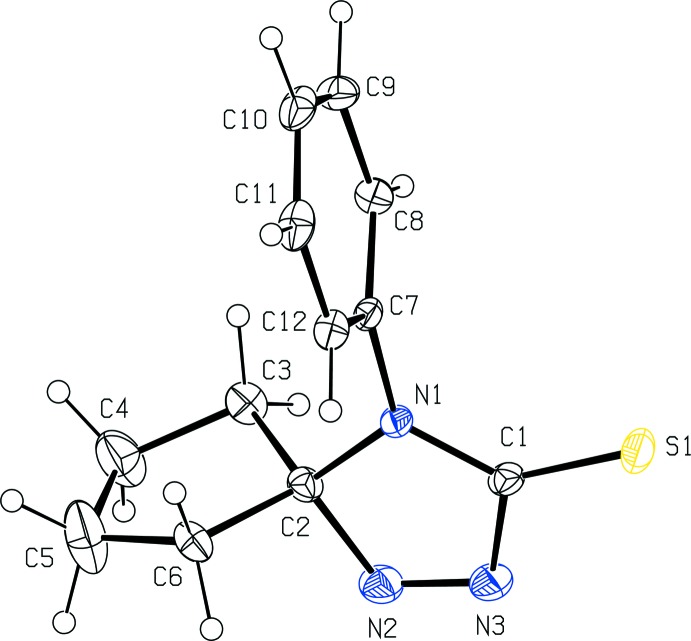
The five-membered cyclopentane ring (C2–C6) of the title compound (I, Fig. 1), adopts a half-chair conformation with the puckering parameters (Cremer & Pople, 1975) of Q(2) = 0.250 (3) Å and φ(2) = 191.9 (9)°. The dihedral angle between the mean plane of the 4,5-dihydro-3H-1,2,4-triazole-3-thione ring (N1–N3/C1/C2) and the phenyl ring (C7–C12) is 85.49 (14)° with the S1 atom 0.046 (1) Å out of the former plane.
All bond lengths and bond angles in (I) are comparable with those for the similar compound "4-Phenyl-1,2,4-triazaspiro[4.5]dec-1-ene-3-thione" that we have reported previously (Akkurt et al., 2013). The crystal structure is stabilized only by van der Waals interactions.
2. Experimental
The title compound was prepared according to our previous reported method (Akkurt et al. 2013). Orange block crystals suitable for X-ray diffraction were obtained from ethylacetate solution of I at room temperature (m.p. 455 – 457 K).
3. Refinement
The crystal used proved to be twinned by a 180° rotation about c* (CELL_NOW, Sheldrick, 2008a) and the final structure was refined as a 2-component twin with a refined value of the minor twin fraction of 0.12203 (18). All H atoms were placed in geometrically idealized positions and constrained to ride on their parent atoms with O—H = 0.85 Å, N—H = 0.88 Å, C—H = 0.95 Å and 0.98 Å, with Uiso(H) = 1.5 Uiso(C) for methyl H atoms and Uiso(H) = 1.2 Uiso(C,N,O) for other H atoms. Restraints (DELU instructions in SHELXL-97) were used to reduce the components of the anisotropic displacement parameters of all atoms along the chemical bonds.
Figures
Fig. 1.
Perspective view of the title molecule with 30% displacement ellipsoids.
Crystal data
| C12H13N3S | F(000) = 976 |
| Mr = 231.32 | Dx = 1.330 Mg m−3 |
| Monoclinic, C2/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -C 2yc | Cell parameters from 8834 reflections |
| a = 11.4780 (12) Å | θ = 2.4–29.1° |
| b = 12.0452 (12) Å | µ = 0.26 mm−1 |
| c = 17.0439 (17) Å | T = 150 K |
| β = 101.3060 (14)° | Block, orange |
| V = 2310.7 (4) Å3 | 0.15 × 0.13 × 0.12 mm |
| Z = 8 |
Data collection
| Bruker SMART APEX CCD diffractometer | 29725 independent reflections |
| Radiation source: fine-focus sealed tube | 21519 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.050 |
| Detector resolution: 8.3660 pixels mm-1 | θmax = 29.2°, θmin = 2.4° |
| φ and ω scans | h = −15→15 |
| Absorption correction: multi-scan (TWINABS; Sheldrick, 2009) | k = −16→16 |
| Tmin = 0.96, Tmax = 0.97 | l = −23→23 |
| 29725 measured reflections |
Refinement
| Refinement on F2 | 44 restraints |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.059 | H-atom parameters constrained |
| wR(F2) = 0.168 | w = 1/[σ2(Fo2) + (0.0674P)2 + 1.7625P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.05 | (Δ/σ)max < 0.001 |
| 29725 reflections | Δρmax = 0.62 e Å−3 |
| 146 parameters | Δρmin = −0.65 e Å−3 |
Special details
| Geometry. Bond distances, angles etc. have been calculated using the rounded fractional coordinates. All su's are estimated from the variances of the (full) variance-covariance matrix. The cell e.s.d.'s are taken into account in the estimation of distances, angles and torsion angles |
| Refinement. Refinement on F2 for ALL reflections except those flagged by the user for potential systematic errors. Weighted R-factors wR and all goodnesses of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The observed criterion of F2 > σ(F2) is used only for calculating -R-factor-obs etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R-factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| S1 | 0.33502 (7) | 0.94435 (6) | 1.03564 (4) | 0.0358 (2) | |
| N1 | 0.42067 (18) | 0.80829 (17) | 0.93431 (12) | 0.0221 (6) | |
| N2 | 0.5566 (2) | 0.7153 (2) | 1.02559 (14) | 0.0355 (8) | |
| N3 | 0.5050 (2) | 0.7828 (2) | 1.06315 (14) | 0.0364 (8) | |
| C1 | 0.4168 (2) | 0.8470 (2) | 1.00689 (15) | 0.0259 (8) | |
| C2 | 0.5122 (2) | 0.7230 (2) | 0.93878 (15) | 0.0252 (7) | |
| C3 | 0.4680 (2) | 0.6091 (2) | 0.90472 (18) | 0.0321 (9) | |
| C4 | 0.5715 (3) | 0.5557 (3) | 0.8784 (3) | 0.0627 (14) | |
| C5 | 0.6575 (4) | 0.6450 (3) | 0.8686 (3) | 0.0695 (16) | |
| C6 | 0.6150 (2) | 0.7526 (2) | 0.89633 (17) | 0.0321 (9) | |
| C7 | 0.3486 (2) | 0.8458 (2) | 0.86065 (14) | 0.0214 (7) | |
| C8 | 0.2459 (2) | 0.7876 (2) | 0.82843 (16) | 0.0289 (8) | |
| C9 | 0.1807 (3) | 0.8195 (3) | 0.75431 (17) | 0.0374 (9) | |
| C10 | 0.2175 (3) | 0.9075 (3) | 0.71427 (16) | 0.0389 (10) | |
| C11 | 0.3174 (3) | 0.9671 (2) | 0.74808 (16) | 0.0358 (9) | |
| C12 | 0.3844 (2) | 0.9370 (2) | 0.82186 (16) | 0.0280 (8) | |
| H3A | 0.44130 | 0.56350 | 0.94620 | 0.0380* | |
| H3B | 0.40080 | 0.61790 | 0.85890 | 0.0380* | |
| H4A | 0.60970 | 0.50130 | 0.91890 | 0.0750* | |
| H4B | 0.54480 | 0.51630 | 0.82710 | 0.0750* | |
| H5A | 0.66400 | 0.65090 | 0.81160 | 0.0840* | |
| H5B | 0.73710 | 0.62700 | 0.90030 | 0.0840* | |
| H6A | 0.58690 | 0.80240 | 0.85030 | 0.0380* | |
| H6B | 0.67980 | 0.79020 | 0.93380 | 0.0380* | |
| H8 | 0.22050 | 0.72710 | 0.85660 | 0.0350* | |
| H9 | 0.11060 | 0.78020 | 0.73130 | 0.0450* | |
| H10 | 0.17400 | 0.92750 | 0.66290 | 0.0470* | |
| H11 | 0.34040 | 1.02950 | 0.72060 | 0.0430* | |
| H12 | 0.45330 | 0.97790 | 0.84520 | 0.0340* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| S1 | 0.0424 (4) | 0.0413 (4) | 0.0254 (4) | 0.0049 (3) | 0.0109 (3) | −0.0062 (3) |
| N1 | 0.0225 (10) | 0.0259 (11) | 0.0175 (10) | 0.0026 (9) | 0.0027 (8) | 0.0011 (8) |
| N2 | 0.0318 (13) | 0.0438 (14) | 0.0288 (13) | 0.0038 (11) | 0.0009 (10) | 0.0069 (11) |
| N3 | 0.0341 (13) | 0.0498 (15) | 0.0227 (12) | 0.0028 (12) | −0.0006 (10) | 0.0044 (11) |
| C1 | 0.0259 (13) | 0.0331 (14) | 0.0181 (12) | −0.0035 (11) | 0.0030 (10) | 0.0014 (10) |
| C2 | 0.0227 (12) | 0.0273 (13) | 0.0252 (13) | 0.0043 (10) | 0.0040 (11) | 0.0036 (10) |
| C3 | 0.0322 (15) | 0.0253 (14) | 0.0392 (16) | 0.0011 (11) | 0.0082 (13) | 0.0029 (12) |
| C4 | 0.042 (2) | 0.050 (2) | 0.101 (3) | −0.0047 (16) | 0.026 (2) | −0.026 (2) |
| C5 | 0.070 (3) | 0.049 (2) | 0.107 (3) | −0.008 (2) | 0.060 (3) | −0.016 (2) |
| C6 | 0.0243 (13) | 0.0354 (16) | 0.0385 (16) | 0.0022 (11) | 0.0110 (12) | 0.0045 (12) |
| C7 | 0.0243 (12) | 0.0245 (13) | 0.0151 (11) | 0.0056 (10) | 0.0030 (9) | −0.0012 (9) |
| C8 | 0.0264 (13) | 0.0304 (14) | 0.0291 (14) | 0.0015 (11) | 0.0038 (11) | 0.0004 (11) |
| C9 | 0.0290 (15) | 0.0458 (18) | 0.0329 (16) | 0.0075 (13) | −0.0051 (12) | −0.0081 (13) |
| C10 | 0.0472 (18) | 0.0466 (18) | 0.0201 (13) | 0.0245 (15) | −0.0004 (13) | −0.0006 (12) |
| C11 | 0.0524 (18) | 0.0318 (16) | 0.0253 (14) | 0.0142 (14) | 0.0130 (13) | 0.0076 (11) |
| C12 | 0.0329 (14) | 0.0266 (14) | 0.0252 (14) | 0.0027 (11) | 0.0075 (11) | −0.0001 (10) |
Geometric parameters (Å, º)
| S1—C1 | 1.636 (3) | C10—C11 | 1.380 (5) |
| N1—C1 | 1.331 (3) | C11—C12 | 1.387 (4) |
| N1—C2 | 1.461 (3) | C3—H3A | 0.9900 |
| N1—C7 | 1.434 (3) | C3—H3B | 0.9900 |
| N2—N3 | 1.253 (3) | C4—H4A | 0.9900 |
| N2—C2 | 1.470 (3) | C4—H4B | 0.9900 |
| N3—C1 | 1.470 (3) | C5—H5A | 0.9900 |
| C2—C3 | 1.537 (3) | C5—H5B | 0.9900 |
| C2—C6 | 1.542 (3) | C6—H6A | 0.9900 |
| C3—C4 | 1.495 (4) | C6—H6B | 0.9900 |
| C4—C5 | 1.491 (6) | C8—H8 | 0.9500 |
| C5—C6 | 1.494 (5) | C9—H9 | 0.9500 |
| C7—C8 | 1.389 (3) | C10—H10 | 0.9500 |
| C7—C12 | 1.385 (3) | C11—H11 | 0.9500 |
| C8—C9 | 1.390 (4) | C12—H12 | 0.9500 |
| C9—C10 | 1.371 (5) | ||
| C1—N1—C2 | 110.7 (2) | C4—C3—H3A | 111.00 |
| C1—N1—C7 | 125.8 (2) | C4—C3—H3B | 111.00 |
| C2—N1—C7 | 123.6 (2) | H3A—C3—H3B | 109.00 |
| N3—N2—C2 | 111.6 (2) | C3—C4—H4A | 110.00 |
| N2—N3—C1 | 110.0 (2) | C3—C4—H4B | 110.00 |
| S1—C1—N1 | 130.9 (2) | C5—C4—H4A | 110.00 |
| S1—C1—N3 | 122.94 (19) | C5—C4—H4B | 110.00 |
| N1—C1—N3 | 106.1 (2) | H4A—C4—H4B | 108.00 |
| N1—C2—N2 | 101.58 (19) | C4—C5—H5A | 110.00 |
| N1—C2—C3 | 115.3 (2) | C4—C5—H5B | 110.00 |
| N1—C2—C6 | 114.9 (2) | C6—C5—H5A | 110.00 |
| N2—C2—C3 | 110.3 (2) | C6—C5—H5B | 110.00 |
| N2—C2—C6 | 109.9 (2) | H5A—C5—H5B | 108.00 |
| C3—C2—C6 | 104.8 (2) | C2—C6—H6A | 111.00 |
| C2—C3—C4 | 105.9 (2) | C2—C6—H6B | 111.00 |
| C3—C4—C5 | 107.8 (3) | C5—C6—H6A | 111.00 |
| C4—C5—C6 | 109.0 (3) | C5—C6—H6B | 111.00 |
| C2—C6—C5 | 106.0 (2) | H6A—C6—H6B | 109.00 |
| N1—C7—C8 | 119.1 (2) | C7—C8—H8 | 121.00 |
| N1—C7—C12 | 119.6 (2) | C9—C8—H8 | 121.00 |
| C8—C7—C12 | 121.3 (2) | C8—C9—H9 | 120.00 |
| C7—C8—C9 | 118.9 (2) | C10—C9—H9 | 120.00 |
| C8—C9—C10 | 120.2 (3) | C9—C10—H10 | 120.00 |
| C9—C10—C11 | 120.4 (3) | C11—C10—H10 | 120.00 |
| C10—C11—C12 | 120.7 (2) | C10—C11—H11 | 120.00 |
| C7—C12—C11 | 118.4 (2) | C12—C11—H11 | 120.00 |
| C2—C3—H3A | 111.00 | C7—C12—H12 | 121.00 |
| C2—C3—H3B | 111.00 | C11—C12—H12 | 121.00 |
| C2—N1—C1—S1 | 177.9 (2) | N2—N3—C1—N1 | 1.5 (3) |
| C2—N1—C1—N3 | −2.4 (3) | N1—C2—C3—C4 | 153.1 (3) |
| C7—N1—C1—S1 | −0.4 (4) | N2—C2—C3—C4 | −92.6 (3) |
| C7—N1—C1—N3 | 179.3 (2) | C6—C2—C3—C4 | 25.7 (3) |
| C1—N1—C2—N2 | 2.4 (3) | N1—C2—C6—C5 | −149.9 (3) |
| C1—N1—C2—C3 | 121.7 (2) | N2—C2—C6—C5 | 96.3 (3) |
| C1—N1—C2—C6 | −116.2 (2) | C3—C2—C6—C5 | −22.2 (3) |
| C7—N1—C2—N2 | −179.3 (2) | C2—C3—C4—C5 | −19.6 (4) |
| C7—N1—C2—C3 | −60.0 (3) | C3—C4—C5—C6 | 5.6 (5) |
| C7—N1—C2—C6 | 62.1 (3) | C4—C5—C6—C2 | 10.6 (4) |
| C1—N1—C7—C8 | −96.5 (3) | N1—C7—C8—C9 | −175.6 (2) |
| C1—N1—C7—C12 | 85.5 (3) | C12—C7—C8—C9 | 2.5 (4) |
| C2—N1—C7—C8 | 85.5 (3) | N1—C7—C12—C11 | 175.7 (2) |
| C2—N1—C7—C12 | −92.6 (3) | C8—C7—C12—C11 | −2.3 (4) |
| C2—N2—N3—C1 | 0.1 (3) | C7—C8—C9—C10 | −0.4 (4) |
| N3—N2—C2—N1 | −1.5 (3) | C8—C9—C10—C11 | −1.9 (5) |
| N3—N2—C2—C3 | −124.2 (2) | C9—C10—C11—C12 | 2.1 (5) |
| N3—N2—C2—C6 | 120.7 (2) | C10—C11—C12—C7 | 0.0 (4) |
| N2—N3—C1—S1 | −178.8 (2) |
Footnotes
Supporting information for this paper is available from the IUCr electronic archives (Reference: RZ5108).
References
- Akkurt, M., Mague, J. T., Mohamed, S. K., Hassan, A. A. & Albayati, M. R. (2013). Acta Cryst. E69, o1259. [DOI] [PMC free article] [PubMed]
- Billah, S. M. R., Christie, R. M. & Morgan, K. M. (2008). Coloration Technol. 124, 229–233.
- Brandenburg, K. & Putz, H. (2012). DIAMOND Crystal Impact GbR, Bonn, Germany.
- Bruker (2013). APEX2 and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Chande, M. S., Verma, R. S., Barve, P. A., Khanwelkar, R. R., Vaidya, R. B. & Ajaikumar, K. B. (2005). Eur. J. Med. Chem. 40, 1143–1148. [DOI] [PubMed]
- Chin, Y.-W., Salim, A. A., Su, B.-N., Mi, Q., Chai, H.-B., Riswan, S., Kardono, L. B. S., Ruskandi, A., Farnsworth, N. R., Swanson, S. M. & Kinghorn, A. D. (2008). J. Nat. Prod. 3, 390–395. [DOI] [PMC free article] [PubMed]
- Cremer, D. & Pople, J. A. (1975). J. Am. Chem. Soc. 97, 1354–1358.
- Farrugia, L. J. (2012). J. Appl. Cryst. 45, 849–854.
- Hu, H., Guo, H., Li, E., Liu, X., Zhou, Y. & Che, Y. (2006). J. Nat. Prod. 69, 1672–1675. [DOI] [PubMed]
- Kamiński, K., Obniska, J. & Dybała, M. (2008). Eur. J. Med. Chem. 43, 53–61. [DOI] [PubMed]
- Méhes, G., Nomura, H., Zhang, Q., Nakagawa, T. & Adachi, C. (2012). Angew. Chem. Int. Ed. 51, 11311–11315. [DOI] [PubMed]
- Nakao, K., Ikeda, K., Kurokawa, T., Togashi, Y., Umeuchi, H., Honda, T., Okano, K. & Mochizuki, H. (2008). Nihon Shinkei Seishin Yakurigaku Zasshi, 28, 75–83. [PubMed]
- Obniska, J. & Kamiński, K. (2006). Acta Pol. Pharm. 63, 101–108. [PubMed]
- Obniska, J., Kamiński, K. & Tatarczynska, E. (2006). Pharmacol. Rep. 58, 207–214. [PubMed]
- Park, H. B., Jo, N. H., Hong, J. H., Choi, J. H., Cho, J.-H., Yoo, K. H. & Oh, C.-H. (2007). Arch. Pharm. 340, 530–537. [DOI] [PubMed]
- Pawar, M. J., Burungale, A. B. & Karale, B. K. (2009). ARKIVOC, XIII, 97–107.
- Rongbao, W., Yang, L. & Ya, L. (2009). Chin. J. Org. Chem. 12, 476–487.
- Sar, S., Blunt, J. & Munro, M. (2006). Org. Lett. 8, 2059–2069. [DOI] [PubMed]
- Sarma, B. K., Manna, D., Minoura, M. & Mugesh, G. (2010). J. Am. Chem. Soc. 132, 5364–5374. [DOI] [PubMed]
- Sheldrick, G. M. (2008a). CELL_NOW University of Göttingen, Germany.
- Sheldrick, G. M. (2008b). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sheldrick, G. M. (2009). TWINABS University of Göttingen, Germany.
- Shimakawa, S., Yoshida, Y. & Niki, E. (2003). Lipids , 38, 225–231. [DOI] [PubMed]
- Thadhaney, B., Sain, D., Pernawat, G. & Talesara, G. L. (2010). Indian J. Chem. Sect. B, 49, 368–373.
- Wang, W.-L., Zhu, T.-J., Tao, H.-W., Lu, Z.-Y., Fang, Y.-C., Gu, Q.-Q. & Zhu, W.-M. (2007). Chem. Biodivers. 4, 2913–2919. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536814005418/rz5108sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536814005418/rz5108Isup2.hkl
Supporting information file. DOI: 10.1107/S1600536814005418/rz5108Isup3.cml
CCDC reference: 990878
Additional supporting information: crystallographic information; 3D view; checkCIF report