Abstract
In the title compound, C18H24N6O·H2O, the piperidine ring adopts a chair conformation with an N—C—C—C torsion angle of 39.5 (5)° between the cis-related substituents. The pyrrole N—H group forms a water-mediated intermolecular hydrogen bond to one of the N atoms of the annelated pyrimidine ring. The water molecule connects two organic molecules and is disorderd over two positions (occupancies of 0.48 and 0.52). The crystal packing shows zigzag chains of alternating organic and water molecules running parallel to the a axis.
Related literature
For the biological activity and structure–activity relationships of tofacitinib {systematic name: 3-[(3R,4R)-4-methyl-3-[methyl(7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]piperidin-1-yl]-3-oxopropanenitrile} derivatives, see: Flanagan et al. (2010 ▶). For a general overview on the JAK–STAT pathway, see: Shuai & Liu (2003 ▶). The use of oxetanes as carbonyl bioisosteres has been reviewed extensively by Wuitschik et al. (2010 ▶). For a recent application of this concept towards tofacitinib-derived JAK3 inhibitors, see: Gehringer et al. (2014 ▶).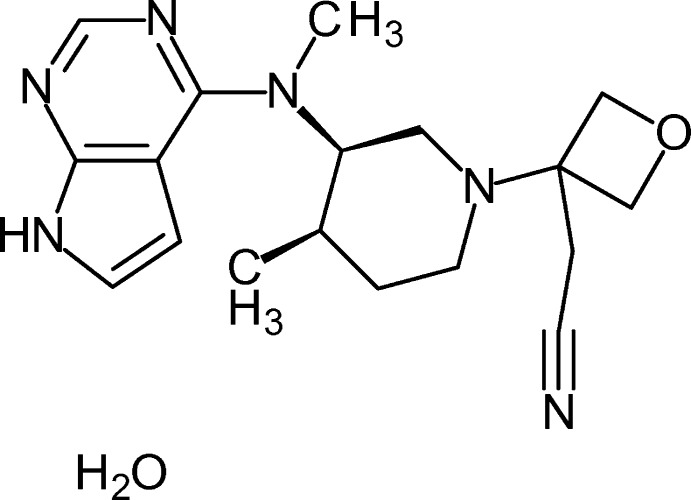
Experimental
Crystal data
C18H24N6O·H2O
M r = 358.45
Orthorhombic,
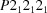
a = 6.6088 (6) Å
b = 10.1483 (8) Å
c = 26.813 (2) Å
V = 1798.3 (3) Å3
Z = 4
Mo Kα radiation
μ = 0.09 mm−1
T = 193 K
0.29 × 0.27 × 0.06 mm
Data collection
Stoe IPDS 2T diffractometer
6672 measured reflections
4184 independent reflections
1716 reflections with I > 2σ(I)
R int = 0.079
Refinement
R[F 2 > 2σ(F 2)] = 0.062
wR(F 2) = 0.150
S = 0.90
4184 reflections
246 parameters
H-atom parameters constrained
Δρmax = 0.19 e Å−3
Δρmin = −0.22 e Å−3
Data collection: X-AREA (Stoe & Cie, 2010 ▶); cell refinement: X-AREA; data reduction: X-RED (Stoe & Cie, 2010 ▶); program(s) used to solve structure: SIR97 (Altomare et al., 1999 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: PLATON (Spek, 2009 ▶); software used to prepare material for publication: PLATON.
Supplementary Material
Crystal structure: contains datablock(s) I, Global. DOI: 10.1107/S1600536814004449/bt6965sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536814004449/bt6965Isup2.hkl
Supporting information file. DOI: 10.1107/S1600536814004449/bt6965Isup3.cml
CCDC reference: 988870
Additional supporting information: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1⋯O1L | 0.88 | 1.90 | 2.783 (8) | 178 |
| N1—H1⋯O2L | 0.88 | 2.06 | 2.816 (7) | 144 |
| O1L—H1L2⋯N8i | 0.84 | 2.27 | 2.868 (8) | 129 |
| O2L—H2L2⋯N8i | 0.84 | 2.20 | 2.733 (7) | 121 |
| O2L—H2L2⋯N25ii | 0.84 | 2.43 | 3.026 (10) | 129 |
Symmetry codes: (i) 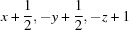 ; (ii)
; (ii) 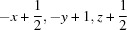 .
.
Acknowledgments
The authors thank Michael Forster for fruitful discussions and suggestions and acknowledge support by the Deutsche Forschungsgemeinschaft and the Open Access Publishing Fund of Tuebingen University.
supplementary crystallographic information
1. Comment
Janus Kinases (JAKs) are non-receptor protein tyrosine kinases mediating signalling through the JAK-STAT (Signal Transducer and Activator of Transcription) pathway. Being crucial signal transducers for a variety of cytokines, growth factors, and interferons, JAKs are involved in numerous pathologies including malignancies, myeloproliferative disorders and autoimmune diseases (Shuai & Liu, 2003). Recently, Tofacitinib (CP690,550; 3-[(3R,4R)-4-methyl-3-[methyl(7H-pyrrolo[2,3-d] pyrimidin-4-yl)amino]piperidin-1-yl]-3-oxopropanenitrile) a small-molecule pan-JAK inhibitor was approved by the US Food and Drug Administration for the treatment of rheumatoid athritis (Flanagan et al., 2010). The compound also shows promising results in late stage clinical trials for psoriasis, transplant rejection, and other disorders of the immune system. In search for novel JAK inhibitors, the title compound was prepared as a Tofacitinib bioisostere with altered physicochemical properties (Wuitschik et al., 2010).
In the crystal structure of the title compound, C18H24N6O, the exocyclic amino substituent is oriented almost coplanar to the heteroaromatic ring system with a torsion angle of 0.6 (2)° for C11-N10-C5-C4. The piperidine ring adopts a chair conformation with a torsion angle of 39.5 (5) ° between the cis substituents. One of the protons (H23B) of the methylene group between the oxetane ring and the nitrile function is involved in an intermolecular C—H···π interaction while the other methylene proton forms an intermolecular C—H···N interaction with the nitrile group. The oxygen atom of the oxetane ring makes two intermolecular C—H···O contacts with two H atoms (H13A and H15B) of the piperidine ring. The heterocyclic pyrrol N—H forms an intermolecular water mediated hydrogen bond to one of the nitrogen atoms (N8) in the 6-membered pyrmidine heterocycle with a length of 2.78 (2) Å for the N—H···O and 2.86 (8) Å for the O—H···N contact. The water molecule connecting two molecules is disorderd over two positions (s.o.f. 0.48/0.52). The crystal packing is shows N—H···OH···N hydrogen bonds resulting in infinite chains parallel to the a axis.
2. Experimental
In an HPLC-vial, (3R,4R)-N,4-dimethyl-N-{7H-pyrrolo[2,3-d]pyrimidin-4- yl}piperidin-3-amine (50.0 mg, 204 µmol) and (oxetan-3-ylidene)acetonitrile (19.9 mg, 210 µmol) were dissolved in dry ethanol (400 µl) and the mixture stirred at 323 K during 48 h. The solvent was evaporated under reduced pressure and the product purified by column chromatography (SiO2, ethyl acetate + 2% methanol). The product was obtained as off-white solid (57.0 mg, 82%). Crystals of the title compound were obtained by slow evaporation of a solution in chloroform + 10% methanol at 298 K.
3. Refinement
Site occupation factors of the disordered water molecule were fixed assuming similar isotropic displacement parameters for alternative positions of the oxygen atom. Hydrogen atoms attached to carbons were placed at calculated positions with C—H = 0.95 Å (aromatic) or 0.99–1.00 Å (sp3 C-atom). All H atoms were refined with isotropic displacement parameters (set at 1.2–1.5 times of the Ueq of the parent atom). One of the H atoms of the disordered water molecule could be position to make a hydrogen bond to an N atom. The other one was positioneded with idealized geometric with respect to the first one. The absolute configuration was assigned according to the synthesis.
Figures
Fig. 1.
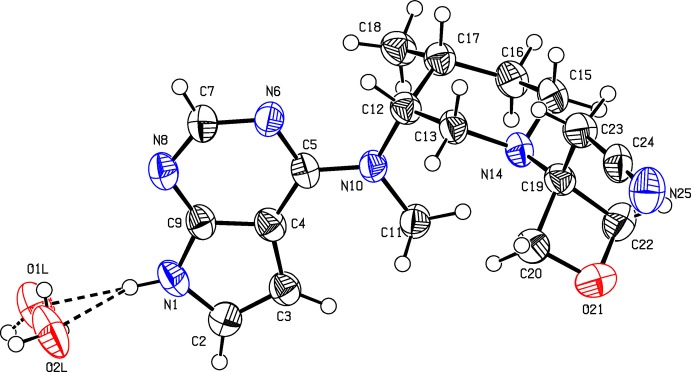
Crystal structure of the title compound with labeling and displacement ellipsoids drawn at the 50% probability level. Water molecules are disorderd with s.o.f. 0.52/0.48.
Crystal data
| C18H24N6O·H2O | F(000) = 768 |
| Mr = 358.45 | Dx = 1.324 Mg m−3 |
| Orthorhombic, P212121 | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: P 2ac 2ab | Cell parameters from 6886 reflections |
| a = 6.6088 (6) Å | θ = 2.5–27.8° |
| b = 10.1483 (8) Å | µ = 0.09 mm−1 |
| c = 26.813 (2) Å | T = 193 K |
| V = 1798.3 (3) Å3 | Plate, colourless |
| Z = 4 | 0.29 × 0.27 × 0.06 mm |
Data collection
| Stoe IPDS 2T diffractometer | 1716 reflections with I > 2σ(I) |
| Radiation source: sealed Tube | Rint = 0.079 |
| Graphite monochromator | θmax = 28.0°, θmin = 3.2° |
| Detector resolution: 6.67 pixels mm-1 | h = −7→8 |
| rotation method scans | k = −11→13 |
| 6672 measured reflections | l = −29→35 |
| 4184 independent reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.062 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.150 | H-atom parameters constrained |
| S = 0.90 | w = 1/[σ2(Fo2) + (0.0523P)2] where P = (Fo2 + 2Fc2)/3 |
| 4184 reflections | (Δ/σ)max < 0.001 |
| 246 parameters | Δρmax = 0.19 e Å−3 |
| 0 restraints | Δρmin = −0.22 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | Occ. (<1) | |
| N1 | 0.1280 (5) | 0.1062 (4) | 0.42472 (12) | 0.0485 (10) | |
| H1 | 0.1160 | 0.1276 | 0.4564 | 0.058* | |
| C2 | 0.2919 (7) | 0.0433 (4) | 0.40332 (16) | 0.0485 (12) | |
| H2 | 0.4106 | 0.0163 | 0.4205 | 0.058* | |
| C3 | 0.2563 (7) | 0.0264 (4) | 0.35393 (15) | 0.0439 (11) | |
| H3 | 0.3446 | −0.0139 | 0.3305 | 0.053* | |
| C4 | 0.0579 (7) | 0.0814 (4) | 0.34361 (15) | 0.0423 (11) | |
| C5 | −0.0680 (7) | 0.1060 (4) | 0.30217 (14) | 0.0391 (10) | |
| N6 | −0.2486 (6) | 0.1667 (4) | 0.30983 (12) | 0.0449 (9) | |
| C7 | −0.2941 (7) | 0.2044 (4) | 0.35573 (15) | 0.0476 (11) | |
| H7 | −0.4211 | 0.2473 | 0.3591 | 0.057* | |
| N8 | −0.1909 (6) | 0.1916 (4) | 0.39726 (12) | 0.0470 (9) | |
| C9 | −0.0105 (7) | 0.1291 (4) | 0.38869 (14) | 0.0413 (10) | |
| N10 | −0.0216 (5) | 0.0778 (3) | 0.25423 (11) | 0.0396 (8) | |
| C11 | 0.1748 (6) | 0.0150 (4) | 0.24320 (14) | 0.0451 (11) | |
| H11A | 0.1885 | 0.0029 | 0.2071 | 0.068* | |
| H11B | 0.1815 | −0.0709 | 0.2598 | 0.068* | |
| H11C | 0.2847 | 0.0712 | 0.2554 | 0.068* | |
| C12 | −0.1610 (7) | 0.1075 (4) | 0.21317 (14) | 0.0415 (10) | |
| H12 | −0.2699 | 0.1634 | 0.2281 | 0.050* | |
| C13 | −0.0683 (7) | 0.1906 (4) | 0.17183 (13) | 0.0419 (11) | |
| H13A | −0.1782 | 0.2339 | 0.1528 | 0.050* | |
| H13B | 0.0160 | 0.2606 | 0.1870 | 0.050* | |
| N14 | 0.0562 (5) | 0.1135 (3) | 0.13743 (11) | 0.0401 (9) | |
| C15 | −0.0684 (7) | 0.0153 (4) | 0.11233 (14) | 0.0437 (11) | |
| H15A | 0.0130 | −0.0318 | 0.0870 | 0.052* | |
| H15B | −0.1832 | 0.0588 | 0.0953 | 0.052* | |
| C16 | −0.1472 (7) | −0.0818 (4) | 0.15091 (15) | 0.0474 (11) | |
| H16A | −0.0314 | −0.1284 | 0.1663 | 0.057* | |
| H16B | −0.2331 | −0.1483 | 0.1341 | 0.057* | |
| C17 | −0.2698 (7) | −0.0137 (4) | 0.19154 (14) | 0.0424 (11) | |
| H17 | −0.3970 | 0.0189 | 0.1755 | 0.051* | |
| C18 | −0.3324 (7) | −0.1107 (5) | 0.23178 (15) | 0.0506 (11) | |
| H18A | −0.4324 | −0.1724 | 0.2182 | 0.076* | |
| H18B | −0.3920 | −0.0627 | 0.2599 | 0.076* | |
| H18C | −0.2135 | −0.1598 | 0.2432 | 0.076* | |
| C19 | 0.1743 (7) | 0.1946 (4) | 0.10400 (14) | 0.0421 (10) | |
| C20 | 0.3401 (7) | 0.2731 (5) | 0.12998 (15) | 0.0528 (13) | |
| H20A | 0.3255 | 0.3696 | 0.1259 | 0.063* | |
| H20B | 0.3571 | 0.2497 | 0.1656 | 0.063* | |
| O21 | 0.4946 (5) | 0.2159 (3) | 0.09772 (13) | 0.0672 (10) | |
| C22 | 0.3494 (7) | 0.1203 (5) | 0.07878 (17) | 0.0539 (12) | |
| H22A | 0.3701 | 0.0303 | 0.0920 | 0.065* | |
| H22B | 0.3400 | 0.1190 | 0.0419 | 0.065* | |
| C23 | 0.0478 (7) | 0.2774 (5) | 0.06772 (15) | 0.0472 (12) | |
| H23A | −0.0318 | 0.2179 | 0.0460 | 0.057* | |
| H23B | −0.0486 | 0.3322 | 0.0869 | 0.057* | |
| C24 | 0.1729 (8) | 0.3629 (5) | 0.03667 (16) | 0.0484 (12) | |
| N25 | 0.2739 (7) | 0.4295 (4) | 0.01284 (15) | 0.0658 (12) | |
| O1L | 0.0901 (13) | 0.1801 (9) | 0.5242 (3) | 0.077 (2) | 0.48 |
| H1L1 | 0.1783 | 0.2403 | 0.5036 | 0.115* | 0.48 |
| H1L2 | 0.1406 | 0.1662 | 0.5525 | 0.115* | 0.48 |
| O2L | 0.1719 (14) | 0.2748 (9) | 0.5074 (2) | 0.085 (2) | 0.52 |
| H2L1 | 0.0459 | 0.2619 | 0.5042 | 0.128* | 0.52 |
| H2L2 | 0.1751 | 0.3371 | 0.5283 | 0.128* | 0.52 |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| N1 | 0.062 (3) | 0.052 (3) | 0.0307 (18) | −0.009 (2) | −0.0069 (18) | 0.0009 (18) |
| C2 | 0.048 (3) | 0.051 (3) | 0.047 (3) | 0.001 (2) | 0.002 (2) | −0.001 (2) |
| C3 | 0.049 (3) | 0.044 (3) | 0.038 (2) | −0.002 (2) | −0.001 (2) | 0.002 (2) |
| C4 | 0.049 (3) | 0.041 (3) | 0.037 (2) | −0.006 (2) | −0.002 (2) | 0.0023 (19) |
| C5 | 0.053 (3) | 0.033 (2) | 0.032 (2) | −0.003 (2) | 0.0015 (19) | −0.0006 (19) |
| N6 | 0.046 (2) | 0.052 (2) | 0.0373 (19) | 0.0060 (19) | 0.0052 (17) | −0.0011 (17) |
| C7 | 0.056 (3) | 0.050 (3) | 0.037 (2) | 0.002 (2) | 0.005 (2) | −0.000 (2) |
| N8 | 0.059 (3) | 0.048 (2) | 0.0340 (18) | −0.004 (2) | 0.0029 (19) | −0.0006 (16) |
| C9 | 0.052 (3) | 0.039 (3) | 0.032 (2) | −0.004 (2) | 0.001 (2) | 0.0042 (19) |
| N10 | 0.041 (2) | 0.047 (2) | 0.0304 (17) | 0.0066 (18) | 0.0005 (15) | −0.0002 (16) |
| C11 | 0.046 (3) | 0.051 (3) | 0.038 (2) | 0.006 (2) | 0.003 (2) | 0.001 (2) |
| C12 | 0.044 (3) | 0.043 (3) | 0.037 (2) | 0.005 (2) | −0.003 (2) | 0.001 (2) |
| C13 | 0.048 (3) | 0.047 (3) | 0.031 (2) | 0.008 (2) | −0.0015 (19) | −0.001 (2) |
| N14 | 0.050 (2) | 0.037 (2) | 0.0333 (17) | 0.0038 (19) | −0.0005 (16) | −0.0040 (16) |
| C15 | 0.055 (3) | 0.038 (3) | 0.039 (2) | −0.000 (2) | −0.003 (2) | −0.005 (2) |
| C16 | 0.059 (3) | 0.042 (3) | 0.041 (2) | −0.001 (2) | −0.002 (2) | 0.002 (2) |
| C17 | 0.046 (3) | 0.042 (3) | 0.040 (2) | 0.002 (2) | −0.001 (2) | 0.001 (2) |
| C18 | 0.053 (3) | 0.055 (3) | 0.044 (2) | 0.001 (3) | −0.007 (2) | 0.006 (2) |
| C19 | 0.044 (3) | 0.047 (3) | 0.035 (2) | −0.005 (2) | −0.002 (2) | 0.003 (2) |
| C20 | 0.050 (3) | 0.061 (3) | 0.048 (3) | −0.004 (3) | −0.000 (2) | −0.002 (2) |
| O21 | 0.045 (2) | 0.081 (3) | 0.076 (2) | −0.002 (2) | 0.0006 (18) | −0.005 (2) |
| C22 | 0.054 (3) | 0.055 (3) | 0.052 (3) | 0.010 (3) | 0.009 (2) | 0.002 (2) |
| C23 | 0.051 (3) | 0.051 (3) | 0.040 (2) | −0.001 (2) | −0.001 (2) | 0.012 (2) |
| C24 | 0.060 (3) | 0.048 (3) | 0.037 (2) | 0.013 (3) | 0.002 (2) | −0.004 (2) |
| N25 | 0.084 (3) | 0.059 (3) | 0.054 (2) | −0.003 (2) | 0.014 (2) | 0.006 (2) |
| O1L | 0.113 (7) | 0.072 (6) | 0.045 (4) | 0.005 (5) | −0.013 (4) | 0.001 (4) |
| O2L | 0.130 (7) | 0.078 (6) | 0.049 (4) | 0.010 (5) | −0.020 (5) | −0.024 (4) |
Geometric parameters (Å, º)
| N1—C9 | 1.351 (5) | C15—H15A | 0.9900 |
| N1—C2 | 1.381 (5) | C15—H15B | 0.9900 |
| N1—H1 | 0.8800 | C16—C17 | 1.524 (6) |
| C2—C3 | 1.356 (6) | C16—H16A | 0.9900 |
| C2—H2 | 0.9500 | C16—H16B | 0.9900 |
| C3—C4 | 1.452 (6) | C17—C18 | 1.519 (6) |
| C3—H3 | 0.9500 | C17—H17 | 1.0000 |
| C4—C9 | 1.378 (5) | C18—H18A | 0.9800 |
| C4—C5 | 1.410 (6) | C18—H18B | 0.9800 |
| C5—N10 | 1.352 (5) | C18—H18C | 0.9800 |
| C5—N6 | 1.359 (5) | C19—C20 | 1.523 (6) |
| N6—C7 | 1.323 (5) | C19—C23 | 1.534 (6) |
| C7—N8 | 1.312 (5) | C19—C22 | 1.538 (6) |
| C7—H7 | 0.9500 | C20—O21 | 1.458 (5) |
| N8—C9 | 1.369 (6) | C20—H20A | 0.9900 |
| N10—C12 | 1.467 (5) | C20—H20B | 0.9900 |
| N10—C11 | 1.476 (5) | O21—C22 | 1.456 (6) |
| C11—H11A | 0.9800 | C22—H22A | 0.9900 |
| C11—H11B | 0.9800 | C22—H22B | 0.9900 |
| C11—H11C | 0.9800 | C23—C24 | 1.459 (7) |
| C12—C13 | 1.521 (6) | C23—H23A | 0.9900 |
| C12—C17 | 1.538 (6) | C23—H23B | 0.9900 |
| C12—H12 | 1.0000 | C24—N25 | 1.145 (6) |
| C13—N14 | 1.463 (5) | O1L—H1L1 | 1.0100 |
| C13—H13A | 0.9900 | O1L—H1L2 | 0.8390 |
| C13—H13B | 0.9900 | O1L—H2L1 | 1.0319 |
| N14—C19 | 1.445 (5) | O2L—H1L1 | 0.3669 |
| N14—C15 | 1.458 (5) | O2L—H2L1 | 0.8478 |
| C15—C16 | 1.520 (6) | O2L—H2L2 | 0.8441 |
| C9—N1—C2 | 108.3 (3) | H15A—C15—H15B | 108.3 |
| C9—N1—H1 | 125.8 | C15—C16—C17 | 112.0 (4) |
| C2—N1—H1 | 125.8 | C15—C16—H16A | 109.2 |
| C3—C2—N1 | 109.2 (4) | C17—C16—H16A | 109.2 |
| C3—C2—H2 | 125.4 | C15—C16—H16B | 109.2 |
| N1—C2—H2 | 125.4 | C17—C16—H16B | 109.2 |
| C2—C3—C4 | 107.1 (4) | H16A—C16—H16B | 107.9 |
| C2—C3—H3 | 126.4 | C18—C17—C16 | 111.0 (4) |
| C4—C3—H3 | 126.4 | C18—C17—C12 | 112.2 (3) |
| C9—C4—C5 | 115.8 (4) | C16—C17—C12 | 112.6 (4) |
| C9—C4—C3 | 105.3 (4) | C18—C17—H17 | 106.9 |
| C5—C4—C3 | 138.7 (4) | C16—C17—H17 | 106.9 |
| N10—C5—N6 | 116.0 (4) | C12—C17—H17 | 106.9 |
| N10—C5—C4 | 125.3 (4) | C17—C18—H18A | 109.5 |
| N6—C5—C4 | 118.6 (4) | C17—C18—H18B | 109.5 |
| C7—N6—C5 | 118.1 (4) | H18A—C18—H18B | 109.5 |
| N8—C7—N6 | 130.0 (4) | C17—C18—H18C | 109.5 |
| N8—C7—H7 | 115.0 | H18A—C18—H18C | 109.5 |
| N6—C7—H7 | 115.0 | H18B—C18—H18C | 109.5 |
| C7—N8—C9 | 110.8 (4) | N14—C19—C20 | 113.7 (3) |
| N1—C9—N8 | 123.3 (4) | N14—C19—C23 | 114.2 (4) |
| N1—C9—C4 | 110.1 (4) | C20—C19—C23 | 113.3 (4) |
| N8—C9—C4 | 126.5 (4) | N14—C19—C22 | 113.6 (4) |
| C5—N10—C12 | 121.8 (3) | C20—C19—C22 | 85.2 (3) |
| C5—N10—C11 | 118.8 (3) | C23—C19—C22 | 113.6 (3) |
| C12—N10—C11 | 119.4 (3) | O21—C20—C19 | 91.4 (3) |
| N10—C11—H11A | 109.5 | O21—C20—H20A | 113.4 |
| N10—C11—H11B | 109.5 | C19—C20—H20A | 113.4 |
| H11A—C11—H11B | 109.5 | O21—C20—H20B | 113.4 |
| N10—C11—H11C | 109.5 | C19—C20—H20B | 113.4 |
| H11A—C11—H11C | 109.5 | H20A—C20—H20B | 110.7 |
| H11B—C11—H11C | 109.5 | C22—O21—C20 | 90.6 (3) |
| N10—C12—C13 | 114.1 (3) | O21—C22—C19 | 90.9 (3) |
| N10—C12—C17 | 114.3 (3) | O21—C22—H22A | 113.5 |
| C13—C12—C17 | 110.9 (3) | C19—C22—H22A | 113.5 |
| N10—C12—H12 | 105.5 | O21—C22—H22B | 113.5 |
| C13—C12—H12 | 105.5 | C19—C22—H22B | 113.5 |
| C17—C12—H12 | 105.5 | H22A—C22—H22B | 110.8 |
| N14—C13—C12 | 112.9 (4) | C24—C23—C19 | 112.2 (4) |
| N14—C13—H13A | 109.0 | C24—C23—H23A | 109.2 |
| C12—C13—H13A | 109.0 | C19—C23—H23A | 109.2 |
| N14—C13—H13B | 109.0 | C24—C23—H23B | 109.2 |
| C12—C13—H13B | 109.0 | C19—C23—H23B | 109.2 |
| H13A—C13—H13B | 107.8 | H23A—C23—H23B | 107.9 |
| C19—N14—C15 | 114.1 (3) | N25—C24—C23 | 178.8 (5) |
| C19—N14—C13 | 113.0 (3) | H1L1—O1L—H1L2 | 111.5 |
| C15—N14—C13 | 109.8 (3) | H1L1—O1L—H2L1 | 52.4 |
| N14—C15—C16 | 108.8 (3) | H1L2—O1L—H2L1 | 135.9 |
| N14—C15—H15A | 109.9 | H1L1—O2L—H2L1 | 86.4 |
| C16—C15—H15A | 109.9 | H1L1—O2L—H2L2 | 153.9 |
| N14—C15—H15B | 109.9 | H2L1—O2L—H2L2 | 102.0 |
| C16—C15—H15B | 109.9 | ||
| C9—N1—C2—C3 | −0.2 (5) | C12—C13—N14—C19 | −169.0 (3) |
| N1—C2—C3—C4 | −0.2 (5) | C12—C13—N14—C15 | 62.3 (4) |
| C2—C3—C4—C9 | 0.5 (5) | C19—N14—C15—C16 | 167.6 (4) |
| C2—C3—C4—C5 | 174.8 (5) | C13—N14—C15—C16 | −64.4 (4) |
| C9—C4—C5—N10 | 174.1 (4) | N14—C15—C16—C17 | 58.3 (5) |
| C3—C4—C5—N10 | 0.2 (8) | C15—C16—C17—C18 | −175.4 (4) |
| C9—C4—C5—N6 | −3.7 (6) | C15—C16—C17—C12 | −48.6 (5) |
| C3—C4—C5—N6 | −177.5 (5) | N10—C12—C17—C18 | 39.5 (5) |
| N10—C5—N6—C7 | −175.4 (4) | C13—C12—C17—C18 | 170.1 (4) |
| C4—C5—N6—C7 | 2.6 (6) | N10—C12—C17—C16 | −86.7 (4) |
| C5—N6—C7—N8 | −0.7 (7) | C13—C12—C17—C16 | 44.0 (5) |
| N6—C7—N8—C9 | 0.1 (7) | C15—N14—C19—C20 | −166.0 (4) |
| C2—N1—C9—N8 | −179.3 (4) | C13—N14—C19—C20 | 67.7 (5) |
| C2—N1—C9—C4 | 0.5 (5) | C15—N14—C19—C23 | 61.8 (5) |
| C7—N8—C9—N1 | 178.2 (4) | C13—N14—C19—C23 | −64.6 (5) |
| C7—N8—C9—C4 | −1.5 (6) | C15—N14—C19—C22 | −70.7 (5) |
| C5—C4—C9—N1 | −176.4 (4) | C13—N14—C19—C22 | 162.9 (3) |
| C3—C4—C9—N1 | −0.7 (5) | N14—C19—C20—O21 | 123.8 (4) |
| C5—C4—C9—N8 | 3.3 (6) | C23—C19—C20—O21 | −103.5 (4) |
| C3—C4—C9—N8 | 179.1 (4) | C22—C19—C20—O21 | 10.2 (3) |
| N6—C5—N10—C12 | −1.8 (6) | C19—C20—O21—C22 | −10.7 (3) |
| C4—C5—N10—C12 | −179.6 (4) | C20—O21—C22—C19 | 10.6 (3) |
| N6—C5—N10—C11 | 178.5 (4) | N14—C19—C22—O21 | −124.0 (4) |
| C4—C5—N10—C11 | 0.6 (6) | C20—C19—C22—O21 | −10.2 (3) |
| C5—N10—C12—C13 | 125.6 (4) | C23—C19—C22—O21 | 103.2 (4) |
| C11—N10—C12—C13 | −54.6 (5) | N14—C19—C23—C24 | 176.8 (4) |
| C5—N10—C12—C17 | −105.3 (4) | C20—C19—C23—C24 | 44.4 (5) |
| C11—N10—C12—C17 | 74.5 (5) | C22—C19—C23—C24 | −50.7 (5) |
| N10—C12—C13—N14 | 79.8 (4) | C19—C23—C24—N25 | −12 (26) |
| C17—C12—C13—N14 | −51.0 (5) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1···O1L | 0.88 | 1.90 | 2.783 (8) | 178 |
| N1—H1···O2L | 0.88 | 2.06 | 2.816 (7) | 144 |
| O1L—H1L2···N8i | 0.84 | 2.27 | 2.868 (8) | 129 |
| O2L—H2L2···N8i | 0.84 | 2.20 | 2.733 (7) | 121 |
| O2L—H2L2···N25ii | 0.84 | 2.43 | 3.026 (10) | 129 |
Symmetry codes: (i) x+1/2, −y+1/2, −z+1; (ii) −x+1/2, −y+1, z+1/2.
Footnotes
Supporting information for this paper is available from the IUCr electronic archives (Reference: BT6965).
References
- Altomare, A., Burla, M. C., Camalli, M., Cascarano, G. L., Giacovazzo, C., Guagliardi, A., Moliterni, A. G. G., Polidori, G. & Spagna, R. (1999). J. Appl. Cryst. 32, 115–119.
- Flanagan, M. E., Blumenkopf, T. A., Brissette, W. H., Brown, M. F., Casavant, J. M., Shang-Poa, C., Doty, J. L., Elliott, E. A., Fisher, M. B., Hines, M., Kent, C., Kudlacz, E. M., Lillie, B. M., Magnuson, K. S., McCurdy, S. P., Munchhof, M. J., Perry, B. D., Sawyer, P. S., Strelevitz, T. J., Subramanyam, C., Sun, J., Whipple, D. A. & Changelian, P. S. (2010). J. Med. Chem. 53, 8468–8484. [DOI] [PubMed]
- Gehringer, M., Pfaffenrot, E., Bauer, S. & Laufer, S. A. (2014). ChemMedChem, 9, 277–281. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Shuai, K. & Liu, B. (2003). Nat. Rev. Immunol. 3, 900–911. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Stoe & Cie (2010). X-RED and X-AREA Stoe & Cie, Darmstadt, Germany.
- Wuitschik, G., Carreira, E. M., Wagner, B., Fischer, H., Parrilla, I., Schuler, F., Rogers-Evans, M. & Mueller, K. J. (2010). J. Med. Chem. 53, 3227–3246. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, Global. DOI: 10.1107/S1600536814004449/bt6965sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536814004449/bt6965Isup2.hkl
Supporting information file. DOI: 10.1107/S1600536814004449/bt6965Isup3.cml
CCDC reference: 988870
Additional supporting information: crystallographic information; 3D view; checkCIF report


