Summary
Stem cells possess the capacity to generate two cells of distinct fate upon division; one cell retaining stem cell identity and the other cell destined to differentiate. These cell fates are established by cell-type-specific genetic networks. To comprehensively identify components of these networks, we performed a large-scale RNAi screen in Drosophila female germline stem cells (GSCs) covering ~25% of the genome. The screen identified 366 genes that affect GSC maintenance, differentiation or other processes involved in oogenesis. Comparison of GSC regulators with neural stem cell self-renewal factors identifies common and cell-type-specific self-renewal genes. Importantly, we identify the histone methyltransferase Set1 as a GSC specific self-renewal factor. Loss of Set1 in neural stem cells does not affect cell fate decisions, suggesting a differential requirement of H3K4me3 in different stem cell lineages. Altogether, our study provides a resource that will help to further dissect the networks underlying stem cell self-renewal.
Introduction
Stem cells play essential roles during animal development and homeostasis. Embryonic stem cells develop into all types of tissues and organs, while adult stem cells continuously replace dying and damaged cells. One of the key questions in stem cell biology is to understand the molecular basis of how stem cell self-renewal is controlled. Although mammalian cell culture approaches have provided insight in this process (Ding et al., 2009; Hu et al., 2009), it is desirable to study stem cells in their native environment.
Drosophila germline stem cells (GSCs) are a model of choice to identify genes involved in stem cell self-renewal (Spradling et al., 2011; Xie et al., 2008). In the Drosophila ovary, two or three GSCs are located in the most anterior part of the germarium, where they interact with the stem cell niche. A GSC divides asymmetrically to produce another self-renewing GSC and a cystoblast committed to differentiate. The cystoblast divides 4 times synchronously to form a 16-cell cyst. Of these, one cell will differentiate into an oocyte whereas the remaining cells will adopt a nurse cell fate. The activity of GSCs are controlled both by extrinsic and intrinsic factors. Decapentaplegic (Dpp) and Glass bottom boat (Gbb) produced from niche activate BMP signaling in the GSC to repress the transcription of a key differentiation gene, bag of marbles (bam), thereby maintaining GSC identity (Chen and McKearin, 2003; McKearin and Spradling, 1990; Song et al., 2004; Xie and Spradling, 1998). Besides cell-to-cell signaling, stem cell intrinsic programs are important for binary fate decisions. Nanos and Pumilio, components of a translational repression complex, are important for GSC maintenance (Forbes and Lehmann, 1998; Lin and Spradling, 1997; Wang and Lin, 2004). Similarly, components of the microRNA machinery are required for GSC maintenance (Forstemann et al., 2005; Jin and Xie, 2007; Park et al., 2007; Yang et al., 2007), suggesting that translational control is essential to maintain stem cell identity.
GSC self-renewal and differentiation are further controlled at the level of chromatin structure, transcription and splicing. The chromatin remodeling factor Iswi and the DNA-associated protein Stonewall are required for GSC maintenance through bam dependent- and independent-pathways (Maines et al., 2007; Xi and Xie, 2005). Similarly, Scrawny (Scny), a histone (H2B) deubiquitinase (Buszczak et al., 2009) and the histone H3K9 trimethylase Eggless (Egg) have been shown to be required for GSC maintenance (Wang et al., 2011). Conversely, the female-specific RNA-binding protein Sex-lethal (Sxl), as well as the U1 snRNP protein Sans-fille (Snf) that controls sxl alternative splicing, are essential for GSC differentiation (Chau et al., 2009; Schupbach, 1985) in part through regulation of Nanos levels (Chau et al., 2012).
Historically, genes regulating GSCs have been identified via genetic screens for female sterility in homozygous mutant animals (Cooley et al., 1988; Perrimon et al., 1986; Schupbach and Wieschaus, 1991). However, most genes relevant to oogenesis are also required during animal development, making it impossible to recover homozygous mutant animals. While the phenotypes of these genes can be analyzed by clonal mosaic analysis approaches, as done for maternal effect phenotypes (Perrimon et al., 1989; Perrimon et al., 1996), systematic screens for GSC self-renewal and differentiation have not been done. Recently, transgenic RNAi in Drosophila has been widely used to study gene function in somatic tissues, including other stem cell system such as neuroblasts (Nbs) (Dietzl et al., 2007; Neumuller et al., 2011). Here, we systematically analyzed GSC self-renewal using transgenic RNAi optimized for germline expression (Ni et al., 2009; Ni et al., 2011). We screened a collection of 3491 germline-enriched genes and identified 366 that cause female fertility defects, allowing us to construct a network of the genes regulating GSC self-renewal. Cross correlation with regulators of Nb self-renewal revealed GSC specific as well as commonly required regulators of self-renewal. We demonstrate a GSC specific role for the histone methyltransferase Set1 in GSCs and identify scrawny and domino as commonly required regulators in GSCs and Nbs. Our data thus constitute a useful resource for future studies of stem cell self-renewal.
Results
GSC self-renewal screen
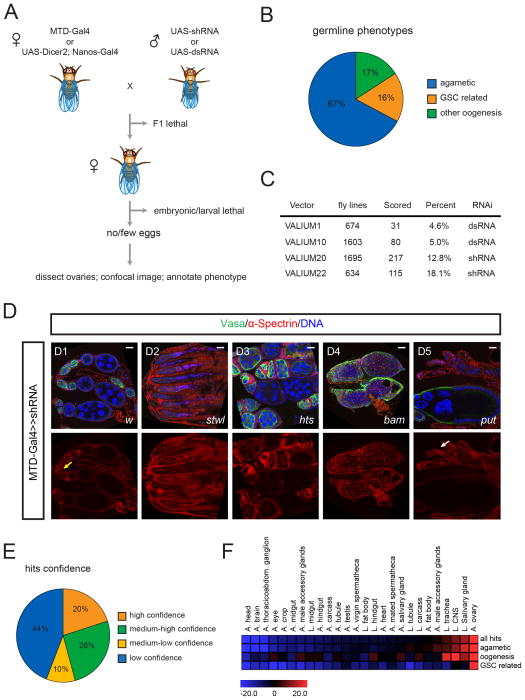
To systematically analyze the function of individual genes in the female germline, we screened the existing TRiP (Transgenic RNAi Project) collection of long dsRNA (VALIUM 1 and 10 vectors) and short shRNA (VALIUM20 and 22 vectors) lines (Ni et al., 2009; Ni et al., 2011). To express shRNAs or dsRNAs, we used a maternal triple driver MTD-Gal4 or UAS-dcr2; nanos-Gal4 to produce strong expression in the germarium and throughout oogenesis (Figure 1A, 2A) (Petrella et al., 2007). To identify potential stem cell phenotypes, ovaries of F1 females that laid no eggs were dissected and stained for three markers: The α-Spectrin antibody labels the spectrosome and the fusome, cytoplasmic organelles present in stem cells and cystocytes respectively; the Vasa antibody labels all germ cells; and DAPI was used to label nuclei for monitoring oocyte and nurse cell formation (Figure 1D1). We took confocal images of the ovaries, annotated the phenotypes and integrated all information into an online database (http://www.flyrnai.org/RSVP.html).
Figure 1. Transgenic RNAi screen.
(A) Workflow of the germline RNAi screen. (See Table S1)
(B) F1 females with no eggs or few eggs were dissected and ovaries were analyzed by confocal microscopy. The phenotypes were divided into three categories: agametic, GSC-related, and other oogenesis.
(C) Summary of the screen results.
(D) D1-D5, ovaries expressing shRNAs targeting w, stwl, hts, bam, or put by MTD-Gal4 stained for α-Spectrin, Vasa and DAPI. Yellow arrow indicates germline stem cells. White arrow points to empty germarium. (See Figure S1).
(E) Confidence of identified 366 genes from the screen. High confidence genes are identified by >2 independent RNAi lines; medium-high confident genes are identified by one RNAi line but they co-complex with high confidence hits; medium-low confidence genes are identified by one RNAi line but they co-complex with other low confidence hits; low confidence hits are identified by one RNAi only (Complexes are identified by COMPLEAT and listed in table S2).
(F) Heat map showing over- and underrepresentation of tissue-specific gene sets (as defined by their expression levels in the listed tissues) in three phenotypic categories found in the screen (Figure 1B). Color code represents Z-score with colors from blue (underrepresented) to red (overrepresented).
Scale bars: 20 μm.
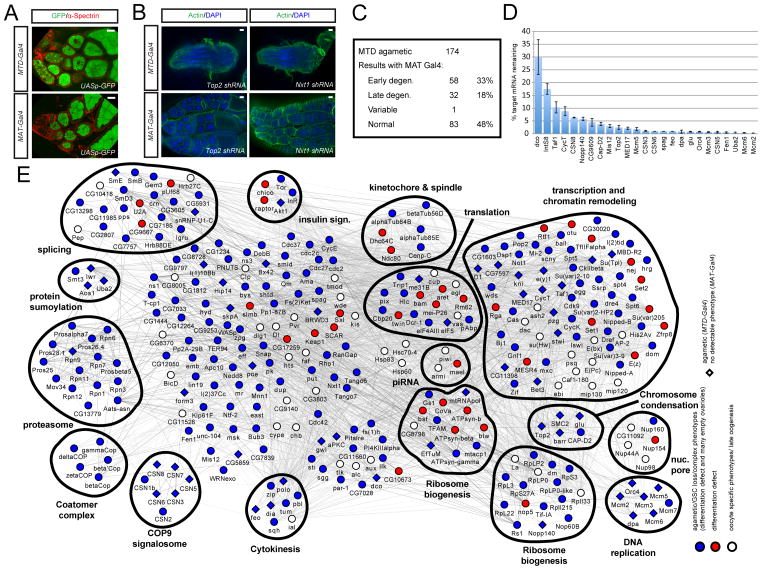
Figure 2. Quality control and regulatory network for genes identified from the screen.
(A) Expression patterns of UASp-GFP using the MTD-Gal4 or MAT-Gal4 drivers.
(B) Examples of the MAT screen. Top2 or Nxt1 shRNA is induced by MTD-Gal4 or MAT-Gal4 and the ovaries are stained for actin and DAPI. Both shRNAs result in agametic phenotypes with MTD-Gal4; but Top2 shRNA produce normal eggs with MAT-Gal4 while Nxt1 is still defective with MAT-Gal4.
(C) Summary of the secondary MAT-Gal4 screen of 174 agametic lines. 83 (48%) of the RNAi lines produce normal eggs with MAT-Gal4, indicating that these genes have GSC- or cystocyte-specific functions.
(D) qPCR experiments assessing the knockdown efficiency of a select set of shRNAs in 0–4 hr eggs laid from MAT-Gal4/shRNA females. Twenty-five genes are ranked according to the degree of knockdown. x-axis: genes tested; y-axis: % target mRNA remaining. Data are means ± s.e.m.
(See Figure S2)
(E) Network of genes identified by the germline screen. Genes are shown as nodes and the node color and shape indicate the observed phenotype in the screen. Red circle: differentiation defect; blue circle: agametic/stem cell loss; white circle: other oogenesis defects; blue diamond: agametic (MAT normal). Overall, red genes represent those required for GSC differentiation, and blue ones are those required for GSC maintenance. The edges denote the interactions of the genes and are represented in different grey tones: light edges are text mining/ genetic interaction based and dark edges represent PPI data.
Scale bars: 20 μm.
In total, we screened 4608 transgenic lines, representing 3491 germline enriched genes or ~25% of the Drosophila genome (Table S1). Among them, 444 lines, targeting 366 genes, showed oogenesis defects. The phenotypes were divided into three categories: agametic (67%), GSC related (16%) and other oogenesis (17%) (Figure 1B). In agametic ovaries, no or very few Vasa-positive germ cells are present, suggesting a defect in cell survival or GSC maintenance (Figure 1D2). GSC-related phenotypes include those that block stem cell differentiation and those that show bona fide stem cell maintenance phenotypes. For example bam shRNA ovarioles were filled with extra stem-cell like cells (Figure 1D4) (McKearin and Spradling, 1990). In punt (encoding the type II TGF-β receptor) shRNA ovaries, GSCs were lost from the germarium and differentiating egg chambers could be observed at later stages of oogenesis (Figure 1D5). (Xie and Spradling, 1998). Finally, many lines were associated with other oogenesis defects such as fusome structure (Figure 1D3), oocyte fate specification (Figure S1), nurse cell number, oocyte nuclear localization, and egg polarity.
Quality control
Four lines of evidence suggest that our screen has identified stem cell regulators with high confidence. First, we found many previously identified genes required for GSC differentiation (bam, sxl, otu, Mei-P26, mael, twin and aret) and GSC maintenance (punt, dcr-1, iswi, scny, stwl and egg) (Figure S1) (Buszczak et al., 2009; Findley et al., 2003; Jin and Xie, 2007; Maines et al., 2007; McKearin and Spradling, 1990; Morris et al., 2005; Page et al., 2000; Parisi et al., 2001; Pek et al., 2009; Schupbach, 1985; Wang et al., 2011; Xi and Xie, 2005; Xie and Spradling, 1998). Second, many of the identified hits (96 genes) whose gene products are subunits of protein complexes show a high degree of phenotypic similarity (see below). Third, we were able to confirm the phenotype of 73 genes by two independent RNAi (Figure 1E, Table S1). Lastly, we obtained evidence for efficient knockdown of a select set of genes by qPCR analyses or antibody staining.
The set of genes associated with GSC phenotypes is significantly enriched for genes expressed in adult ovaries and in the larval CNS, while most other tissues are underrepresented (Figure 1F). Since both tissues contain stem cells and differentiated cell types, they might use similar molecular mechanisms to regulate self-renewal. Human orthologs were found for 96% of our identified gene set, suggesting that those genes might have conserved functions in mammalian stem cell systems (Table S1).
Among our identified RNAi lines, agametic phenotypes (67%) represent the largest category. Those genes could be required for general cell viability, or they might have specific roles in stem cell maintenance. To distinguish these possibilities, we screened 174 agametic lines using maternal-tub-Gal4 (MAT-Gal4), which induces transgene expression outside the GSC compartment starting from stage 1 egg chambers in the posterior germarium (Figure 2A). Thus, this experiment can distinguish cell essential genes from GSC regulators. For example, Nxt1 shRNA is agametic with MTD-Gal4, and is also defective in oogenesis with MAT-Gal4, suggesting a general requirement in cell survival (Figure 2B). However, Top2 shRNA is agametic with MTD-Gal4 but produces normal eggs with MAT-Gal4, arguing it has GSC-specific functions (Figure 2B). Therefore, this secondary screen allows us to identify genes specific for a function in the germarium, including GSCs (Figure 2C, Table S1).
To test the efficiency of RNA knockdown, we selected a set of shRNAs for quantitative RT-PCR analysis and antibody staining. For quantitative PCR, we chose 25 shRNA lines that are agametic with MTD-Gal4, but produce normal eggs with MAT-Gal4. We prepared RNA from 0–4 hour eggs laid from MAT-Gal4/shRNA females, where most of the mRNAs are deposited during oogenesis. Compared to control shRNA, RNA levels for the target gene are reduced to <10% in 22 of the 25 lines (Figure 2D), suggesting that the shRNAs are highly efficient in knocking down gene expressed during oogenesis. Furthermore, we evaluated the knockdown efficiency at the protein level using antibodies, such as Akt1, Hts, HP1, Osa and Brm (see below). For example, expression of Akt1 shRNA with MAT-Gal4 produces normal eggs although its protein level is significantly reduced in the germline cells (Figure S2A). Further, hts shRNA induces an almost complete depletion of the fusome and ring canal specific forms of the Hts protein in their respective region when expressed by MTD-Gal4 or MAT-Gal4 (Figure S2B). These proof-of-principle experiments show that the shRNA lines achieve efficient knockdown in our screen.
Gene network regulating GSCs
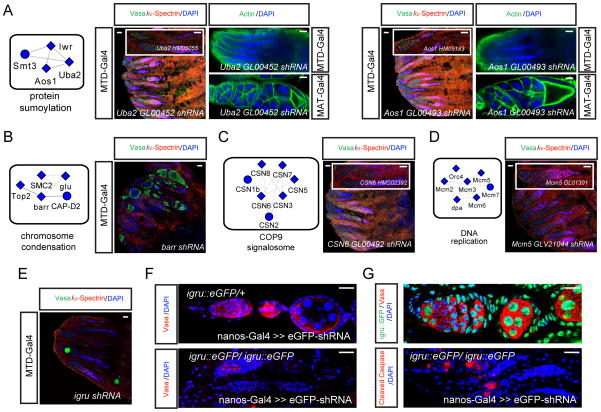
To better visualize our screening results, we generated a gene-interaction network querying publicly available databases containing protein-protein interactions, yeast two-hybrid interactions, genetic interactions, and text-mining data (Figure 2E). We divided the phenotypes into different categories including differentiation defects, GSC loss, late oogenesis, agametic, agametic (MAT normal), agametic (MAT defective) and arranged them into functional groups (Figure 2E). To identify protein complexes in this network we used COMPLEAT to perform a complex-enrichment analysis (Figure S3) (Vinayagam et al., 2013). This analysis allowed us to identify several protein complexes required for GSC maintenance, such as complexes involved in mRNA splicing, the COP9 signalosome (CSN), protein sumoylation, DNA replication and condensin complexes. CSN is a highly conserved, eight-subunit protein complex that is involved in diverse cellular and developmental processes. The most studied CSN function is regulation of protein degradation, but recent data suggest that CSN also regulates transcription. RNAi targeting of CSN1b and CSN2/alien using nanos-Gal4 generate an empty germarium phenotype. In addition, CSN3, CSN5, CSN6, CSN7 and CSN8 shRNA resulted in a complete loss of all germline cells with MTD-Gal4 (Figure 3C), but produced normal eggs with MAT-Gal4 suggesting that CSN function is only required early in the GSC lineage. Similarly, SUMO protein has been detected in the nuclei of the GSC and cystocytes, suggesting a role in GSC regulation (Hashiyama et al., 2009). Interestingly, knockdown of the two SUMO-activating enzymes Uba2 and Aos1 with two independent RNAi constructs using MTD-Gal4 resulted in a loss of GSCs whereas expression of these shRNAs with MAT-Gal4 resulted in the production of normal eggs (Figure 3A). Consistently, knockdown of the SUMO protein Smt3, and SUMO-conjugating enzyme Lwr, led to agametic ovaries, arguing that sumoylation is required for GSC maintenance. Finally, seven components of a DNA replication complex, Mcm2, Mcm3, Mcm4/dpa, Mcm5, Mcm6, Mcm7, Orc4 and five components of the condensin complex, SMC2, barr, glu, Top2, CAP-D2 generated an agametic phenotype when knocked down by MTD-Gal4, and most of them produced normal eggs (despite signs of lower DNA content in nurse cells upon knockdown of Mcm-complex genes) with MAT-Gal4 (Figure 3B, 3D, S2C). Thus, both DNA replication and condensin complexes are predominantly required in the germarium, potentially reflecting their roles in actively dividing GSC and cystocytes and to a lesser extent for endoreplication of nurse cells. Importantly, we have confirmed the knockdown efficiency of many of these shRNAs using qPCR (Figure 2D). Besides defined molecular complexes, we identified many other interesting genes required for GSC maintenance including phosphatidylinositol 4-kinase (PI4KIIIα), glutamine synthetase 1 (GS1) and heterogeneous nuclear ribonucleoprotein Hrb98DE (Figure S4A).
Figure 3. Genes and complexes required for GSC maintenance.
(A–D) Identified protein complexes required for GSC maintenance (Figure S3, Table S2). Complexes are enlarged from Figure 2E. RNAi against one representative gene from each complex is shown and other genes in the complex have a similar phenotype when knocked down by RNAi. (A) Knockdown of Uba2 or Aos1 by two independent RNAi constructs using MTD-Gal4 or UAS-dcr2; nanos-Gal4 results in a depletion of the germline. MAT-Gal4 mediated knockdown of these genes does not induce an obvious phenotype at later stages of oogenesis. (B-D) Knockdown of members of the respective complex members results in a depletion of the germline. Independent shRNA constructs are shown in inserts.
(E) Knockdown of igru (CG11266) with MTD-Gal4 results in a depletion of the germline.
(F) Trap-mediated loss-of-function validates the loss of vasa-positive germline cells upon loss of igru function.
(G) igru::GFP is expressed throughout the germline including high level expression in GSCs and low expression levels in the cystocyte region (top panel). Remaining germline cells stain positive for Vasa and cleaved Caspase (lower panel).
Scale bars: 20 μm.
Furthermore, our screen identified genes involved in GSC regulatory processes for which no phenotypic data are available to date. For example, knockdown of CG11266 (referred to as igru thereafter (see Experimental Procedures for details)), a proposed splicing factor, resulted in an almost complete depletion of germline cells (Figure 3E) suggesting a role for igru in GSC maintenance. We selected this gene for independent validation experiments as a functional igru::eGFP protein trap line permits localization and high stringency loss-of-function studies. We took advantage of the recently developed ‘trap-mediated loss-of-function’ method that uses well characterized eGFP shRNA lines to knockdown eGFP trapped genes (Neumuller et al., 2012). Consistent with the shRNA phenotype we found that GFP-mediated knockdown of igru with two independent eGFP specific shRNAs resulted in indistinguishable phenotypes, confirming the requirement of igru in GSC maintenance (Figure 3F and data not shown). The few remaining Vasa positive germline cells strongly stain for cleaved Caspase suggesting a requirement for igru in germline cell survival (Figure 3G). Consistent with this role and the proposed molecular function of igru, we found strong nuclear igru::eGFP expression in GSCs and polyploid nurse cells. Lower levels of igru expression are conversely detectable in cystocytes (Figure 3G). Together, these results demonstrate that igru is required for GSC maintenance and suggest that our screen can phenotypically annotate previously unstudied genes.
Genes important for stem cell differentiation
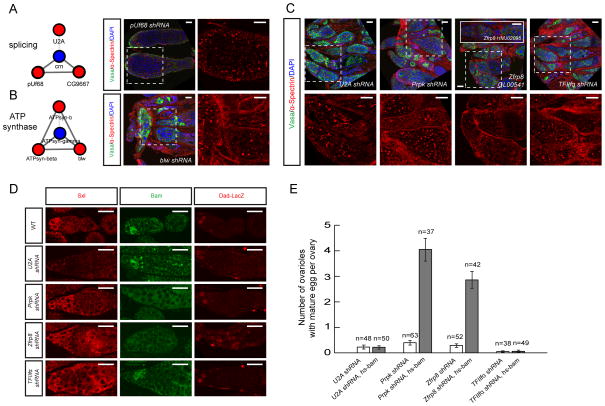
RNAi knockdown of genes required for GSC or cystoblast differentiation results in the accumulation of undifferentiated cells containing extra spectrosomes and/or fusomes. From our network analysis, we found several genes important for GSC differentiation including components of the RNA splicing machinery and mitochondrial ATP synthase complex. For example, three genes involved in RNA splicing were identified: pUf68 (the homolog of human PUF60), U2A and CG9667 (the ISY1 splicing factor homolog) (Figure 4A). Although previous reports suggest that pUf68 mutants display striking defects in the splicing of otu, they only show late oogenesis and egg morphology phenotypes (Van Buskirk and Schupbach, 2002). The strong differentiation defects associated with pUf68 knockdown (Figure 4A) resemble the strongest otu phenotype, potentially reflecting its role in otu splicing. Interestingly, knockdown of three genes encoding subunits of ATP synthase complex, blw/ATPsyn-α, ATPsyn-β, ATPsyn-b, as well as cytochrome c oxidase subunit Va (CoVa), also caused differentiation defects (Figure 4B). These data suggest that mitochondrial dysfunction is associated with GSC differentiation defects. Other interesting genes involved in GSC differentiation include Prpk, homolog of P53 regulating kinase; zfrp8 (phenotype confirmed by two independent shRNAs), zinc finger protein involved in Drosophila hematopoietic stem cell regulation (Minakhina and Steward, 2010); the transcription initiation factor TfIIFα, SCAR, WAVE/SCAR complex component; slmb, F-box/WD40 E3 ubiquitin ligase; keap1, regulator of cellular redox state; CG10426, inositol polyphosphate-5-phosphatase; Ndc80, a kinetochore protein; Dhc64c, dynein heavy chain 64C; bsf, a mRNA binding protein and Ccn, a potential growth factor (Figure 4C and S4B).
Figure 4. Genes and complexes required for GSC differentiation.
(A–B) Identified protein complexes required for GSC differentiation. RNAi against one representative gene from each complex is shown. Ovaries expressing RNAi targeting pUf68 and blw are stained for α-Spectrin, Vasa and DAPI. Areas marked by the dashed squares are enlarged.
(C) shRNAs against U2A, Prpk, zfrp8 or TFIIfα expressed by MTD-Gal4 lead to strong differentiation defects. Ovaries are stained for α-Spectrin, Vasa and DAPI and Zfrp8 phenotype is confirmed by two independent shRNAs.
(D) Ovaries expressing U2A, Prpk, zfrp8 or TFIIfα shRNAs are stained for Sxl, Bam or Dad-lacZ. Sxl and Bam experiments are driven by MTD-Gal4, and Dad-lacZ experiments are driven by nanos-Gal4.
(E) Quantification of hs-bam rescue experiments. shRNAs against U2A, Prpk, zfrp8 or TFIIfα expressed using nanos-Gal4 with or without hs-bam expression. The number of ovarioles with mature egg per ovary is shown and n is number of ovaries examined. Data are mean ± s.e.m. Representative ovary images are shown in Figure S4C.
Scale bars: 20 μm.
To study the underlying mechanisms we selected four genes associated with strong differentiation defects, U2A, Prpk, Zfrp8 and TFIIfα. To characterize the loss-of-function phenotypes in greater detail, we stained these ovaries with available molecular markers Sxl, Bam and Dad-LacZ. In wild-type germarium, the Sxl protein accumulates to high levels in GSCs and cystoblasts; Bam is expressed in cystoblasts and early cysts; and Dad-lacZ, a reporter of Dpp signal activation, is confined to the 2–3 GSCs (Figure 4D). Sxl accumulation is normal in Prpk, Zrfp8 and TFIIfα shRNA mutant ovaries, however, it is strongly reduced in U2A shRNA ovaries (Figure 4D). U2A encodes the Drosophila U2 snRNP and the reduction of Sxl protein likely reflects its role in sxl splicing (Nagengast and Salz, 2001). Next, Bam protein expression is largely normal in U2A and TFIIfα shRNA ovaries, but reduced significantly in Prpk and Zfrp8 shRNA ovaries, suggesting that these two genes control differentiation in a bam-dependent manner. To confirm these results, we overexpressed bam from a heat shock inducible promoter in the shRNA knock-down background (Ohlstein and McKearin, 1997). Heat-shock induced bam (hs-bam) is able to rescue Prpk and zfrp8 shRNA, generating normally developed egg chambers and reversing the fertility phenotype (see Figure 4E for quantification and S4C for representative images). On the other hand, hs-bam has no obvious effect in U2A and TFIIfα shRNA ovaries (Figure 4E and S4C). Lastly, Dad-lacZ is expressed normally in U2A, Prpk and TFIIfα shRNA ovaries, but is ectopically induced in Zfrp8 ovaries. These results indicate that Zfrp8 regulates bam expression through controlling Dpp signaling. Together, we have identified factors that control multiple steps of GSC differentiation.
Transcriptional network for GSC regulation
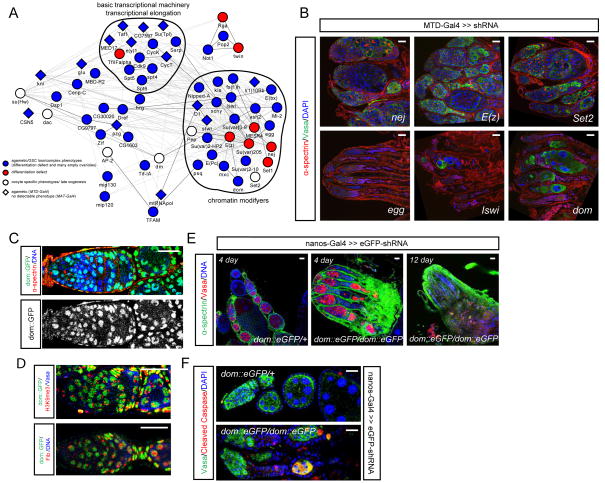
As stem cell self-renewal is controlled by key transcription factors and by epigenetic regulation, we decided to analyze in particular these two classes of genes. To systematically study transcriptional regulation in GSC self-renewal we built a subnetwork containing 65 experimentally verified or computationally predicted transcription factors and chromatin regulators (Figure 5A). Among those we identified several previously known GSC transcriptional regulators (Figure 5B). Twin, a subunit of the CCR4-NOT complex, is required for GSC differentiation (Morris et al., 2005). The ATP-dependent chromatin remodeling factor ISWI (Xi and Xie, 2005), the DNA-associated protein Stonewall (Maines et al., 2007), the histone H3K9 trimethylase Eggless (Wang et al., 2011), the histone H2B ubiquitin protease Scny (Buszczak et al., 2009), and the transcription elongation factor Spt6 (Neumuller et al., 2012) are essential for GSC maintenance. Besides these known factors our screen identified several additional transcriptional regulators of GSC self-renewal. For example, knockdown of CCR4-NOT subunit Rga, Paf1 complex subunit Rtf1, Drosophila HP1/Su(var)205, Polycomb group protein E(z), histone acetyltransferase nej/dCBP and histone H3K36 methylase Set2 are associated with defects in differentiation; while PIAS homolog Su(var)2–10 or components of the basic transcriptional machinery, including the TATA box binding protein (TBP)-associated factor Taf1, e(y)1/TafII40, or the mediator component MED17 and the transcription elongation factor Spt4, Su(Tpl), are important for GSC maintenance (Figure 5B, S5A). As an example, we probed the Su(var)205 shRNA ovaries with HP1 antibody and the heterochromatic marker H3K9me3. As shown in Figure S5B and S5C, HP1 staining is abolished from these germline cells and H3K9me3 staining is also reduced. These results suggest the shRNA effectively knocks down Su(var)205 protein level, which is important to maintain heterochromatin structure in the germline. Together, these results provide a first step toward generating a complete transcription factor network regulating GSC self-renewal.
Figure 5. Transcription factor network regulating GSC self-renewal.
(A) Network for transcription factors and chromatin regulators identified from our screen. Nodes and edges are represented the same as those in Figure 2E.
(B) shRNAs are expressed using MTD-Gal4 and ovaries stained for α-Spectrin, Vasa and DAPI. shRNAs against nej, E(z) and Set2 cause differentiation defects, while shRNAs against egg, Iswi and dom result in agametic or stem-cell-loss phenotype. (See Figure S5 for additional examples).
(C) dom-eGFP protein trap ovaries stained for α-Spectrin and DAPI show the expression pattern of Dom.
(D) dom-eGFP ovaries stained for Fibrillarin or H3K9me3.
(E) Ovaries expressing eGFP shRNA using nanos-Gal4 in the dom-eGFP heterozygous or homozygous background are stained for α-Spectrin,Vasa and DAPI. Days indicate time after eclosion. Germ cells are lost in dom loss-of-function after 12 days compared to wild type (data not shown).
(F) Ovaries expressing eGFP shRNA using nanos-Gal4 in the dom-eGFP heterozygous or homozygous background are stained for cleaved Caspase, Vasa and DAPI.
Scale bars: 20 μm.
Next, we focused our attention on the ATP-dependent chromatin remodeling factor Domino (Dom). Dom was reported not to be required in GSCs (Xi and Xie, 2005), but our data suggested that dom is potentially required for GSC maintenance since dom shRNA induced by MTD-Gal4 generates a loss-of-stem-cell phenotype (Figure 5B). To clarify whether dom is required for GSC maintenance and to confirm the specificity of the knockdown, we used trap-mediated loss-of-function method. We used a homozygous viable dom-eGFP protein trap line and found that Dom-eGFP is expressed ubiquitously in the germline (Figure 5C), consistent with a potential requirement in GSCs. Dom is a nuclear protein that presumably localizes to active sites of Pol II transcription as we did not detect Dom-eGFP in nuclear domains positive for the heterochromatic marker H3K9me3 and the nucleolar marker Fibrillarin (Figure 5D). Trap-mediated loss of dom function in GSC resulted in a complete loss of GSCs 12 days after eclosion (Figure 5E). Prior to the loss of all germline cells we noticed a high number of Vasa and α-spectrin double positive cells throughout the ovarioles, suggesting a cystocyte differentiation defect. This phenotype was only transitory as increased numbers of apoptotic cells lead to a progressive loss of germline cells (Figure 5F). Taken together, our results demonstrate a requirement for dom in GSC self-renewal and also in cystocyte differentiation.
Comparison of germline and neural stem cell self-renewal
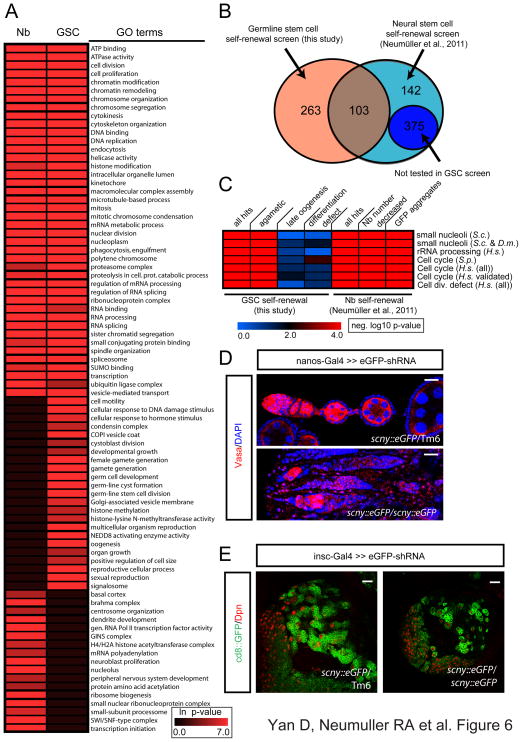
To understand if different stem cell systems use similar mechanisms to regulate self-renewal and differentiation, we compared the results of our screen with a previous analysis of neural stem cell self-renewal (Neumuller et al., 2011). GSCs are regulated through a niche-dependent mechanism and Nbs control self-renewal through intrinsic asymmetric cell division. As both stem cell types are mitotically active, we expected basic cellular processes to be commonly required. However, cell-type-specific regulators might be rare modulators of these commonly required cellular networks.
Importantly, the genes we identified in our GSC screen are similarly enriched in ovaries and the larval CNS (Figure 1F). Of the 366 genes identified in the GSC screen 103 genes were reported to be required for Nb self-renewal (Figure 6B). A GO term analysis for these overlapping genes suggests that GSCs and Nbs share a requirement for many cellular processes in the regulation of self-renewal (Figure 6A). In addition, we compared our gene interaction network (Figure 2E) with the gene network for Nb self-renewal (Neumuller et al., 2011). From these analyses, we found common as well as distinct regulators in these two cell types. Importantly, many basic cellular processes such as DNA replication, cell division, histone modification and splicing are commonly required in Nbs and GSCs. Additionally, many transcription factors, chromatin remodeling genes, the proteasome as well as ribosome genes are associated with stem cell maintenance or differentiation defects in both systems. To gain further insight into the extent of overlap between genes regulating stem cell maintenance in these two systems, we performed gene set enrichment analyses. We used previously published data for nucleolar size regulation (Neumuller et al., 2013), rRNA processing (Tafforeau et al., 2013) and cell division (Hayles et al., 2013; Kittler et al., 2007) from a diverse set of species and found these genes commonly enriched in both GSC and Nb maintenance gene-sets. Conversely, these genes were not significantly enriched in the gene-sets associated with GSC differentiation or late oogenesis defects (Figure 6C). These data suggest a common requirement for cell growth and cell division for maintenance of Nbs and GSCs.
Figure 6. Comparison of GSC and the neural stem cell RNAi screens.
(A) Heat map displaying overrepresentation of selected GO terms in genes identified in the GSC and the Nb screen.
(B) Number of genes identified in the GSC and Nb screen. 103 genes were found in both screens. Note that 375 identified genes in Nb screen were not tested in the germline screen (See Table S1 for the detailed list of genes).
(C) Comparative gene set enrichment between GSCs and Nbs of genes associated with small nucleoli, rRNA processing defects and cell division defects in the respective phenotypic categories.
(D) Ovaries expressing eGFP shRNA using nanos-Gal4 in the protein trap scny-eGFP heterozygous or homozygous background stained for Vasa and DAPI. (scny shRNA phenotypes are shown in Figure S6C).
(E) Larval brains expressing eGFP shRNA using insc-Gal4≫CD8::GFP in the scny-eGFP heterozygous or homozygous background stained for Nb marker Dpn (Note: the eGFP shRNA does not target CD8::GFP).
Scale bars: 20 μm.
Genes only required in GSC regulation, but not in Nbs, include the COP9 signalosome complex, the protein sumoylation complex, several mitochondrial genes and histone methyltransferases. It will be interesting to further study why these protein complexes are preferentially required in GSC, but not Nbs. Conversely, knockdown of brahma (brm), osa or moira is associated with an expansion of stem-cell like cells in type II Nb lineages (Neumuller et al., 2011). osa and brm are expressed at all stages in the GSC lineage. When expressed from MTD-Gal4, osa as well as brm shRNA constructs effectively deplete the respective proteins without inducing detectable phenotypes (Figure S6A, B). We next searched amongst the stem cell differentiation factors in GSC and Nb lineages for overlap. Interestingly, our analysis revealed an almost mutually exclusive set of differentiation factors (Ccn is the only factor shared between Nbs and GSCs), providing a comprehensive comparison of context dependent differentiation and tumorigenesis in two stem cell lineages. However, as both studies (GSC as well as Nb self-renewal) do not cover the entire genome, we cannot formally exclude more potentially shared differentiation factors.
scny, encoding a ubiquitin-specific protease, was recently shown to be a common factor regulating self-renewal in germline, epithelial and intestinal stem cell maintenance (Buszczak et al., 2009). However scny has no described function in larval Nbs raising the possibility that its function might be dispensable in this developmental cell type. To examine if scny is a general regulator of stem cell maintenance we used trap-mediated loss-of-function to knockdown egfp::scny in a homozygous viable and fertile GFP trap line. eGFP shRNA mediated knockdown in GSCs resulted in a depletion of Vasa-positive cells from the germline (Figure 6D, S6C), confirming previous results (Buszczak et al., 2009). Next, we studied scny function in Nbs by using a Nb-specific Gal4 line, insc-Gal4 (Neumuller et al., 2011). While eGFP shRNAs generated no phenotype in a heterozygous scny-eGFP background, they strongly reduced the number of Deadpan (Dpn) positive Nbs in a homozygous scny-eGFP background (Figure 6E). We did not detect evidence for enhanced cell death upon scny loss-of-function in Nbs (data not shown). These data suggest that Scny is an essential stem cell maintenance factor in most if not all Drosophila stem cell types.
Set1 H3K4 methyltransferase is required for cystocyte, but not neuroblast differentiation
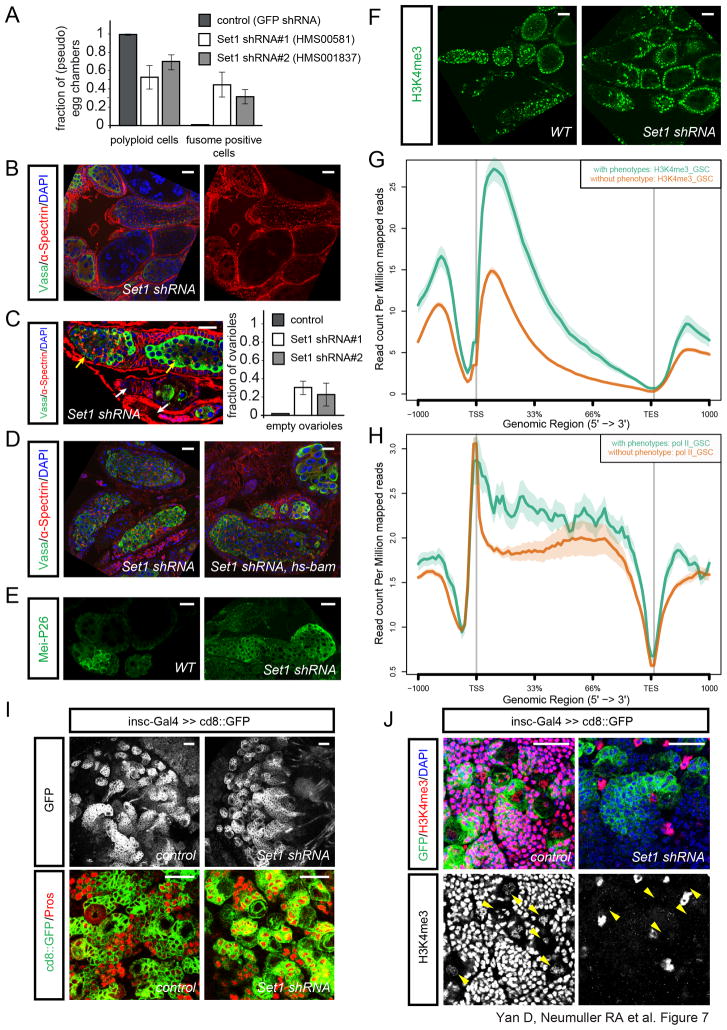
Methylation of histone 3 lysine 4 (H3K4) is a histone modification associated with active transcription. However, its function in adult stem cell regulation remains to be determined. In yeast, all mono-, di- and tri-methylation of H3K4 is catalyzed by a single Set1 enzyme, while in Drosophila there are four known H3K4 methyltransferases: Trx, Ash1, Trr and Set1 (Hallson et al., 2012). Drosophila Set1 is located in the centric heterochromatin region and has been difficult to characterize by traditional genetic methods (Ardehali et al., 2011). Our systematic comparison of Nb and GSC self-renewal suggested that Set1 might be a GSC-specific self-renewal factor. Set1 shRNA expressed by MTD-Gal4 leads to the co-occurrence of pseudo-egg chambers filled with fusome-and spectrosome-containing cells, pseudo-egg chambers containing >15 nurse cells as well as empty ovarioles, suggesting a role for Set1 in stem cell maintenance (as recently reported by (Xuan et al., 2013)) as well as differentiation (Figure 7A–7C). This result was confirmed using two additional independent shRNA lines (Figure 7A, S7C), indicating that Set1 is required during multiple processes in oogenesis. To show that the ectopic undifferentiated cells in Set1 shRNA ovaries retained their proliferative potential, we stained for the mitotic marker phosphorylated-histone H3 (pH3). In wild-type ovaries, pH3 positive cells were restricted to the anterior tip of the germarium, but were detected throughout Set1 shRNA ovaries (Figure S7B). Next, we found that hs-bam failed to fully rescue the Set1 shRNA phenotype (Figure 7D), suggesting that Set1 likely regulates GSC differentiation independent of Bam. Further, we used an antibody against Mei-P26, which is expressed at low levels in GSCs, upregulated in cystocytes and absent afterwards (Liu et al., 2009; Neumuller et al., 2008). Compared to wild type, Set1 shRNA ovaries have strong Mei-P26 staining throughout egg chamber, indicating that the germline development is blocked at the cystocyte stage (Figure 7E).
Figure 7. Set1 regulates GSC but not Nb self-renewal.
(A) Quantification of the Set1 loss of function phenotype in the germline by MTD-Gal4 (bars represent the mean+/− S.D. of the observed frequencies (n=175 pseudo egg chambers (HMS00581), n=105 pseudo egg chmbers (HMS001837)).
(B) Ovaries expressing Set1 shRNA (HMS00581) by MTD-Gal4 are labeled by α-Spectrin, Vasa and DAPI staining.
(C) Co-occurrence of pseudo egg chambers filled with undifferentiated fusome containing cells (yellow arrows) and empty ovarioles (white arrows) in MTD/Set1 shRNA ovaries. Quantification of the empty ovariole phenotype (bars represent the mean+/− S.D. of the observed frequencies (n=74 ovarioles (HMS00581), n=55 ovarioles (HMS001837)).
(D) Overexpressing bam using hs-bam fails to fully rescue the differentiation defects in Set1 shRNA/nanos-Gal4 background, as shown by α-Spectrin, Vasa and DAPI staining.
(E) Mei-P26 antibody staining in WT and Set1 shRNA/MTD-Gal4 ovaries.
(F) H3K4me3 staining in WT and Set1 shRNA/MTD-Gal4 ovaries.
(G) H3K4me3 ChIP from FACS purified GSCs showing increased levels of lysine4 tri-methylation at genes with a phenotype (green) in our screen over genes without a detectable phenotype (brown). x-axis depicts 1000 bp upstream of the transcriptional start site (TSS), the length of the gene bodies in percentage, the transcriptional end site (TES) and 1000 bp downstream. (See Figure S7 for further ChIP results and phenotypic characterization of Set1 loss-of-function)
(H) Pol II ChIP from FACS purified GSCs showing an increased association of Pol II at genes with a phenotype (green) in our screen over genes without a detectable phenotype (brown). x-axis depicts 1000 bp upstream of the transcriptional start site (TSS), the length of the gene bodies in percentage, the transcriptional end site (TES) and 1000 bp downstream.
(I–J) Larval brains expressing Set1 shRNA, or no RNAi (control) using insc-Gal4≫CD8::GFP stained by neuronal marker Pros (I), or H3K4me3 and DAPI (J). Yellow arrowheads point to Nbs.
Scale bars: 20 μm.
Since Set1 is a major H3K4 methyltransferase, we determined its in vivo function using antibodies specific to different forms of methylated H3K4. In wild-type ovaries, all three modifications, mono-, di- and trimethyl H3K4, were detected both in germaria and egg chambers (Figure 7F, S7D). Set1 shRNA had no effect on the pattern of H3K4me1, but almost completely abolished H3K4me2 and H4K4me3 staining in the germline, suggesting that Set1 acts as a major H3K4 di- and trimethyltransferase in the germline (Figure 7F and S7D, S7E). Consistently, by using Chromatin Immunoprecipitation (ChIP) from FACS purified GSCs, we found an increased occurrence of trimethylated H3K4 on genomic regions covering genes that our screen identified as being associated with GSC maintenance or differentiation defects. Conversely, H3K4me3 levels are lower over genomic regions covering genes that did not result in a phenotype upon loss-of-function (Figure 7G). Importantly a similar result was obtained in ChIP experiments for Pol II suggesting that H3K4me3 is predominantly associated with actively transcribed genes that are functionally required in GSCs (Figure 7H). As we found H3K4me3 at genes regulating both GSC maintenance and differentiation as well as genes that control both processes, such as Mei-P26 (Li et al., 2012) (Figure S7F), we postulate that altered expression of a yet to be determined set of key regulatory genes is underlying the observed phenotypes.
As we have shown that Scny, which deubiquitylates histone H2B that is required for H3K4 methylation, is required for stem cell maintenance in both GSCs and Nbs, we tested whether Set1 controls self-renewal in Nb lineages as well. Surprisingly, knockdown of Set1 by insc-Gal4 had no effect on Nb differentiation or maintenance (Figure 7I), although it reduced H3K4me3 to undetectable levels in those cells (Figure 7J). The neuronal markers Prospero and Elav were expressed normally upon Set1 knockdown and Nb numbers as well as Dpn positive progeny in Type II lineages were similar to wild type (Figure 7J and data not shown). These results indicate that unlike GSCs, neural stem cell differentiation and maintenance does not require Set1-mediated methylation of H3K4. It will be interesting to examine other stem cell systems and whether this mechanism applies to mammalian stem cells.
Discussion
Most genes required for GSC self-renewal have previously been identified using homozygous mutant animals or mosaic analyses. Because of the limitations associated with both approaches (female sterile mutations usually represent hypomorphic alleles of zygotic lethals (Perrimon et al., 1986) and production of germline mosaics is relatively cumbersome (Perrimon et al., 1989; Perrimon et al., 1996)), no large-scale screens have yet been performed to systematically identify genes involved in GSC self-renewal. Here, using transgenic RNAi in the Drosophila female germline, we screened 25% of the fly genome and identified 366 genes associated with specific stem cell defects. Based on our screen, we constructed a genetic network governing GSC self-renewal and identified several protein complexes essential for GSC regulation, such as the COP9 signalosome, protein sumyolation, the ATP synthase complex as well as chromatin remodeling and transcription factors. This study significantly expands the factors known to be required for GSC self-renewal and will serve as a resource for future studies in GSC biology.
Detailed analysis of our data and a screen in Nbs allowed us to identify genes that control stem cell self-renewal in both systems. Our systematic analysis provides evidence that basic cellular processes as cell division, growth regulation or splicing are commonly required in GSCs and Nbs. The molecular context in which these processes are embedded might however differ significantly. For example, alternative splicing has been shown to be a key regulatory step in both Nbs and GSCs. In Nbs, alternative splicing of the transcription factor lola has been suggested to be required for regulating self-renewal (Neumuller et al., 2011). GSCs conversely control this process through the alternative splicing of sxl suggesting that basic cellular machineries are acting on different targets in GSCs and Nbs (Chau et al., 2009). We further evaluated the role of dom and scny in stem cell maintenance. Dom is an ATP-dependent chromatin remodeling factor that has previously been implicated in somatic stem cell maintenance in the female ovary (Xi and Xie, 2005). A requirement in GSCs had not been documented potentially due to the use of hypomorphic alleles. Our analysis demonstrates a role of dom in GSCs. As shRNA-mediated knockdown of dom in Nbs also induces stem cell loss (data not shown), we propose that dom is commonly required in different stem cells to control their maintenance. Similarly, the histone H2B deubiquitinase Scny has been suggested to be a general regulator of stem cell maintenance in adult stem cell lineages. We expand this function to Nbs and thus provide further evidence for a general role for histone deubiquitination in stem cell maintenance in Drosophila. Consistently, H2B monoubiquitination has been shown to significantly increase upon differentiation of human mesenchymal stem cells (Karpiuk et al., 2012). Altogether, these data establish histone ubiquitination as a common regulatory mechanism in stem cell biology.
Consistent with previous studies that found Nb and GSC daughter cell differentiation to be controlled by different mechanisms (intrinsic versus extrinsic asymmetric cell division) we did not obtain evidence for extensive overlap in genes controlling differentiation with our systematic approach. For example, loss of the brm complex is associated with tumor formation in type II Nb lineages and differentiation defects in intestinal stem cells (Jin et al., 2013; Zeng et al., 2013). Conversely, in the germline, loss of brm complex members is not associated with differentiation defects. These observations are in agreement with data on mutations in the SWI/SNF complex in different human cancers ranging from 0 to 75% in frequency (Shain and Pollack, 2013), underlining the value of Drosophila as a model system to study context-dependent tumorigenesis. Similarly, the gene barricade (barc) has been shown to result in an increased number of intermediate neural progenitor cells upon knockdown in Nb lineages (Neumuller et al., 2011), whereas depletion of barc results in a loss of GSCs phenotype in the germline (Figure S6D). These data suggest fundamental differences in lineage specific tumorigenesis and suggest that an almost mutually exclusive set of differentiation genes operate in these two stem cell lineages. Interestingly, germline specific genes can be ectopically expressed in certain brain tumor mutants and functionally sustain tumor growth (Janic et al., 2010). This ectopic expression might contribute to sustained tumor growth and it will be interesting to determine if genes required for tumor maintenance are shared between Nb and GSC tumors. Together, our data provide systematic evidence that Nb and GSC lineages share an extensive stem cell maintenance network while genetic programs regulating differentiation differ between these two cell types.
Importantly, we were able to identify several candidate genes with a specific requirement in GSCs. Our data demonstrate that Set1 and histone H3K4 tri-methylation are important for germline differentiation and GSC maintenance, but appear not to be required for Nb self-renewal. Xuan et al. recently documented a similar requirement of Set1 in GSC maintenance (Xuan et al., 2013). Our study, using multiple independent shRNA constructs suggests that Set1 is required at multiple steps in the early GSC lineage, including cystocyte differentiation. Set1 is required for the bulk H3K4 trimethylation, a histone modification that has been associated with active sites of transcription. Consistent with the phenotypic spectrum of Set1 loss-of-function, we find H3K4me3 on genes that promote both GSC maintenance and differentiation. A recent report suggests that H3K4 methylation is dispensable for active transcription in somatic tissues (Hodl and Basler, 2012). Consistently, we find that in Nb lineages, key differentiation genes like Pros or Elav are normally induced and that H3K4me3 is not required for lineage progression. Conversely, our data suggest that H3K4 methylation is required for GSC self-renewal and it will be interesting to determine if this differentiation defect is indeed linked to insufficient levels of active gene transcription of key differentiation genes.
Experimental Procedures
RNAi screen and Drosophila strains
UAS-RNAi lines are generated by TRiP and are available at the Bloomington Drosophila stock center (BDSC). For the RNAi experiments we used a maternal triple driver MTD-Gal4 (BDSC 31777) or UAS-dcr2; nanos-Gal4 (BDSC 25751) to drive expression of UAS-RNAi transgenes in GSCs, MAT-Gal4 (BDSC 7063) for germline expression outside the germarium, and insc-Gal4 for expression in larval Nbs. For trap-mediated loss of function analyses we used UAS-shRNAs targeting eGFP as previously described (Neumuller et al., 2012). Protein trap lines scny::GFP, dom::GFP and CG11266::GFP are described in (Buszczak et al., 2007). We chose the name inselgruppe (German for ‘group of islands’, abbreviated: igru) due to the few remaining, scattered Vasa positive cells observed in the ovaries upon knockdown.
Immunofluorescence and antibodies
Larval brains and female ovaries were stained as previously described (Neumuller et al., 2008). Briefly, tissues were dissected in PBS, fixed in 4% paraformaldehyde in PBST (PBS + 0.1% Triton X-100). After blocking in 1% normal donkey serum in PBST for 1 hour, the samples were incubated with the primary antibody in the same solution at 4 °C overnight. After three washes in PBST, samples were incubated with the secondary antibody for 2 hour at room temperature, washed in PBST for three times, and subsequently mounted in VECTASHIELD. The following antibodies were used: mouse anti-α-Spectrin (3A9, DSHB), rabbit anti-Vasa (Santa Cruz Biotechology), mouse anti-Bam (DSHB), mouse anti-Sxl (M18, DSHB), mouse anti-LacZ (Promega), mouse anti-Osa (DSHB), guinea pig anti-Brm (gift from P. Harte), rabbit anti-Akt (Cell Signaling), mouse anti-HP1 (C1A9, DSHB), rabbit anti-Mei-P26 (gift from P. Lasko), mouse anti-Hts (1B1, DSHB), mouse anti-Hts RC (DSHB), mouse anti-Prospero (MR1A, DSHB), mouse anti-Elav (9F8A9, DSHB), guinea pig anti-Deadpan (gift from J. Skeath), rabbit anti-phospho-Histone H3 (Millipore), rabbit anti-H3K4me3 (Cell Signaling), mouse anti-H3K4me2 (Active Motif), rabbit anti-H3K4me1 (Active Motif), mouse anti-H3K9me3 (Abcam), mouse anti-Fibrillarin (Abcam), rabbit anti-GFP (Abcam), rabbit anti-cleaved caspase (Cell Signaling), Alexa 488-phalloidin (Molecular Probes), DAPI (Molecular Probes). All images were taken on a Leica SP5 microscope.
qPCR
Total RNA was extracted from from 0–4 hr old eggs derived from MAT-Gal4/shRNA females using TRIzol (Invitrogen), and purified through RNeasy MinElute Cleanup Kit (Qiagen). cDNA was generated from 1 microgram of purified RNA using iScript cDNA Synthesis Kit (Bio-Rad). qPCR analysis was performed twice with technical triplicates in iQ SYBR Green Supermix (Bio-Rad), using a CFX96 Real-Time PCR detection system (Bio-Rad). Query transcript detection was normalized to the expression of three reference genes: alpha-tubulin, rp49 and nuclear fallout. Fold change was calculated in comparison to an shRNA knockdown targeting the white gene. Primers are selected using FlyPrimerBank.
FACS Isolation and chromatin immunoprecipitation (ChIP)
GFP-positive GSCs and cystoblasts (CBs) are isolated from the ovaries of vasa-GFP/+; nos-gal4/UASp-tkvCA and vasa-GFP/+; bamΔ86/bamΔ86 using FACS according to the previously published procedure (Song et al., 2004). The ChIP experiments were performed based on the published protocol (Zeitlinger et al., 2007) and the antibodies used are: H3K4me3 (Abcam, ab8580) and PolII (Abcam, ab5131).
Bioinformatics analyses are described in the Supplemental Experimental Procedures.
Supplementary Material
Highlights.
RNAi screen identifies 366 genes required for germline stem cell (GSC) regulation
Comparison of GSC with neural stem cells identifies common and distinct regulators
COP9, sumoylation, spliceosome and ATP synthase complexes are required in GSCs
Identification of a set of transcription factors essential for GSC self-renewal
Acknowledgments
We thank M. Buszczak, D. Chen, D. Glover, A. Greenleaf, P. Harte, G. Karpen, T. Kerppola, J. Knoblich, P. Lasko, J. Lis, P. Macdonald, K. McCall, D. McKearin, D. Montell, A. Nakamura, H. Nakato, D. Price, M. Przewloka, G. Rogers, S. Rogers, T. Schupbach, A. Shilatifard, J. Skeath, E. Wahle and F. Winston for antibodies and fly stocks. D.Y. is supported by Damon Runyon Cancer Research fellowship. R.A.N is supported by EMBO and Human Frontier Science Program (HFSP) Long-Term fellowships. This work was supported by NIH/NIGMS R01-GM084947, NIH/NIGMS R01-GM067761 to N.P. and NIH GM043301 to L.C.; by the Stowers Institute for Medical Research to T.X.. N.P. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ardehali MB, Mei A, Zobeck KL, Caron M, Lis JT, Kusch T. Drosophila Set1 is the major histone H3 lysine 4 trimethyltransferase with role in transcription. EMBO J. 2011;30:2817–2828. doi: 10.1038/emboj.2011.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buszczak M, Paterno S, Lighthouse D, Bachman J, Planck J, Owen S, Skora AD, Nystul TG, Ohlstein B, Allen A, et al. The carnegie protein trap library: a versatile tool for Drosophila developmental studies. Genetics. 2007;175:1505–1531. doi: 10.1534/genetics.106.065961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buszczak M, Paterno S, Spradling AC. Drosophila stem cells share a common requirement for the histone H2B ubiquitin protease scrawny. Science. 2009;323:248–251. doi: 10.1126/science.1165678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau J, Kulnane LS, Salz HK. Sex-lethal facilitates the transition from germline stem cell to committed daughter cell in the Drosophila ovary. Genetics. 2009;182:121–132. doi: 10.1534/genetics.109.100693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau J, Kulnane LS, Salz HK. Sex-lethal enables germline stem cell differentiation by down-regulating Nanos protein levels during Drosophila oogenesis. Proc Natl Acad Sci U S A. 2012;109:9465–9470. doi: 10.1073/pnas.1120473109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, McKearin DM. A discrete transcriptional silencer in the bam gene determines asymmetric division of the Drosophila germline stem cell. Development. 2003;130:1159–1170. doi: 10.1242/dev.00325. [DOI] [PubMed] [Google Scholar]
- Cooley L, Kelley R, Spradling A. Insertional mutagenesis of the Drosophila genome with single P elements. Science. 1988;239:1121–1128. doi: 10.1126/science.2830671. [DOI] [PubMed] [Google Scholar]
- Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- Ding L, Paszkowski-Rogacz M, Nitzsche A, Slabicki MM, Heninger AK, de Vries I, Kittler R, Junqueira M, Shevchenko A, Schulz H, et al. A genome-scale RNAi screen for Oct4 modulators defines a role of the Paf1 complex for embryonic stem cell identity. Cell Stem Cell. 2009;4:403–415. doi: 10.1016/j.stem.2009.03.009. [DOI] [PubMed] [Google Scholar]
- Findley SD, Tamanaha M, Clegg NJ, Ruohola-Baker H. Maelstrom, a Drosophila spindle-class gene, encodes a protein that colocalizes with Vasa and RDE1/AGO1 homolog, Aubergine, in nuage. Development. 2003;130:859–871. doi: 10.1242/dev.00310. [DOI] [PubMed] [Google Scholar]
- Forbes A, Lehmann R. Nanos and Pumilio have critical roles in the development and function of Drosophila germline stem cells. Development. 1998;125:679–690. doi: 10.1242/dev.125.4.679. [DOI] [PubMed] [Google Scholar]
- Forstemann K, Tomari Y, Du T, Vagin VV, Denli AM, Bratu DP, Klattenhoff C, Theurkauf WE, Zamore PD. Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol. 2005;3:e236. doi: 10.1371/journal.pbio.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallson G, Hollebakken RE, Li T, Syrzycka M, Kim I, Cotsworth S, Fitzpatrick KA, Sinclair DA, Honda BM. dSet1 is the main H3K4 di- and tri-methyltransferase throughout Drosophila development. Genetics. 2012;190:91–100. doi: 10.1534/genetics.111.135863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashiyama K, Shigenobu S, Kobayashi S. Expression of genes involved in sumoylation in the Drosophila germline. Gene Expr Patterns. 2009;9:50–53. doi: 10.1016/j.gep.2008.08.001. [DOI] [PubMed] [Google Scholar]
- Hayles J, Wood V, Jeffery L, Hoe KL, Kim DU, Park HO, Salas-Pino S, Heichinger C, Nurse P. A genome-wide resource of cell cycle and cell shape genes of fission yeast. Open Biol. 2013;3:130053. doi: 10.1098/rsob.130053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodl M, Basler K. Transcription in the absence of histone H3.2 and H3K4 methylation. Curr Biol. 2012;22:2253–2257. doi: 10.1016/j.cub.2012.10.008. [DOI] [PubMed] [Google Scholar]
- Hu G, Kim J, Xu Q, Leng Y, Orkin SH, Elledge SJ. A genome-wide RNAi screen identifies a new transcriptional module required for self-renewal. Genes Dev. 2009;23:837–848. doi: 10.1101/gad.1769609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janic A, Mendizabal L, Llamazares S, Rossell D, Gonzalez C. Ectopic expression of germline genes drives malignant brain tumor growth in Drosophila. Science. 2010;330:1824–1827. doi: 10.1126/science.1195481. [DOI] [PubMed] [Google Scholar]
- Jin Y, Xu J, Yin MX, Lu Y, Hu L, Li P, Zhang P, Yuan Z, Ho MS, Ji H, et al. Brahma is essential for Drosophila intestinal stem cell proliferation and regulated by Hippo signaling. Elife. 2013;2:e00999. doi: 10.7554/eLife.00999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z, Xie T. Dcr-1 maintains Drosophila ovarian stem cells. Curr Biol. 2007;17:539–544. doi: 10.1016/j.cub.2007.01.050. [DOI] [PubMed] [Google Scholar]
- Karpiuk O, Najafova Z, Kramer F, Hennion M, Galonska C, Konig A, Snaidero N, Vogel T, Shchebet A, Begus-Nahrmann Y, et al. The histone H2B monoubiquitination regulatory pathway is required for differentiation of multipotent stem cells. Mol Cell. 2012;46:705–713. doi: 10.1016/j.molcel.2012.05.022. [DOI] [PubMed] [Google Scholar]
- Kittler R, Pelletier L, Heninger AK, Slabicki M, Theis M, Miroslaw L, Poser I, Lawo S, Grabner H, Kozak K, et al. Genome-scale RNAi profiling of cell division in human tissue culture cells. Nat Cell Biol. 2007;9:1401–1412. doi: 10.1038/ncb1659. [DOI] [PubMed] [Google Scholar]
- Li Y, Maines JZ, Tastan OY, McKearin DM, Buszczak M. Mei-P26 regulates the maintenance of ovarian germline stem cells by promoting BMP signaling. Development. 2012;139:1547–1556. doi: 10.1242/dev.077412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Spradling AC. A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development. 1997;124:2463–2476. doi: 10.1242/dev.124.12.2463. [DOI] [PubMed] [Google Scholar]
- Liu N, Han H, Lasko P. Vasa promotes Drosophila germline stem cell differentiation by activating mei-P26 translation by directly interacting with a (U)-rich motif in its 3′ UTR. Genes Dev. 2009;23:2742–2752. doi: 10.1101/gad.1820709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maines JZ, Park JK, Williams M, McKearin DM. Stonewalling Drosophila stem cell differentiation by epigenetic controls. Development. 2007;134:1471–1479. doi: 10.1242/dev.02810. [DOI] [PubMed] [Google Scholar]
- McKearin DM, Spradling AC. bag-of-marbles: a Drosophila gene required to initiate both male and female gametogenesis. Genes Dev. 1990;4:2242–2251. doi: 10.1101/gad.4.12b.2242. [DOI] [PubMed] [Google Scholar]
- Minakhina S, Steward R. Hematopoietic stem cells in Drosophila. Development. 2010;137:27–31. doi: 10.1242/dev.043943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JZ, Hong A, Lilly MA, Lehmann R. twin, a CCR4 homolog, regulates cyclin poly(A) tail length to permit Drosophila oogenesis. Development. 2005;132:1165–1174. doi: 10.1242/dev.01672. [DOI] [PubMed] [Google Scholar]
- Nagengast AA, Salz HK. The Drosophila U2 snRNP protein U2A’ has an essential function that is SNF/U2B” independent. Nucleic Acids Res. 2001;29:3841–3847. doi: 10.1093/nar/29.18.3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumuller RA, Betschinger J, Fischer A, Bushati N, Poernbacher I, Mechtler K, Cohen SM, Knoblich JA. Mei-P26 regulates microRNAs and cell growth in the Drosophila ovarian stem cell lineage. Nature. 2008;454:241–245. doi: 10.1038/nature07014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumuller RA, Gross T, Samsonova AA, Vinayagam A, Buckner M, Founk K, Hu Y, Sharifpoor S, Rosebrock AP, Andrews B, et al. Conserved regulators of nucleolar size revealed by global phenotypic analyses. Sci Signal. 2013;6:ra70. doi: 10.1126/scisignal.2004145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumuller RA, Richter C, Fischer A, Novatchkova M, Neumuller KG, Knoblich JA. Genome-wide analysis of self-renewal in Drosophila neural stem cells by transgenic RNAi. Cell Stem Cell. 2011;8:580–593. doi: 10.1016/j.stem.2011.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumuller RA, Wirtz-Peitz F, Lee S, Kwon Y, Buckner M, Hoskins RA, Venken KJ, Bellen HJ, Mohr SE, Perrimon N. Stringent analysis of gene function and protein-protein interactions using fluorescently tagged genes. Genetics. 2012;190:931–940. doi: 10.1534/genetics.111.136465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni JQ, Liu LP, Binari R, Hardy R, Shim HS, Cavallaro A, Booker M, Pfeiffer BD, Markstein M, Wang H, et al. A Drosophila resource of transgenic RNAi lines for neurogenetics. Genetics. 2009;182:1089–1100. doi: 10.1534/genetics.109.103630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni JQ, Zhou R, Czech B, Liu LP, Holderbaum L, Yang-Zhou D, Shim HS, Tao R, Handler D, Karpowicz P, et al. A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat Methods. 2011;8:405–407. doi: 10.1038/nmeth.1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlstein B, McKearin D. Ectopic expression of the Drosophila Bam protein eliminates oogenic germline stem cells. Development. 1997;124:3651–3662. doi: 10.1242/dev.124.18.3651. [DOI] [PubMed] [Google Scholar]
- Page SL, McKim KS, Deneen B, Van Hook TL, Hawley RS. Genetic studies of mei-P26 reveal a link between the processes that control germ cell proliferation in both sexes and those that control meiotic exchange in Drosophila. Genetics. 2000;155:1757–1772. doi: 10.1093/genetics/155.4.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi MJ, Deng W, Wang Z, Lin H. The arrest gene is required for germline cyst formation during Drosophila oogenesis. Genesis. 2001;29:196–209. doi: 10.1002/gene.1024. [DOI] [PubMed] [Google Scholar]
- Park JK, Liu X, Strauss TJ, McKearin DM, Liu Q. The miRNA pathway intrinsically controls self-renewal of Drosophila germline stem cells. Curr Biol. 2007;17:533–538. doi: 10.1016/j.cub.2007.01.060. [DOI] [PubMed] [Google Scholar]
- Pek JW, Lim AK, Kai T. Drosophila maelstrom ensures proper germline stem cell lineage differentiation by repressing microRNA-7. Dev Cell. 2009;17:417–424. doi: 10.1016/j.devcel.2009.07.017. [DOI] [PubMed] [Google Scholar]
- Perrimon N, Engstrom L, Mahowald AP. Zygotic lethals with specific maternal effect phenotypes in Drosophila melanogaster. I. Loci on the X chromosome. Genetics. 1989;121:333–352. doi: 10.1093/genetics/121.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrimon N, Lanjuin A, Arnold C, Noll E. Zygotic lethal mutations with maternal effect phenotypes in Drosophila melanogaster. II. Loci on the second and third chromosomes identified by P-element-induced mutations. Genetics. 1996;144:1681–1692. doi: 10.1093/genetics/144.4.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrimon N, Mohler D, Engstrom L, Mahowald AP. X-linked female-sterile loci in Drosophila melanogaster. Genetics. 1986;113:695–712. doi: 10.1093/genetics/113.3.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrella LN, Smith-Leiker T, Cooley L. The Ovhts polyprotein is cleaved to produce fusome and ring canal proteins required for Drosophila oogenesis. Development. 2007;134:703–712. doi: 10.1242/dev.02766. [DOI] [PubMed] [Google Scholar]
- Schupbach T. Normal female germ cell differentiation requires the female X chromosome to autosome ratio and expression of sex-lethal in Drosophila melanogaster. Genetics. 1985;109:529–548. doi: 10.1093/genetics/109.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schupbach T, Wieschaus E. Female sterile mutations on the second chromosome of Drosophila melanogaster. II. Mutations blocking oogenesis or altering egg morphology. Genetics. 1991;129:1119–1136. doi: 10.1093/genetics/129.4.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shain AH, Pollack JR. The spectrum of SWI/SNF mutations, ubiquitous in human cancers. PLoS One. 2013;8:e55119. doi: 10.1371/journal.pone.0055119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Wong MD, Kawase E, Xi R, Ding BC, McCarthy JJ, Xie T. Bmp signals from niche cells directly repress transcription of a differentiation-promoting gene, bag of marbles, in germline stem cells in the Drosophila ovary. Development. 2004;131:1353–1364. doi: 10.1242/dev.01026. [DOI] [PubMed] [Google Scholar]
- Spradling A, Fuller MT, Braun RE, Yoshida S. Germline stem cells. Cold Spring Harb Perspect Biol. 2011;3:a002642. doi: 10.1101/cshperspect.a002642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tafforeau L, Zorbas C, Langhendries JL, Mullineux ST, Stamatopoulou V, Mullier R, Wacheul L, Lafontaine DL. The complexity of human ribosome biogenesis revealed by systematic nucleolar screening of Pre-rRNA processing factors. Mol Cell. 2013;51:539–551. doi: 10.1016/j.molcel.2013.08.011. [DOI] [PubMed] [Google Scholar]
- Van Buskirk C, Schupbach T. Half pint regulates alternative splice site selection in Drosophila. Dev Cell. 2002;2:343–353. doi: 10.1016/s1534-5807(02)00128-4. [DOI] [PubMed] [Google Scholar]
- Vinayagam A, Hu Y, Kulkarni M, Roesel C, Sopko R, Mohr SE, Perrimon N. Protein complex-based analysis framework for high-throughput data sets. Sci Signal. 2013;6:rs5. doi: 10.1126/scisignal.2003629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Pan L, Wang S, Zhou J, McDowell W, Park J, Haug J, Staehling K, Tang H, Xie T. Histone H3K9 trimethylase Eggless controls germline stem cell maintenance and differentiation. PLoS Genet. 2011;7:e1002426. doi: 10.1371/journal.pgen.1002426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Lin H. Nanos maintains germline stem cell self-renewal by preventing differentiation. Science. 2004;303:2016–2019. doi: 10.1126/science.1093983. [DOI] [PubMed] [Google Scholar]
- Xi R, Xie T. Stem cell self-renewal controlled by chromatin remodeling factors. Science. 2005;310:1487–1489. doi: 10.1126/science.1120140. [DOI] [PubMed] [Google Scholar]
- Xie T, Song X, Jin Z, Pan L, Weng C, Chen S, Zhang N. Interactions between stem cells and their niche in the Drosophila ovary. Cold Spring Harb Symp Quant Biol. 2008;73:39–47. doi: 10.1101/sqb.2008.73.014. [DOI] [PubMed] [Google Scholar]
- Xie T, Spradling AC. decapentaplegic is essential for the maintenance and division of germline stem cells in the Drosophila ovary. Cell. 1998;94:251–260. doi: 10.1016/s0092-8674(00)81424-5. [DOI] [PubMed] [Google Scholar]
- Xuan T, Xin T, He J, Tan J, Gao Y, Feng S, He L, Zhao G, Li M. dBre1/dSet1-dependent pathway for histone H3K4 trimethylation has essential roles in controlling germline stem cell maintenance and germ cell differentiation in the Drosophila ovary. Dev Biol. 2013;379:167–181. doi: 10.1016/j.ydbio.2013.04.015. [DOI] [PubMed] [Google Scholar]
- Yang L, Chen D, Duan R, Xia L, Wang J, Qurashi A, Jin P, Chen D. Argonaute 1 regulates the fate of germline stem cells in Drosophila. Development. 2007;134:4265–4272. doi: 10.1242/dev.009159. [DOI] [PubMed] [Google Scholar]
- Zeitlinger J, Zinzen RP, Stark A, Kellis M, Zhang H, Young RA, Levine M. Whole-genome ChIP-chip analysis of Dorsal, Twist, and Snail suggests integration of diverse patterning processes in the Drosophila embryo. Genes Dev. 2007;21:385–390. doi: 10.1101/gad.1509607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X, Lin X, Hou SX. The Osa-containing SWI/SNF chromatin-remodeling complex regulates stem cell commitment in the adult Drosophila intestine. Development. 2013;140:3532–3540. doi: 10.1242/dev.096891. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.