Abstract
In the cation of the title molecular salt, C5H6N3O2 +·C2HO4 −, the dihedral angle between the aromatic ring and the nitro group is 3.5 (3)°; in the anion, the dihedral angle between the CO2 and CO2H planes is 10.5 (2)°. In the crystal, the anions are linked into [100] chains by O—H⋯O hydrogen bonds. The cations cross-link the chains by way of N—H⋯O hydrogen bonds and the structure is consolidated by C—H⋯O interactions.
Related literature
For the crystal structures of related pyridine derivatives, see: Babu et al. (2014 ▶); Anderson et al. (2005 ▶); Karle et al. (2003 ▶). For simple organic–inorganic salts containing strong intermolecular hydrogen bonds, see: Fu et al. (2011 ▶); Sethuram et al. (2013a
▶,b
▶); Shihabuddeen Syed et al. (2013 ▶); Showrilu et al. (2013 ▶); Huq et al. (2013 ▶). For the structure of oxalic acid, see: Derissen & Smith (1974 ▶). For graph-set analysis, see: Bernstein et al.(1995 ▶). 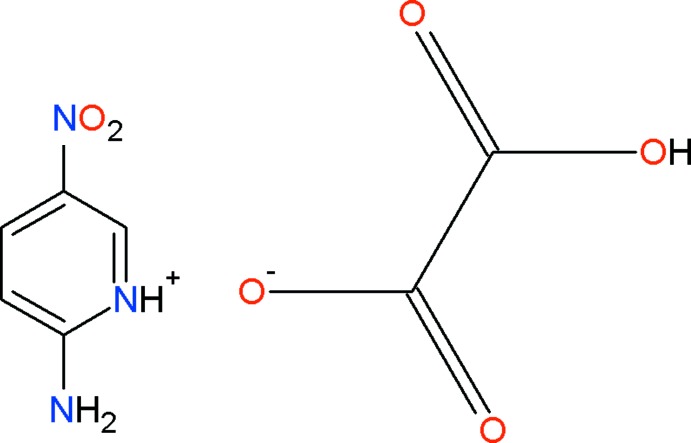
Experimental
Crystal data
C5H6N3O2 +·C2HO4 −
M r = 229.16
Triclinic,
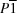
a = 5.5609 (2) Å
b = 9.2012 (4) Å
c = 9.2305 (4) Å
α = 90.245 (2)°
β = 98.500 (2)°
γ = 100.038 (2)°
V = 459.74 (3) Å3
Z = 2
Mo Kα radiation
μ = 0.15 mm−1
T = 293 K
0.35 × 0.30 × 0.30 mm
Data collection
Bruker Kappa APEXII CCD diffractometer
Absorption correction: multi-scan (SADABS; Sheldrick, 2004 ▶) T min = 0.950, T max = 0.957
10142 measured reflections
1615 independent reflections
1417 reflections with I > 2σ(I)
R int = 0.020
Refinement
R[F 2 > 2σ(F 2)] = 0.039
wR(F 2) = 0.110
S = 1.07
1615 reflections
153 parameters
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.28 e Å−3
Δρmin = −0.32 e Å−3
Data collection: APEX2 (Bruker, 2004 ▶); cell refinement: APEX2 and SAINT (Bruker, 2004 ▶); data reduction: SAINT and XPREP (Bruker, 2004 ▶); program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 for Windows (Farrugia, 2012 ▶) and Mercury (Macrae et al., 2008 ▶); software used to prepare material for publication: WinGX (Farrugia, 2012 ▶) and PLATON (Spek, 2009 ▶).
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S160053681400525X/jj2183sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S160053681400525X/jj2183Isup2.hkl
Supporting information file. DOI: 10.1107/S160053681400525X/jj2183Isup3.cml
CCDC reference: 990527
Additional supporting information: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C2—H2⋯O5i | 0.93 | 2.48 | 3.323 (2) | 152 |
| C3—H3⋯O2ii | 0.93 | 2.37 | 3.296 (2) | 178 |
| C5—H5⋯O1iii | 0.93 | 2.42 | 3.186 (2) | 140 |
| C5—H5⋯O4iv | 0.93 | 2.44 | 2.970 (2) | 116 |
| N1—H1⋯O6iv | 0.86 | 1.94 | 2.7697 (18) | 160 |
| O4—H4⋯O5v | 0.82 | 1.64 | 2.4486 (16) | 170 |
| N2—H2A⋯O3v | 0.89 (3) | 2.16 (3) | 2.959 (2) | 149 (2) |
| N2—H2A⋯O5v | 0.89 (3) | 2.36 (3) | 3.007 (2) | 130 (2) |
| N2—H2B⋯O3vi | 0.89 (3) | 1.99 (3) | 2.870 (2) | 173 (2) |
Symmetry codes: (i) 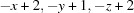 ; (ii)
; (ii) 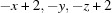 ; (iii)
; (iii) 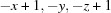 ; (iv)
; (iv) 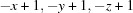 ; (v)
; (v) 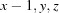 ; (vi)
; (vi) 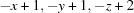 .
.
Acknowledgments
MAR, DPA and SJX would like to thank the Board of Research in Nuclear Sciences, Department of Atomic Energy (BRNS-DAE) (file No. 2012/34/63/BRNS/2865; dated 01/03/2013), for funding this major research project.
supplementary crystallographic information
1. Comment
Simple organic–inorganic salts containing strong intermolecular hydrogen bonds have attracted an attention as materials which display ferroelectric-paraelectric phase transitions (Fu et al., 2011; Sethuram, et al., 2013a,b; Huq, et al., 2013; Shihabuddeen Syed, et al., 2013; Showrilu, et al., 2013). As part of our ongoing investigations of pyridine derivatives (Babu et al., 2014), the title compound was synthesized and we report herein on its crystal structure.
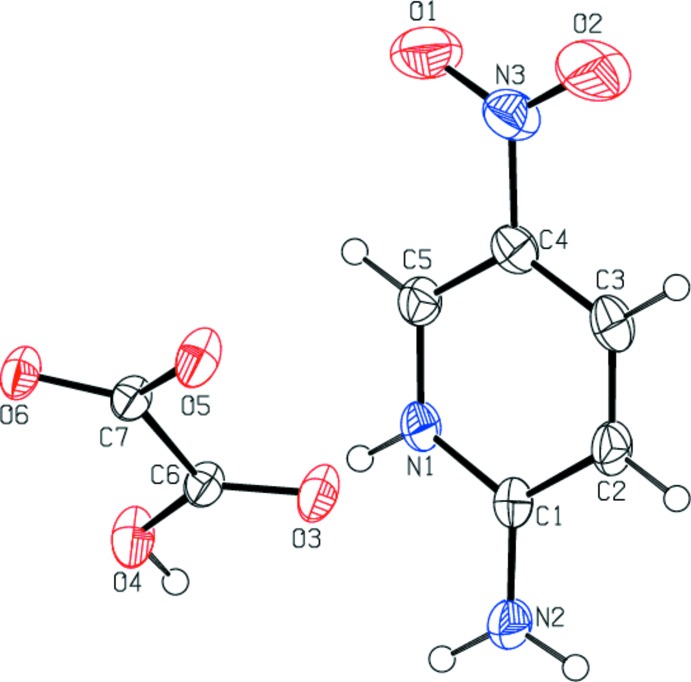
In the title salt, (C5H6N3O2)+, (C2HO4)-, the asymmetric unit consists of an independent 2-amino-5-nitropyridinium cation, and oxalic actetate anion, which lie on an inversion symmetry. A proton transfer from the carboxyl group of oxalic acid to atom N1 of 2-amino-5-nitro pyridinine resulted in the formation of a salt. This protonation lead to the widening of the C5-N1-C1 angle of the pyridine ring to 122.79 (14)°, compared to 115.25 (13)° in the unprotonated aminopyridine (Anderson et al., 2005). This type of protonation is observed in various aminopyridine acid complexes (Babu et al., 2014; Karle et al., 2003).
The bond lengths and bond angles of the aminopyridine are comparable to the values reported earlier for aminopyridine (Babu et al., 2014; Anderson et al., 2005). The bond lengths and bond angles of the oxalate are comparable to the values reported for oxalic acid (Derissen & Smith, 1974). The non hydrogen pyridine ring, C1/C2/C3/C4/C5/N1, is planar with a maximum deviation of 0.006 (1)Å from the least squares plane for the C3 atom, with the endocyclic angles covering range of 117.88 (16) - 122.79 (14)°. The hydrogen oxalate anion O3/O4/O5/O6/C6/C7, is less planar with a maximum deviation of -0.131 (1)Å for the O3 and O6 atoms.
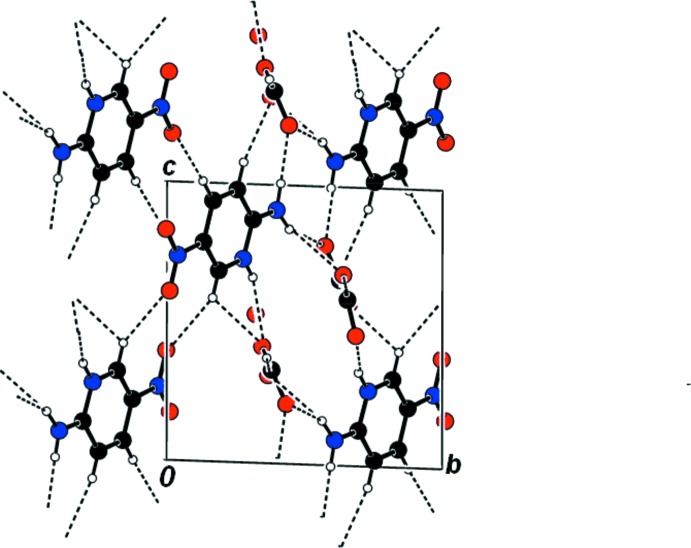
The crystal packing is consolidated by intermolecular N—H···O and O—H···O hydrogen bonds and weak C—H···O intermolecular interactions (Table 1 and Fig. 2). In the crystal structure, the 2-Amino-5-nitropyridinium unit is bound to acetate anions by five distinct N—H···O hydrogen bonds. The ion pairs are joined by two N—H···O hydrogen bonds in which the N atom of the 2-amino-5-nitroPyridinium unit acts as a bifurcated donor, thus generating R12(5) ring motifs (Bernstein et al., 1995). The hydroxyl group hydrogen atom is also hydrogen-bonded to the carboxylate oxygen atom through strong intermolecular O—H···O hydrogen bonds, with the O···O distance of 2.4486 (16)Å, which is from a chain, C(5), running along the b axis (Bernstein et al., 1995). The structure is further stabilized by weak C—H···O intermolecular inteactions, forming a three-dimensional network.
2. Experimental
Crystals of the title compound were obtained by slow evaporation of a 1:1 mol. mixture of 2-amino-5-nitropyridine and oxalic acid in methanol at room temperature.
3. Refinement
The amino group NH2 H atoms of the pyridine derivatives were located in difference Fourier maps and refined in the riding mode approximation. The OH, NH(Protonated) and C-bound H-atoms were placed in calculated positions and treated as riding atoms: O-H = 0.82Å, N-H = 0.86Å, C-H = 0.93Å with Uiso(H) = 1.5Ueq(O) and = 1.2Ueq(N,C) for other H atoms.
Figures
Fig. 1.
View of the title compound showing the atom-numbering scheme. Displacement ellipsoids are drawn at the 40% probability level. H atoms are presented as a small spheres of arbitrary radius.
Fig. 2.
The crystal packing of the title compound viewed along a axis. N—H···O, O—H···O hydrogen bonds and weak C—H···O intermolecular inteeractions are shown as dashed lines, forming a three-dimensional network. H atoms not involved in hydrogen bonding have been omitted for clarity.
Crystal data
| C5H6N3O2+·C2HO4− | Z = 2 |
| Mr = 229.16 | F(000) = 236 |
| Triclinic, P1 | Dx = 1.655 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation, λ = 0.71073 Å |
| a = 5.5609 (2) Å | Cell parameters from 2794 reflections |
| b = 9.2012 (4) Å | θ = 2.4–31.1° |
| c = 9.2305 (4) Å | µ = 0.15 mm−1 |
| α = 90.245 (2)° | T = 293 K |
| β = 98.500 (2)° | Block, colourless |
| γ = 100.038 (2)° | 0.35 × 0.30 × 0.30 mm |
| V = 459.74 (3) Å3 |
Data collection
| Bruker Kappa APEXII CCD diffractometer | 1615 independent reflections |
| Radiation source: fine-focus sealed tube | 1417 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.020 |
| Detector resolution: 0.1000 pixels mm-1 | θmax = 25.0°, θmin = 2.2° |
| ω and φ scans | h = −6→6 |
| Absorption correction: multi-scan (SADABS; Sheldrick, 2004) | k = −10→10 |
| Tmin = 0.950, Tmax = 0.957 | l = −10→10 |
| 10142 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.039 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.110 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.07 | w = 1/[σ2(Fo2) + (0.0559P)2 + 0.2329P] where P = (Fo2 + 2Fc2)/3 |
| 1615 reflections | (Δ/σ)max < 0.001 |
| 153 parameters | Δρmax = 0.28 e Å−3 |
| 0 restraints | Δρmin = −0.32 e Å−3 |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > 2sigma(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | 0.3486 (3) | 0.30577 (19) | 0.87248 (18) | 0.0302 (4) | |
| C2 | 0.5136 (3) | 0.2468 (2) | 0.97783 (19) | 0.0361 (4) | |
| H2 | 0.5206 | 0.2695 | 1.0768 | 0.043* | |
| C3 | 0.6606 (3) | 0.1580 (2) | 0.9356 (2) | 0.0380 (4) | |
| H3 | 0.7711 | 0.1201 | 1.0046 | 0.046* | |
| C4 | 0.6446 (3) | 0.12349 (19) | 0.78639 (19) | 0.0338 (4) | |
| C5 | 0.4879 (3) | 0.18053 (19) | 0.68547 (19) | 0.0332 (4) | |
| H5 | 0.4796 | 0.1582 | 0.5863 | 0.040* | |
| C6 | 0.8206 (3) | 0.61691 (18) | 0.66445 (17) | 0.0270 (4) | |
| C7 | 1.0607 (3) | 0.64305 (19) | 0.59536 (17) | 0.0269 (4) | |
| N1 | 0.3441 (3) | 0.26987 (16) | 0.72996 (15) | 0.0323 (4) | |
| H1 | 0.2451 | 0.3057 | 0.6649 | 0.039* | |
| N2 | 0.2009 (3) | 0.39211 (19) | 0.90933 (19) | 0.0408 (4) | |
| N3 | 0.7996 (3) | 0.02858 (19) | 0.73614 (19) | 0.0450 (4) | |
| O1 | 0.7803 (3) | −0.0001 (2) | 0.60645 (19) | 0.0674 (5) | |
| O2 | 0.9490 (4) | −0.0148 (3) | 0.8270 (2) | 0.0882 (7) | |
| O3 | 0.8197 (2) | 0.56345 (16) | 0.78510 (13) | 0.0410 (4) | |
| O4 | 0.6388 (2) | 0.65291 (16) | 0.58166 (14) | 0.0409 (4) | |
| H4 | 0.5159 | 0.6372 | 0.6226 | 0.061* | |
| O5 | 1.2521 (2) | 0.62615 (16) | 0.68401 (13) | 0.0395 (4) | |
| O6 | 1.0537 (2) | 0.67353 (14) | 0.46624 (12) | 0.0342 (3) | |
| H2A | 0.114 (5) | 0.439 (3) | 0.842 (3) | 0.060 (7)* | |
| H2B | 0.208 (4) | 0.407 (2) | 1.005 (3) | 0.051 (6)* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.0316 (9) | 0.0336 (9) | 0.0242 (8) | 0.0042 (7) | 0.0024 (7) | 0.0058 (7) |
| C2 | 0.0424 (10) | 0.0428 (10) | 0.0217 (8) | 0.0085 (8) | −0.0013 (7) | 0.0051 (7) |
| C3 | 0.0379 (10) | 0.0426 (10) | 0.0313 (9) | 0.0111 (8) | −0.0070 (7) | 0.0093 (8) |
| C4 | 0.0324 (9) | 0.0333 (9) | 0.0351 (10) | 0.0082 (7) | 0.0002 (7) | 0.0039 (7) |
| C5 | 0.0387 (10) | 0.0347 (9) | 0.0253 (9) | 0.0074 (7) | 0.0010 (7) | 0.0015 (7) |
| C6 | 0.0212 (8) | 0.0392 (9) | 0.0216 (8) | 0.0085 (7) | 0.0027 (6) | 0.0034 (6) |
| C7 | 0.0205 (8) | 0.0390 (9) | 0.0221 (8) | 0.0082 (6) | 0.0022 (6) | 0.0029 (6) |
| N1 | 0.0344 (8) | 0.0391 (8) | 0.0231 (7) | 0.0118 (6) | −0.0027 (6) | 0.0064 (6) |
| N2 | 0.0464 (10) | 0.0537 (10) | 0.0275 (9) | 0.0224 (8) | 0.0064 (7) | 0.0078 (7) |
| N3 | 0.0458 (10) | 0.0423 (9) | 0.0481 (11) | 0.0169 (8) | −0.0003 (8) | 0.0007 (7) |
| O1 | 0.0832 (12) | 0.0683 (11) | 0.0567 (10) | 0.0355 (9) | 0.0044 (9) | −0.0148 (8) |
| O2 | 0.0942 (14) | 0.1202 (17) | 0.0668 (12) | 0.0807 (14) | −0.0069 (10) | 0.0069 (11) |
| O3 | 0.0313 (7) | 0.0720 (9) | 0.0249 (7) | 0.0190 (6) | 0.0090 (5) | 0.0143 (6) |
| O4 | 0.0193 (6) | 0.0712 (9) | 0.0359 (7) | 0.0150 (6) | 0.0074 (5) | 0.0221 (6) |
| O5 | 0.0192 (6) | 0.0751 (10) | 0.0257 (6) | 0.0139 (6) | 0.0014 (5) | 0.0102 (6) |
| O6 | 0.0257 (6) | 0.0560 (8) | 0.0234 (6) | 0.0122 (5) | 0.0054 (5) | 0.0099 (5) |
Geometric parameters (Å, º)
| C1—N2 | 1.315 (2) | C6—O3 | 1.220 (2) |
| C1—N1 | 1.351 (2) | C6—O4 | 1.268 (2) |
| C1—C2 | 1.413 (2) | C6—C7 | 1.545 (2) |
| C2—C3 | 1.347 (3) | C7—O6 | 1.222 (2) |
| C2—H2 | 0.9300 | C7—O5 | 1.2759 (19) |
| C3—C4 | 1.398 (3) | N1—H1 | 0.8600 |
| C3—H3 | 0.9300 | N2—H2A | 0.89 (3) |
| C4—C5 | 1.352 (2) | N2—H2B | 0.89 (3) |
| C4—N3 | 1.448 (2) | N3—O1 | 1.210 (2) |
| C5—N1 | 1.344 (2) | N3—O2 | 1.211 (2) |
| C5—H5 | 0.9300 | O4—H4 | 0.8200 |
| N2—C1—N1 | 119.96 (16) | O3—C6—C7 | 120.04 (14) |
| N2—C1—C2 | 122.16 (16) | O4—C6—C7 | 112.84 (13) |
| N1—C1—C2 | 117.88 (16) | O6—C7—O5 | 126.27 (14) |
| C3—C2—C1 | 120.31 (16) | O6—C7—C6 | 120.06 (14) |
| C3—C2—H2 | 119.8 | O5—C7—C6 | 113.64 (13) |
| C1—C2—H2 | 119.8 | C5—N1—C1 | 122.79 (14) |
| C2—C3—C4 | 118.97 (16) | C5—N1—H1 | 118.6 |
| C2—C3—H3 | 120.5 | C1—N1—H1 | 118.6 |
| C4—C3—H3 | 120.5 | C1—N2—H2A | 121.5 (16) |
| C5—C4—C3 | 120.75 (17) | C1—N2—H2B | 115.1 (15) |
| C5—C4—N3 | 118.45 (16) | H2A—N2—H2B | 123 (2) |
| C3—C4—N3 | 120.79 (16) | O1—N3—O2 | 123.01 (19) |
| N1—C5—C4 | 119.29 (16) | O1—N3—C4 | 119.28 (16) |
| N1—C5—H5 | 120.4 | O2—N3—C4 | 117.68 (17) |
| C4—C5—H5 | 120.4 | C6—O4—H4 | 109.5 |
| O3—C6—O4 | 127.10 (15) | ||
| N2—C1—C2—C3 | 179.55 (17) | O3—C6—C7—O5 | −10.4 (2) |
| N1—C1—C2—C3 | 0.1 (3) | O4—C6—C7—O5 | 171.21 (16) |
| C1—C2—C3—C4 | −0.9 (3) | C4—C5—N1—C1 | −0.1 (3) |
| C2—C3—C4—C5 | 1.3 (3) | N2—C1—N1—C5 | −179.04 (16) |
| C2—C3—C4—N3 | 179.89 (17) | C2—C1—N1—C5 | 0.4 (3) |
| C3—C4—C5—N1 | −0.8 (3) | C5—C4—N3—O1 | −1.9 (3) |
| N3—C4—C5—N1 | −179.43 (16) | C3—C4—N3—O1 | 179.50 (18) |
| O3—C6—C7—O6 | 168.06 (17) | C5—C4—N3—O2 | 175.9 (2) |
| O4—C6—C7—O6 | −10.3 (2) | C3—C4—N3—O2 | −2.7 (3) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C2—H2···O5i | 0.93 | 2.48 | 3.323 (2) | 152 |
| C3—H3···O2ii | 0.93 | 2.37 | 3.296 (2) | 178 |
| C5—H5···O1iii | 0.93 | 2.42 | 3.186 (2) | 140 |
| C5—H5···O4iv | 0.93 | 2.44 | 2.970 (2) | 116 |
| N1—H1···O4iv | 0.86 | 2.47 | 2.9770 (18) | 119 |
| N1—H1···O6iv | 0.86 | 1.94 | 2.7697 (18) | 160 |
| O4—H4···O5v | 0.82 | 1.64 | 2.4486 (16) | 170 |
| N2—H2A···O3v | 0.89 (3) | 2.16 (3) | 2.959 (2) | 149 (2) |
| N2—H2A···O5v | 0.89 (3) | 2.36 (3) | 3.007 (2) | 130 (2) |
| N2—H2B···O3vi | 0.89 (3) | 1.99 (3) | 2.870 (2) | 173 (2) |
Symmetry codes: (i) −x+2, −y+1, −z+2; (ii) −x+2, −y, −z+2; (iii) −x+1, −y, −z+1; (iv) −x+1, −y+1, −z+1; (v) x−1, y, z; (vi) −x+1, −y+1, −z+2.
Footnotes
Supporting information for this paper is available from the IUCr electronic archives (Reference: JJ2183).
References
- Anderson, F. P., Gallagher, J. F., Kenny, P. T. M. & Lough, A. J. (2005). Acta Cryst. E61, o1350–o1353.
- Babu, K. S. S., Peramaiyan, G., NizamMohideen, M. & Mohan, R. (2014). Acta Cryst. E70, o391–o392. [DOI] [PMC free article] [PubMed]
- Bernstein, J., Davis, R. E., Shimoni, L. & Chang, N.-L. (1995). Angew. Chem. Int. Ed. Engl. 34, 1555–1573.
- Bruker (2004). APEX2, SAINT and XPREP Bruker AXS Inc., Madison, Wisconsin, USA.
- Derissen, J. L. & Smith, P. H. (1974). Acta Cryst. B30, 2240–2242.
- Farrugia, L. J. (2012). J. Appl. Cryst. 45, 849–854.
- Fu, D.-W., Zhang, W., Cai, H.-L., Ge, J.-Z., Zhang, Y. & Xiong, R.-G. (2011). Adv. Mater. 23, 5658–5662. [DOI] [PubMed]
- Huq, C. A. M. A., Fouzia, S. & NizamMohideen, M. (2013). Acta Cryst. E69, o1766–o1767. [DOI] [PMC free article] [PubMed]
- Karle, I., Gilardi, R. D., Chandrashekhar Rao, Ch., Muraleedharan, K. M. & Ranganathan, S. (2003). J. Chem. Crystallogr. 33, 727–749.
- Macrae, C. F., Bruno, I. J., Chisholm, J. A., Edgington, P. R., McCabe, P., Pidcock, E., Rodriguez-Monge, L., Taylor, R., van de Streek, J. & Wood, P. A. (2008). J. Appl. Cryst. 41, 466–470.
- Sethuram, M., Bhargavi, G., Dhandapani, M., Amirthaganesan, G. & NizamMohideen, M. (2013a). Acta Cryst. E69, o1301–o1302. [DOI] [PMC free article] [PubMed]
- Sethuram, M., Rajasekharan, M. V., Dhandapani, M., Amirthaganesan, G. & NizamMohideen, M. (2013b). Acta Cryst. E69, o957–o958. [DOI] [PMC free article] [PubMed]
- Sheldrick, G. M. (2004). SADABS University of Göttingen, Germany.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Shihabuddeen Syed, A., Rajarajan, K. & NizamMohideen, M. (2013). Acta Cryst. E69, i33. [DOI] [PMC free article] [PubMed]
- Showrilu, K., Rajarajan, K. & NizamMohideen, M. (2013). Acta Cryst. E69, m469–m470. [DOI] [PMC free article] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S160053681400525X/jj2183sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S160053681400525X/jj2183Isup2.hkl
Supporting information file. DOI: 10.1107/S160053681400525X/jj2183Isup3.cml
CCDC reference: 990527
Additional supporting information: crystallographic information; 3D view; checkCIF report