Abstract
In the title compound, C17H18N4O5·0.47CH3OH, the virtually planar (r.m.s. deviation = 0.128 Å) carbonohydrazide molecule is located on a twofold axis and conformation of its C=N bonds is E. There are short intramolecular O—H⋯N hydrogen bonds between the hydroxy groups and hydrazide N atoms. In the crystal, bifurcated N—H⋯(O,O) hydrogen bonds assemble the carbonohydrazide molecules into a three-dimensional network. There are C 2 symmetric voids in this network, 47% of which are occupied by disordered methanol molecules.
Related literature
For related structures, see: Du & Zhang (2010 ▶); He et al. (2010 ▶); Kong et al. (2010 ▶). For the biological activity of carbonohydrazides, see: Bacchi et al. (1999 ▶); El-Gammal et al. (2012 ▶).
Experimental
Crystal data
C17H18N4O5·0.47CH4O
M r = 373.40
Orthorhombic,
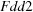
a = 9.4470 (7) Å
b = 17.5850 (9) Å
c = 22.8714 (12) Å
V = 3799.5 (4) Å3
Z = 8
Mo Kα radiation
μ = 0.10 mm−1
T = 293 K
0.1 × 0.08 × 0.05 mm
Data collection
Enraf–Nonius CAD-4 diffractometer
9573 measured reflections
862 independent reflections
658 reflections with I > 2σ(I)
R int = 0.105
2 standard reflections every 120 min intensity decay: 2%
Refinement
R[F 2 > 2σ(F 2)] = 0.044
wR(F 2) = 0.111
S = 1.25
862 reflections
146 parameters
1 restraint
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.17 e Å−3
Δρmin = −0.19 e Å−3
Data collection: CAD-4 EXPRESS (Enraf–Nonius, 1994 ▶); cell refinement: CAD-4 EXPRESS; data reduction: XCAD4 (Harms & Wocadlo, 1995 ▶); program(s) used to solve structure: SHELXS97 (Sheldrick,2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 for Windows (Farrugia, 2012 ▶); software used to prepare material for publication: SHELXL97.
Supplementary Material
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S1600536814004802/gk2603sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536814004802/gk2603Isup2.hkl
Supporting information file. DOI: 10.1107/S1600536814004802/gk2603Isup3.cml
CCDC reference: 989432
Additional supporting information: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O1—H1O⋯N1 | 0.91 (5) | 1.86 (5) | 2.703 (5) | 152 (5) |
| N2—H2N⋯O3i | 0.94 (6) | 2.38 (5) | 3.044 (6) | 128 (4) |
| N2—H2N⋯O1i | 0.94 (6) | 2.33 (6) | 3.204 (6) | 155 (4) |
Symmetry code: (i)  .
.
supplementary crystallographic information
1. Comment
Carbonohydrazide derivatives give rise to a large spectrum of biological properties such as antioxidant (El-Gammal et al., 2012) and anticancer activities (Bacchi et al., 1999). We report here the crystal structure of the title compound synthesized according to literature (He et al., 2010; Du et al., 2010). All parameters are within normal ranges and comparable with the related structures (Kong et al., 2010).
The molecular structure of the title compound is shown in Fig. 1. The complete carbonohydrazide molecule is generated by a twofold crystallographic axis passing throuth the atoms C8 and O2. A three-center O···(N)H···O intermolecular hydrogen bond involving the amido H atoms and the phenoxo and methoxy O atoms is observed (Fig. 2). There are voids in a three dimensional network containing solvent methanol molecules. Only one methanol molecule can be accommodated in a small void that has C2 symmetry. This leads to disorder of methanol molecules. In addition refinement of occupancy factors of methanol O and C atoms converged at 0.234 (1), indicating that 47% of voids are occupied by the solvent.
2. Experimental
In a round bottomed flask, carbonohydrazide (1.0 g, 11.11 mmol) was introduced with methanol (10 ml). o-Vanillin (3.3 g, 22.22 mmol) dissolved in 10 ml of the same solvent was added. Two drops of glacial acetic acid were added while stirring. After one hour under reflux, the precipitate formed that after cooling to room temperature was filtered off and washed with cold methanol. The resulting solid was dried in air. The filtrate was left at room temperature. Slow evaporation of the solvent gave colorless crystals after one day. Yield: 95%; m.p. 378 K. Anal. Calc. for [C17H18N4O5] (%): C, 56.98; H, 5.06, N, 15.63. Found: C, 56.96; H, 5.04; N, 15.60. Selected IR data (cm-1, KBr pellet): 3291, 2942, 1696, 1553, 1200, 1167. 1H-NMR (DMSO-d6) δ: 3.8 (s, 6H, O—CH3); 6.7 – 7.1 (m, 6H, HAromatic); 8.5 (s, 2H, H—C═N); 7.3 (s, 1H, H—N); 11 (s, 2H, H—O) p.p.m. 13C-NMR (DMSO-d6) d: 151.8 (C═O); 147.8, 146.1, 119.5, 119.4, 118.8, 112.8 (CAromatic); 58,7 (–O—CH3).
3. Refinement
H atoms of the NH and OH groups were located in the Fourier difference maps and refined without restraints. Otherg H atoms were geometrically optimized and refined as riding on their carriers with Uiso(H) = 1.2Ueq(C)(1.5 for CH3 groups). Considerable disorder was detected for the solvent methanol molecule. The occupancy factor of the C and O atoms of methanol refined at 0.234 (1). Thus, there are 0.46 methanol molecules per one carbonohydrazide molecule in the crystal. Owing to a negligible anomalous dispersion effect the Friedel pairs were merged and the absolute structure was not determined.
Figures
Fig. 1.

An ORTEP view of the title compound, showing the atom-numbering scheme. Displacement ellipsoids are plotted at the 50% probability level. Only one position of the disordered solvent methanol molecule is shown for clarity. The symmetry code for generating primed atoms is 2-x,-y,z
Fig. 2.
Intramoleculr and intermolecular hydrogen bonds. Solvent methanol molecules are omitted as they do not form hydrogen bonds.
Crystal data
| C17H18N4O5·0.47CH4O | F(000) = 1571.4 |
| Mr = 373.40 | Dx = 1.306 Mg m−3 |
| Orthorhombic, Fdd2 | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: F 2 -2d | Cell parameters from 25 reflections |
| a = 9.4470 (7) Å | θ = 11–15° |
| b = 17.5850 (9) Å | µ = 0.10 mm−1 |
| c = 22.8714 (12) Å | T = 293 K |
| V = 3799.5 (4) Å3 | Prismatic, colorless |
| Z = 8 | 0.1 × 0.08 × 0.05 mm |
Data collection
| Enraf–Nonius CAD-4 diffractometer | Rint = 0.105 |
| Radiation source: fine-focus sealed tube | θmax = 25.0°, θmin = 2.6° |
| Graphite monochromator | h = −11→11 |
| non–profiled ω/2θ scans | k = −1→20 |
| 9573 measured reflections | l = −27→27 |
| 862 independent reflections | 2 standard reflections every 120 min |
| 658 reflections with I > 2σ(I) | intensity decay: 2% |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.044 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.111 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.25 | w = 1/[σ2(Fo2) + (0.0306P)2 + 5.2614P] where P = (Fo2 + 2Fc2)/3 |
| 862 reflections | (Δ/σ)max < 0.001 |
| 146 parameters | Δρmax = 0.17 e Å−3 |
| 1 restraint | Δρmin = −0.19 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | Occ. (<1) | |
| O1 | 0.6805 (4) | 0.1635 (2) | 0.02775 (15) | 0.0559 (10) | |
| O2 | 1.0000 | 0.0000 | 0.0525 (2) | 0.0641 (14) | |
| O3 | 0.4920 (4) | 0.2637 (2) | −0.00424 (17) | 0.0680 (12) | |
| N1 | 0.8123 (4) | 0.0896 (2) | 0.11591 (18) | 0.0471 (10) | |
| N2 | 0.9127 (5) | 0.0436 (2) | 0.1418 (2) | 0.0533 (11) | |
| C1 | 0.6048 (5) | 0.1974 (3) | 0.0728 (2) | 0.0459 (12) | |
| C2 | 0.5016 (5) | 0.2512 (3) | 0.0560 (2) | 0.0512 (13) | |
| C3 | 0.4200 (5) | 0.2864 (3) | 0.0996 (3) | 0.0567 (14) | |
| H3 | 0.3505 | 0.3211 | 0.0888 | 0.068* | |
| C4 | 0.4409 (5) | 0.2704 (3) | 0.1594 (3) | 0.0595 (15) | |
| H4 | 0.3856 | 0.2946 | 0.1874 | 0.071* | |
| C5 | 0.5434 (5) | 0.2189 (3) | 0.1765 (2) | 0.0528 (13) | |
| H5 | 0.5580 | 0.2087 | 0.2159 | 0.063* | |
| C6 | 0.6269 (5) | 0.1813 (3) | 0.1329 (2) | 0.0423 (11) | |
| C7 | 0.7364 (5) | 0.1284 (3) | 0.1531 (2) | 0.0459 (12) | |
| H7 | 0.7519 | 0.1223 | 0.1930 | 0.055* | |
| C8 | 1.0000 | 0.0000 | 0.1068 (3) | 0.0467 (17) | |
| C9 | 0.4052 (7) | 0.3261 (4) | −0.0244 (3) | 0.0779 (19) | |
| H9A | 0.4081 | 0.3283 | −0.0663 | 0.117* | |
| H9B | 0.4405 | 0.3729 | −0.0086 | 0.117* | |
| H9C | 0.3093 | 0.3185 | −0.0118 | 0.117* | |
| O4 | 0.579 (2) | 0.1913 (12) | 0.3182 (9) | 0.098 (9) | 0.234 (11) |
| H1M | 0.5601 | 0.1599 | 0.3453 | 0.148* | 0.234 (11) |
| C10 | 0.703 (3) | 0.2337 (18) | 0.3207 (10) | 0.075 (10) | 0.234 (11) |
| H10A | 0.7221 | 0.2638 | 0.3554 | 0.113* | 0.234 (11) |
| H10B | 0.7221 | 0.2638 | 0.2861 | 0.113* | 0.234 (11) |
| H10C | 0.7632 | 0.1890 | 0.3207 | 0.113* | 0.234 (11) |
| H1O | 0.743 (5) | 0.134 (3) | 0.048 (2) | 0.057 (15)* | |
| H2N | 0.916 (5) | 0.040 (3) | 0.183 (3) | 0.052 (15)* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.062 (2) | 0.055 (2) | 0.051 (2) | 0.0187 (18) | −0.0048 (18) | −0.0067 (17) |
| O2 | 0.077 (4) | 0.069 (3) | 0.047 (3) | 0.019 (3) | 0.000 | 0.000 |
| O3 | 0.076 (3) | 0.063 (2) | 0.066 (3) | 0.026 (2) | −0.0196 (19) | −0.001 (2) |
| N1 | 0.043 (2) | 0.045 (2) | 0.053 (2) | 0.004 (2) | −0.0026 (19) | 0.0038 (19) |
| N2 | 0.054 (3) | 0.059 (2) | 0.047 (3) | 0.020 (2) | −0.001 (2) | 0.004 (2) |
| C1 | 0.038 (3) | 0.044 (2) | 0.056 (3) | 0.000 (2) | −0.002 (2) | −0.005 (2) |
| C2 | 0.046 (3) | 0.042 (2) | 0.065 (3) | 0.004 (2) | −0.010 (3) | 0.001 (3) |
| C3 | 0.039 (3) | 0.049 (3) | 0.082 (4) | 0.006 (2) | −0.001 (3) | −0.005 (3) |
| C4 | 0.047 (3) | 0.052 (3) | 0.080 (4) | 0.002 (3) | 0.017 (3) | −0.008 (3) |
| C5 | 0.046 (3) | 0.053 (3) | 0.059 (3) | −0.002 (2) | 0.014 (2) | 0.002 (3) |
| C6 | 0.039 (3) | 0.035 (2) | 0.052 (3) | −0.001 (2) | 0.004 (2) | −0.002 (2) |
| C7 | 0.047 (3) | 0.044 (3) | 0.046 (3) | −0.001 (2) | 0.000 (2) | 0.006 (2) |
| C8 | 0.047 (4) | 0.040 (4) | 0.053 (5) | 0.003 (3) | 0.000 | 0.000 |
| C9 | 0.082 (4) | 0.058 (3) | 0.094 (5) | 0.015 (3) | −0.025 (4) | 0.014 (3) |
| O4 | 0.117 (19) | 0.087 (15) | 0.091 (17) | 0.004 (12) | 0.030 (13) | 0.010 (12) |
| C10 | 0.07 (2) | 0.10 (3) | 0.057 (16) | 0.021 (19) | −0.001 (11) | −0.018 (15) |
Geometric parameters (Å, º)
| O1—C1 | 1.389 (6) | C4—H4 | 0.9300 |
| O1—H1O | 0.91 (5) | C5—C6 | 1.433 (7) |
| O2—C8 | 1.243 (8) | C5—H5 | 0.9300 |
| O3—C2 | 1.397 (6) | C6—C7 | 1.466 (6) |
| O3—C9 | 1.445 (6) | C7—H7 | 0.9300 |
| N1—C7 | 1.305 (6) | C8—N2i | 1.381 (6) |
| N1—N2 | 1.381 (5) | C9—H9A | 0.9600 |
| N2—C8 | 1.381 (6) | C9—H9B | 0.9600 |
| N2—H2N | 0.94 (6) | C9—H9C | 0.9600 |
| C1—C2 | 1.413 (6) | O4—C10 | 1.39 (3) |
| C1—C6 | 1.418 (7) | O4—H1M | 0.8500 |
| C2—C3 | 1.405 (8) | C10—C10ii | 1.06 (5) |
| C3—C4 | 1.411 (8) | C10—H10A | 0.9700 |
| C3—H3 | 0.9300 | C10—H10B | 0.9700 |
| C4—C5 | 1.382 (7) | C10—H10C | 0.9700 |
| C1—O1—H1O | 102 (3) | N1—C7—C6 | 121.0 (4) |
| C2—O3—C9 | 118.1 (4) | N1—C7—H7 | 119.5 |
| C7—N1—N2 | 113.9 (4) | C6—C7—H7 | 119.5 |
| N1—N2—C8 | 119.1 (5) | O2—C8—N2 | 125.4 (3) |
| N1—N2—H2N | 120 (3) | O2—C8—N2i | 125.4 (3) |
| C8—N2—H2N | 121 (3) | N2—C8—N2i | 109.2 (7) |
| O1—C1—C2 | 116.1 (5) | O3—C9—H9A | 109.5 |
| O1—C1—C6 | 123.9 (4) | O3—C9—H9B | 109.5 |
| C2—C1—C6 | 120.0 (5) | H9A—C9—H9B | 109.5 |
| O3—C2—C3 | 126.5 (5) | O3—C9—H9C | 109.5 |
| O3—C2—C1 | 114.8 (5) | H9A—C9—H9C | 109.5 |
| C3—C2—C1 | 118.7 (5) | H9B—C9—H9C | 109.5 |
| C2—C3—C4 | 121.7 (5) | C10—O4—H1M | 119.9 |
| C2—C3—H3 | 119.2 | C10ii—C10—O4 | 177.6 (17) |
| C4—C3—H3 | 119.2 | C10ii—C10—H10A | 63.1 |
| C5—C4—C3 | 120.2 (5) | O4—C10—H10A | 118.9 |
| C5—C4—H4 | 119.9 | C10ii—C10—H10B | 63.1 |
| C3—C4—H4 | 119.9 | O4—C10—H10B | 114.5 |
| C4—C5—C6 | 119.4 (5) | H10A—C10—H10B | 109.6 |
| C4—C5—H5 | 120.3 | C10ii—C10—H10C | 87.1 |
| C6—C5—H5 | 120.3 | O4—C10—H10C | 93.3 |
| C1—C6—C5 | 120.1 (4) | H10A—C10—H10C | 109.6 |
| C1—C6—C7 | 122.3 (4) | H10B—C10—H10C | 109.6 |
| C5—C6—C7 | 117.5 (4) |
Symmetry codes: (i) −x+2, −y, z; (ii) −x+3/2, −y+1/2, z.
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H1O···N1 | 0.91 (5) | 1.86 (5) | 2.703 (5) | 152 (5) |
| N2—H2N···O3iii | 0.94 (6) | 2.38 (5) | 3.044 (6) | 128 (4) |
| N2—H2N···O1iii | 0.94 (6) | 2.33 (6) | 3.204 (6) | 155 (4) |
Symmetry code: (iii) x+1/4, −y+1/4, z+1/4.
Footnotes
Supporting information for this paper is available from the IUCr electronic archives (Reference: GK2603).
References
- Bacchi, A., Carcelli, M., Pelagatti, P., Pelizzi, C., Pelizzi, G. & Zani, F. (1999). J. Inorg. Biochem. 75, 123–133. [DOI] [PubMed]
- Du, L. & Zhang, W. (2010). Acta Cryst. E66, o2645. [DOI] [PMC free article] [PubMed]
- El-Gammal, O. A., Abu El-Reash, G. M., Ghazy, S. E. & Radwan, A. H. (2012). J. Mol. Struct. 1020, 6–15.
- Enraf–Nonius (1994). CAD-4 EXPRESS Enraf–Nonius, Delft, The Netherlands.
- Farrugia, L. J. (2012). J. Appl. Cryst. 45, 849–854.
- Harms, K. & Wocadlo, S. (1995). XCAD4 University of Marburg, Germany.
- He, Q.-P., Tan, B. & Lu, Z.-H. (2010). Acta Cryst. E66, o2968. [DOI] [PMC free article] [PubMed]
- Kong, L., Qiao, Y., Gao, Z. & Ju, X. (2010). Acta Cryst. E66, o2901. [DOI] [PMC free article] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S1600536814004802/gk2603sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536814004802/gk2603Isup2.hkl
Supporting information file. DOI: 10.1107/S1600536814004802/gk2603Isup3.cml
CCDC reference: 989432
Additional supporting information: crystallographic information; 3D view; checkCIF report



