Abstract
While the methylation of DNA in 5′ promoters suppresses gene expression, the role of DNA methylation in gene bodies is unclear1–5. In mammals, tissue- and cell type-specific methylation is present in a small percentage of 5′ CpG island (CGI) promoters, while a far greater proportion occurs across gene bodies, coinciding with highly conserved sequences5–10. Tissue-specific intragenic methylation might reduce,3 or, paradoxically, enhance transcription elongation efficiency1,2,4,5. Capped analysis of gene expression (CAGE) experiments also indicate that transcription commonly initiates within and between genes11–15. To investigate the role of intragenic methylation, we generated a map of DNA methylation from human brain encompassing 24.7 million of the 28 million CpG sites. From the dense, high-resolution coverage of CpG islands, the majority of methylated CpG islands were revealed to be in intragenic and intergenic regions, while less than 3% of CpG islands in 5′ promoters were methylated. The CpG islands in all three locations overlapped with RNA markers of transcription initiation, and unmethylated CpG islands also overlapped significantly with trimethylation of H3K4, a histone modification enriched at promoters16. The general and CpG-island-specific patterns of methylation are conserved in mouse tissues. An in-depth investigation of the human SHANK3 locus17,18 and its mouse homologue demonstrated that this tissue-specific DNA methylation regulates intragenic promoter activity in vitro and in vivo. These methylation-regulated, alternative transcripts are expressed in a tissue and cell type-specific manner, and are expressed differentially within a single cell type from distinct brain regions. These results support a major role for intragenic methylation in regulating cell context-specific alternative promoters in gene bodies.
Keywords: Intragenic DNA methylation, alternate promoters, comparative epigenomics, SHANK3
To determine if intragenic DNA methylation is functional, we first generated high-resolution methylome maps of human brain frontal cortex gray matter from two individuals. We developed two complementary next-generation sequencing-based approaches to detect methylated and unmethylated DNA. The first, methylated DNA immunoprecipitation and sequencing (MeDIP-seq), uses antibody-based immunoprecipitation of 5-methylcytosine and sequencing to map the methylated fraction of the genome. In the second method, unmethylated CpG sites are identified at single CpG site resolution by sequencing size-selected fragments from parallel DNA digestions with the methyl-sensitive restriction enzymes (MREs) HpaII, Hin6I, and AciI (MRE-seq, Supplementary Fig. S1).
Of the 28 million CpGs in the haploid human genome, MeDIP-seq covered approximately 24 million at 100–300bp resolution, while MRE-seq detected approximately 1.7 million unmethylated sites at single CpG site resolution (Supplementary Figs S2–S3). The two methods detect different fractions of the genome, with more frequent MeDIP-seq reads observed in the commonly methylated CpG-poor fraction (Supplementary Fig. S4). Similar results were obtained with frontal cortex from a second individual (Supplemental Figs S5–S6; Supplemental Excel File 1).
We determined the DNA methylation status of approximately 27,100 of the 27,639 CGIs in the human genome from the combined MRE-seq and MeDIP-seq datasets (Supplementary Figs S7–S8). MRE-seq scores and MeDIP-seq scores (see Supplemental Methods) for CGIs are anti-correlated (Fig. 1a, Pearson correlation = −0.44, p<10−16). An exception is the differentially methylated regions (DMRs) of imprinted genes which have significant MRE-seq and MeDIP-seq signals (Supplementary Fig. S9). In contrast to array-based methods, MRE-seq and especially MeDIP-seq can interrogate the methylation status of a large fraction of repetitive sequences, which comprise more than 40% of the genome (Supplementary Excel File 2). Genome-wide, about 75% of repetitive regions are covered by MeDIP reads, compared to 3% for MRE-seq, consistent with high methylation of repeat sequences. Validation of MRE-seq and MeDIP-seq by standard bisulfite cloning and sequencing of 24 CGI loci (Fig. 1b; Supplementary Fig. S10a–m; Supplemental Excel File 3) supports the accuracy of MeDIP-seq and MRE-seq for determining methylation status. Across gene bodies, including CGIs and non-CGI regions, we found that the average methylation level is decreased at the 5′ ends of genes, including ~300 bp downstream of the transcription start site (TSS), where methylation might inhibit efficient inititation19, but increases in gene bodies as previously reported1,4,20,21 (Supplemental Fig. S11). However, gene bodies are often large and may contain multiple discrete regulatory sequences. This type of analysis might obscure a more specific role for DNA methylation in regulating particular regulatory sequences within gene bodies.
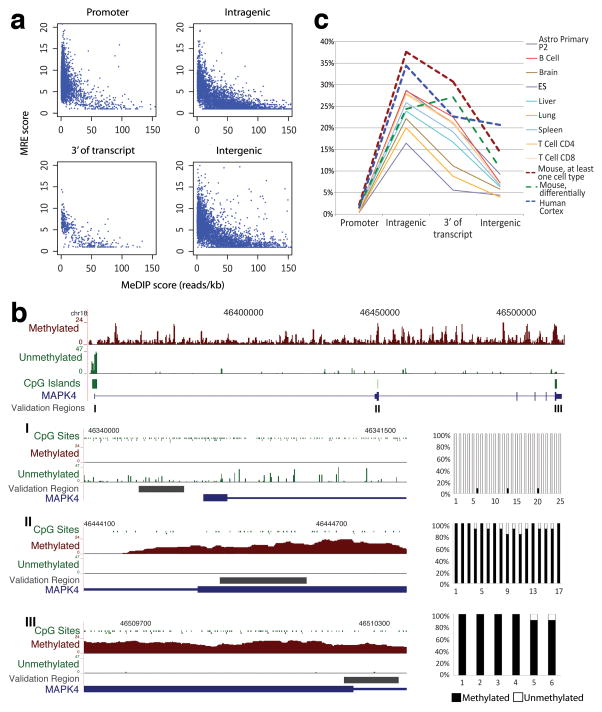
Figure 1. Tissue-specific CpG island methylation is prevalent in gene bodies and rare in 5′ promoter regions.
a, Inverse correlation between MeDIP-seq and MRE-seq in 5′ promoter, intragenic, 3′ and intergenic CGIs. Unmethylated CpGs are shown as an MRE score (a normalized number of reads interrogating each CGI, see Supplementary Methods) on the Y-axis. Methylated regions are shown as reads/kb from MeDIP-seq on the X-axis. b, Top, MAPK4 with methylated regions (MeDIP-seq, dark brown) and unmethylated CpG sites (MRE-seq, green). Zoomed-in views of each CGI are shown below, and percent methylation for each CpG site assessed by bisulfite sequencing is graphed to the right. c, Percent of CGIs that exhibit methylation in a particular tissue, methylation in one or more tissues (mouse16, at least one cell type), or tissue-specific methylation (mouse, differentially).
Since CGIs frequently overlap regulatory DNA sequences, our investigation focused on the DNA methylation status of intragenic CGIs relative to CGIs from canonical 5′ promoter regions, intergenic and 3′ regions. Overall, 16% of all CGIs in the human brain were methylated, while 98% of CGIs associated with annotated 5′ promoters were unmethylated (Fig. 1c; Supplementary Fig. S12). Notably, 34% of all intragenic CGIs were methylated (Fig. 1c). Thus, DNA methylation may serve a broader role in intragenic compared to 5′ promoter CGIs in human brain.
We next addressed whether the general pattern of frequent intragenic CGI methylation and rare 5′ promoter CGI methylation is evolutionarily conserved. Comparison of our DNA methylation profile of human brain with reduced representation bisulfite sequencing-based methylation data from mouse brain and 8 additional tissues16, showed the same general pattern (Fig. 1c). In addition, tissue-specific methylation, defined here as methylation in at least one but not all tissues, is far more common at intragenic CGIs than 5′ promoters (38% vs. 2%). The methylation status of intragenic CGIs in human and mouse brain was concordant for 80% of the orthologous CGIs (Supplementary table 1). Greater than 99% of orthologous 5′ CGIs were unmethylated in human and mouse brain tissue (Supplementary table 1). The relative lack of methylation in 5′ promoter CGIs suggests that DNA methylation at these sites has only a limited role in regulating tissue-specific transcription initiating from the canonical 5′ promoter region. In contrast, the tissue-specific and highly conserved specific pattern of intragenic CGI methylation suggests that it serves a functional role for a significant proportion of genes. The pattern of methylation in intragenic CGIs cannot be accounted for by presence of transposable elements in the CGIs, as just 1.5% of the sequences within these CGIs are annotated as repetitive (Supplementary Excel File 2).
Because many genes have alternative promoters, classically located upstream of the translation start site but also commonly present within genes15, we reasoned that a major function of the frequent, tissue-specific and conserved intragenic methylation may be to regulate the activity of such alternative promoters, as shown in two genes recently5,22. To address this hypothesis genome-wide, we determined whether the CGI loci overlap with sites of transcription initiation and/or with histone methylation marks typically found in association with 5′ promoters.
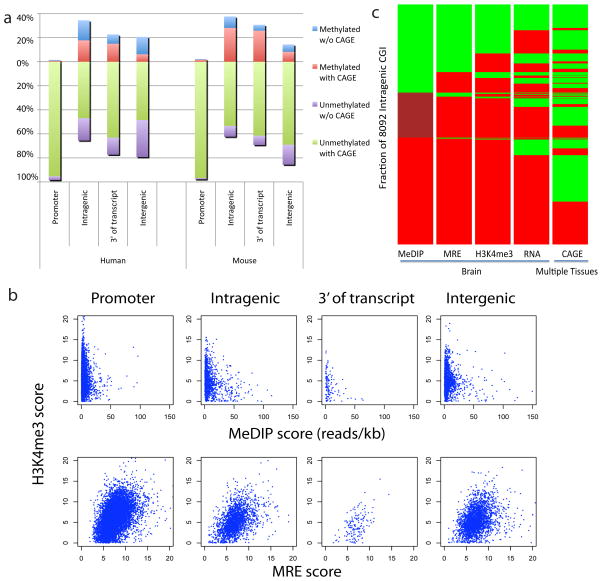
First, we assessed the relationship between the methylation status of CGIs in human brain with CAGE tag datasets from multiple human tissues12,23. CAGE tags are derived from mRNA sequenced in the proximity of the 5′-cap site and those tags that map onto unique genomic regions correspond to potential transcriptional start sites11–15,24, or in a few cases may be derived from posttranscriptionally processed RNAs25. The presence of CAGE tags from one or more tissue types suggests the underlying genomic sequence harbors a promoter, the activity of which depends on the cellular context and epigenetic status. Consistent with this notion, nearly all 5′ promoter CGIs had CAGE tag clusters mapped to them from one or more tissues (Fig. 2a), though 98% of them lack DNA methylation in human brain. CAGE tags from one or multiple tissues also mapped to intragenic, intergenic and 3′ CGIs, a significant proportion of which are methylated in brain tissue. A similar relationship between CAGE tag clusters and CGI methylation status was observed in mouse tissues (Fig. 2a). Together, these data suggest that sites of tissue-specific intragenic methylation overlap with potential alternative CGI promoters embedded within genes, and that this relationship is evolutionarily conserved.
Figure 2. Differentially methylated intragenic CGIs exhibit features of promoters.
a, Methylated CGIs are indicated above the zero line and unmethylated CGIs are below. For human, the methylation data is from frontal cortex, and CAGE tags are derived from multiple tissues11,23. For mouse, the methylation data includes the same set of tissues described in figure 1c, and CAGE data are derived from multiple mouse tissues11,12. 91% of human intragenic CGI CAGE tags mapped outside of exons and are probably not derived from posttranscriptional processing. b, H3K4me3 tissue-ChIP-seq normalized internal coverage (NIC) scores compared to MeDIP- and MRE-seq methylation data at CGIs for human frontal cortex. c, Heatmap view of the status of 8092 intragenic CGIs based on five genome-wide datasets. Each island is coloured according to its status and sorted from top to bottom in the order of increasing signal in MeDIP-seq, then within the three MeDIP-defined subgroups by signals in MRE-seq. This process is performed iteratively based on H3K4me3, RNA-seq TSS and CAGE status. For MeDIP-seq, green indicates unmethylated (0–20 reads/kb), maroon indicates partially methylated (20–50 reads/kb), and red indicates methylated (>50 reads/kb); For MRE-seq, green indicates unmethylated (MRE score 0–5), red indicates methylated (MRE score >5); For H3K4me3 ChIP-seq, green indicates active/with signal, red indicates inactive/without signal. For RNA-seq TSS, green indicates evidence for TSS, red indicates lack of evidence for TSS (see Supplemental Methods). For CAGE, green indicates CAGE tags from one or more tissues that overlap the CGI; red indicates lack of overlapping CAGE tags.
To further test the hypothesis that a significant fraction of intragenic CGIs function as alternate promoters, we generated a map of trimethylation of histone H3 lysine 4 (H3K4me3), an epigenetic mark that coincides with promoters, by ChIP-seq on human brain. Unmethylated 5′ CGI promoters and H3K4me3 overlapped significantly in human brain (Fig. 2b; Supplementary Fig. S13), as observed in mouse16. Interestingly, for intragenic CGIs the degree of DNA methylation correlated inversely with the level of H3K4me3 signal (Pearson correlation −0.46, p<10−10). The strong overlap of H3K4me3 with unmethylated intragenic CGIs, the inverse correlation between H3K4me3 signal and intragenic CGI DNA methylation, and the presence of CAGE tags from one or more tissues suggests that these intragenic sites function as alternative promoters, 34% of which exhibit tissue-specific methylation. In data from mouse tissues11,16, we found a strong inverse correlation between level of DNA methylation and presence of CAGE tags at intragenic CGIs in liver, lung, and brain (Supplementary Fig. S14).
We next performed genome-wide expression profiling using whole-transcriptome shotgun sequencing (WTSS), also known as RNA-seq26, on the human frontal cortex sample for which we had generated MeDIP-seq, MRE-seq and H3K4me3 ChIP-seq datasets. The cDNA library construction protocol employed enriches for full-length mRNAs and tags their 5′ ends, and in conjunction with computational detection and clipping of these 5′ tags, followed by mapping of the adjacent cDNA sequence, allows the inference of putative TSS (Supplemental Methods). Unmethylated, H3K4me3-positive intragenic CGIs were associated with putative TSS significantly more often than methylated, H3K4me3-negative intragenic CGIs. The relationship between DNA methylation, H3K4me3 and transcription initiation sites is further illustrated by a heatmap view of all intragenic CGIs based on five independent experiments (Fig. 2c; Supplementary Fig. S15; Supplementary Table 2). Thus, our RNA-seq data complement the observations made with CAGE tag datasets, and further strengthen the hypothesis that intragenic methylation regulates alternative promoters.
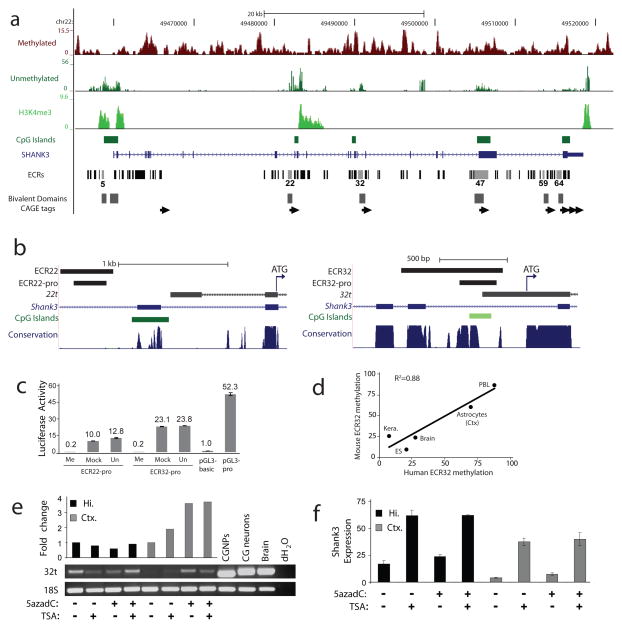
In parallel with the genome-wide analyses, we investigated in-depth a single locus with a 5′ promoter CGI, two conserved intragenic CGIs, one conserved 3′CGI, and one additional intragenic CGI in humans not present in mice. Our prior analysis of this locus, the autism and 22q deletion syndrome gene SHANK317,18, demonstrated evolutionarily conserved and tissue-specific intragenic methylation at one CGI7. The 5′ promoter CGI of SHANK3 was unmethylated, while one intragenic and one 3′ CGI exhibited methylation and two intragenic CGI were predominantly unmethylated (Fig. 3a). Bisulfite sequencing across matched tissues from mice and humans revealed strongly conserved patterns of DNA methylation in SHANK3 (Supplementary Fig. S16). The 5′ CGI was unmethylated in all tissues analysed in both species, irrespective of SHANK3 expression.
Figure 3. Novel transcripts initiate from differentially methylated, evolutionarily conserved intragenic promoters in a cell context-dependent manner.
a, Human frontal cortex MRE-seq, MeDIP-seq and H3K4me3 ChIP-seq at SHANK3 (top). Evolutionarily conserved regions (ECRs) overlap with mouse CAGE tag clusters (arrows), mouse ES H3K4me3 and H3K27me3 bivalent domains27 and human frontal cortex H3K4me3. ECRs with most or all promoter-associated features are shown with light grey bars. b, Diagram of ECR22 (left) and ECR32 (right) mouse genomic regions displaying from top to bottom ECRs, sequences used for promoter assays, 5′ RACE sequences of 22t and 32t with associated ATGs (arrow), known exons, CpG island (dark green) and CpG-rich (light green) regions, and multi-species DNA sequence conservation. c, In vitro methylation of the mouse SHANK3 intragenic promoters abolished their activity in promoter assays. Me, methylated; Mock, mock treated; Un, untreated. d, Bisulfite sequencing of ECR32 in matched tissues/cells from human and mouse. P=0.018; ANOVA regression analysis. e, Increased 32t transcription in cortical, but not hippocampal astrocytes after treatment with 5azadC by transcript-specific RT-PCR (p<0.05, Student’s t-test). Positive controls: untreated primary cultures of cerebellar granule neural progenitor cells (CGNPs), their in vitro differentiated neurons (CG neurons), and whole brain. The 24-bp size difference in CGNPs and CG neurons is due to alternative splicing within the 32t transcript. Hi., hippocampal; Ctx., cortical. f, Increased expression of full-length SHANK3 detected by qRT-PCR in astrocytes treated with TSA alone or in combination with 5azadC (p<0.05, Student’s t-test) but not 5azadC alone.
We first searched for in vivo evidence of promoters embedded within SHANK3 by integrating sequence conservation (ECRs), evidence of transcription initiation in both mouse and human tissues (CAGE tags), the presence of H3K4me3 in human brain as well as overlapping H3K4me3 and H3K27me3 peaks from ChIP-Seq analyses of ES cells27. Five intragenic regions were identified with most or all of these features (Fig. 3a). For two intragenic CGIs, we used 5′-RACE to confirm intragenically initiating transcripts in brain, but not lung, originating from ECR22 (transcript 22t) and ECR32 (transcript 32t) in mouse and human tissue (Fig. 3b and data not shown). Both 22t and 32t are comprised of unique first exons and downstream sequences that correspond to the known exons of the full-length SHANK3, and contain conserved translational start sites in-frame with the full-length SHANK3 protein (Fig. 3b). ECR22 and ECR32 harbor significant promoter activity, which is abolished by in vitro methylation (Fig. 3c and Supplementary Fig. S17). In vivo, the DNA methylation status of ECR22 and ECR32 promoters is inversely correlated with 22t and 32t transcription, respectively, and their expression patterns are similar in matching mouse and human tissues (Supplementary Fig. S18). In particular, the tissue-specific DNA methylation levels of ECR32 are also cell-type and brain-region specific (Supplementary Figs S18–19), and evolutionarily conserved (Fig. 3d). Treatment of primary cortical astrocytes with a DNA methylation inhibitor increased transcripts from the normally methylated ECR32 intragenic promoter (Fig. 3e), but had no effect on the full-length transcript originating from the constitutively unmethylated 5′ promoter CGI (Fig. 3f). Conversely, treatment with a histone deacetylase inhibitor activated the full-length transcript significantly with little change to 32t expression (Fig. 3f). Combined inhibition of DNA methylation and HDAC activity did not increase 32t beyond the effect of blocking DNA methylation alone (Fig. 3e), nor did it increase the full-length transcript expression beyond HDAC inhibition alone (Fig. 3f). Interestingly, primary astrocytes derived from the hippocampus exhibited opposite methylation and expression levels of ECR32 and 32t relative to cortical astrocytes (Supplementary Fig. S19 and Fig. 3e). Additionally, unlike cortical astrocytes, the level of 32t expression in hippocampal astrocytes remained unchanged after HDAC and DNA methylation inhibition (Fig. 3e). In contrast, an increase in expression of the full-length SHANK3 was observed in both astrocyte populations following treatment with an HDAC inhibitor (Fig. 3f). Thus, in addition to the brain-region specific differences between astrocytes, the full-length SHANK3 and 32t appear to be regulated by distinct epigenetic mechanisms within the same cells. Similarly, an intragenic CGI in a second gene, Nfix, also functions as a methylation-regulated intragenic promoter (Supplemental Fig. S20)
Increased gene body methylation correlates with increased transcription genome-wide1,2,4,5, which is seemingly contradictory to our main conclusion. Indeed, in our human brain data, moderately expressed genes exhibited greater gene body methylation on average (Supplementary Fig. S21). However, these correlations use the average methylation level over the entire gene body rather than examining specific CGI sites with potential regulatory function, and involve gene expression measurements that do not discriminate which transcripts are being measured when multiple overlapping transcripts are present. In contrast, the integration of CAGE tags, H3K4me3 peaks and RNA-seq-inferred TSS allow precise mapping of genomic sites of transcription initiation and promoter function.
Despite the stereotype, DNA methylation does not appear to play a major role in gene regulation from 5′ CGI promoters of most autosomal genes, where histone acetylation and histone methylation may be more relevant. Our study also highlights an underappreciated complexity of DNA methylation-associated regulation of alternative promoters within gene bodies, including differences in this regulation within a single cell type from distinct brain regions, and in different regions of the same gene in the same cell. In light of the precision afforded by our approach and the new conclusions drawn from it, it may now be possible to reconcile prior controversies on the role of DNA methylation in the regulation of gene expression during development and cancer28,29. The role of intragenic DNA methylation is but one of many possible important new advances afforded by the synthesis of integrative epigenomics and comparative genomics.
Online-Only Methods
DNA isolation
Cells were lysed in DNA extraction buffer (50 mM Tris pH 8.0, 0.5% sodium dodecyl sulfate, 0.5 mM EDTA pH 8.0, and 1 mg/ml proteinase K) overnight at 55° C. RNA was removed with RNase treatment (40 μg/ml, Roche DNase-free RNase) for 1 hr at 37° C. DNA was purified with 2 phenol/chloroform/isoamyl alcohol extractions followed by 2 chloroform extractions using phase lock gels. DNA was precipitated with sodium acetate and ethanol, washed with 70% ethanol, and resuspended in TE buffer.
MRE-seq
Three parallel digests were performed (HpaII, AciI, and Hin6I; Fermentas), each with 1–5 μg of DNA. Five units of enzyme per microgram DNA were added and incubated at 37° C in Fermentas “Tango” buffer for 3 hrs. A second dose of enzyme was added (5 units of enzyme per microgram DNA) and the DNA was incubated for an additional 3 hrs. Digested DNA was precipitated with sodium acetate and ethanol, and 500 ng of each digest were combined into one tube. Combined DNA was size-selected by electrophoresis on a 1% agarose TBE gel. A 100 – 300 bp gel slice was excised using a sterile scalpel and gel-purified using Qiagen Qiaquick columns, eluting in 30 μl of Qiagen EB buffer. Library construction was performed using the Illumina Genomic DNA Sample Kit (Illumina Inc., USA) with single end adapters, following the manufacturer’s instructions with the following changes. For the end repair reaction, T4 DNA polymerase and T4 polynucleotide kinase were excluded and the klenow DNA polymerase was diluted 1:5 in water and 1 μl used per reaction. For single end oligo adapter ligation, adapters were diluted 1:10 in water and 1 μl used per reaction. After the second size selection, DNA was eluted in 36 μl EB buffer using Qiagen Qiaquick columns, and 13 μl used as template for PCR, using Illumina reagents and cycling conditions with 18 cycles. After cleanup with Qiagen MinElute columns, each library is examined by spectrophotometry (Nanodrop, Thermo Scientific, USA) and Agilent DNA Bioanalyzer (Agilent, USA).
MeDIP-seq
For MeDIP, 5–15 μg DNA isolated as described above was sonicated to ~100–500 bp with a Bioruptor sonicator (Diagenode). Sonicated DNA was end-repaired, A-tailed, and ligated to single-end adapters following the standard Illumina protocol. After agarose size-selection to remove unligated adapters, 2–5 μg of adapter-ligated DNA was used for each immunoprecipitation using a mouse monoclonal anti-methylcytidine antibody (1 mg/ml, Eurogentec, catalog # BI-MECY-0100). For this, DNA was heat denatured at 95° C for 10 minutes, rapidly cooled on ice, and immunoprecipitated with 1 μl primary antibody per microgram of DNA overnight at 4° C with rocking agitation in 500 μl IP buffer (10 mM sodium phosphate buffer, pH 7.0, 140 mM NaCl, 0.05% Triton X-100). To recover the immunoabsorbed DNA fragments, 4 μl of rabbit anti-mouse IgG secondary antibody (2.5 mg/ml, Jackson Immunoresearch) and 100 μl Protein A/G beads (Pierce Biotechnology) were added and incubated for an additional 2 hr at 4° C with agitation. After immunoprecipitation a total of 6 IP washes were performed with ice cold IP buffer. A nonspecific mouse IgG IP (Jackson Immunoresearch) was performed in parallel to methyl DNA IP as a negative control. Washed beads were resuspended in TE with 0.25% SDS and 0.25 mg/ml proteinase K for 2 hrs at 55° C and then allowed to cool to room temperature. MeDIP and supernatant DNA were purified using Qiagen MinElute columns and eluted in 16 μl EB (Qiagen, USA). Fifteen cycles of PCR were performed on 5 μl of the immunoprecipitated DNA using the single end Illumina PCR primers. The resulting reactions are purified over Qiagen MinElute columns, after which a final size selection (192–392 bp) was performed by electrophoresis in 2% agarose. Libraries were QC’d by spectrophotometry and Agilent DNA Bioanalyzer analysis. An aliquot of each library was diluted in EB to 5 ng/μl and 1 μl used as template in 4 independent PCR reactions to confirm enrichment for methylated and de-enrichment for unmethylated sequences, compared to 5 ng of input (sonicated DNA). Two positive controls (SNRPN and MAGEA1 promoters) and 2 negative controls (a CpG-less sequence on Chr15 and GAPDH promoter) were amplified (see Supplementary Materials for primer sequences). Cycling was 95° C for 30 s, 58° C for 30 s, 72° C for 30 s with 30 cycles. PCR products were visualized by 1.8% agarose gel electrophoresis.
ChIP-seq of H3K4me3
Human left hemisphere frontal cortex (Brodmann Area 10) was obtained from the Québec Suicide Brain Bank (QSBB, Montreal, Québec; http://www.douglasrecherche.qc.ca/brain-banks/suicide-bank.asp). All tissue was collected with written informed consent from next of kin. Experimentation with human brain tissue at the Genome Sciences Centre was carried out with approval from the University of British Columbia - British Columbia Cancer Agency Research Ethics Board (REB# H07-01589). For immunoprecipitation of H3K4me3-modified chromatin, human frontal cortex tissue (200–500mg each) from a 57 year old male suspended in chilled douncing buffer (250 μl; 10mM Tris-Cl pH7.5, 4mM MgCl2, 1mM CaCl2), and homogenized by repeated pipetting followed by passing through a 1 ml 26 gauge-syringe 6 times. The homogenate was then incubated with 5U/ml of micrococcal nuclease (Sigma, USA) for 7 min at 37°C (~90% was mononucleosomes after digestion). The reaction was terminated by addition of EDTA (10mM; ~5 μl). To this, 1 ml hypotonic lysis buffer (0.2mM EDTA (pH8.0), 0.1mM benzamidine, 0.1mM PMSF, 1.5mM DTT) with protease inhibitor cocktail was added. The homogenate was incubated on ice for 60 min, with brief vortexing at 10 min intervals. The homogenate was centrifuged at 3000g for 5 min, and the supernatant was transferred to a 1.5 ml non-stick tube. The micrococcal nuclease-digested chromatin fraction was pre-cleared with 100 μl of blocked Protein A/G sepharose beads (Amersham, USA) at 4°C for 2 hrs, and following centrifugation and the supernatant was transferred to fresh tubes. Chromatin immunoprecipitation was carried out either with anti-histone H3 trimethyl K4 (H3K4me3) antibody (ab8580, Abcam), or normal rabbit IgG antibody (12–370, Upstate Biotechnology) to assess fold enrichment. Antibodies were added in manufacturer recommended amounts, and the mixtures incubated at 4°C for 1 hr. To each reaction mixture, 20 μl of Protein A/G beads were added and incubated by rotating at 4°C overnight. Beads were recovered by centrifugation and washed twice with ChIP wash buffer (20 mM Tris-HCl [pH 8.0], 0.1% SDS, 1% Triton X-100, 2 mM EDTA, 150 mM NaCl) and once with ChIP final wash buffer (20 mM Tris-HCl [pH 8.0], 0.1% SDS, 1% Triton X-100, 2 mM EDTA, 500 mM NaCl). DNA-antibody complexes were eluted using 100 μl elution buffer (100 mM NaHCO3, 1% SDS), and incubated with 5 μg of DNAse-free RNAse (Roche, Canada) at 68°C for 2 hrs. The beads were pelleted by centrifugation and the supernatant was collected. Elution was repeated with addition of 100 μl of elution buffer and incubation at 68°C for 5 min. After pooling the two eluates, DNA was recovered using the QIAquick PCR Purification kit (Qiagen, Germany). A ChIP-seq library were constructed as previously described using 11–35 ng of immunoprecipitated DNA.
Categorization of CpG islands
We obtained genomic locations of CpG islands from the UCSC Genome Browser for human (hg18, 27639 islands) and mouse (mm8, 15948 islands). We obtained RefSeq gene definition from the UCSC Genome Browser for human (hg18, 29996 genes) and mouse (mm8, 22307 genes). We grouped CpG islands into four classes based on their distance to RefSeq genes. They are:
promoter islands (if an island ends after 1000bp upstream of a RefGene transcription start site, and starts before 300bp downstream of a RefGene transcription start site);
intragenic islands (if an island starts after 300bp downstream of a RefGene transcription start site and ends before 300bp upstream of a RefGene transcription end site);
3′ transcript islands (if an island ends after 300bp upstream of a RefGene transcription end site and starts before 300bp downstream of a RefGene transcription end site);
intergenic islands (if an island starts after 300bp downstream of a RefGene transcription end site and ends before 1000bp upstream of a RefGene transcription start site.
See Supplemental Fig. S12 for number of different classes of CpG islands in the human and mouse genome.
Definition of Islands with no CpG
We identified 94,239 CpG free regions in the human genome assembly (hg18) that span between 1kb to 3kb. We defined the middle 600bp of these regions to be islands with no CpG.
DNA methylation score for the mouse
We obtained reduced representation bisulfite sequencing data from Meissner et al. 2008. We included data on the following cell types in this analysis: Astro_primary_p2, B cell, Brain, ES cell, Liver, Lung, Spleen, T cell CD4, and T cell CD8. Methylation score for individual CpG site is defined as number of CG/(CG+TG) from bisulfite sequencing reads. A CpG site will have a defined methylation score only when CG+TG is equal or greater than 5; otherwise, the score is undefined. Methylation score for individual CpG island is defined as the average score of all CpG sites with a defined methylation score within this island. The score is multiplied by 1000.
A CpG island is defined as completely methylated if its methylation score is equal or greater than 500; as partially methylated if its methylation score is between 100 and 500; and as unmethylated if its methylation score is less than 100.
MeDIP-seq and methylation score for the human
We sequenced the same sample on Illumina GAI and GAII with a total number of reads about 106 million. Redundant reads were removed, and 47 million reads were mapped to the current human genome assembly (hg18) with MAQ. We extended each mapped reads to 200bp in length. Overall, 24 million CpG sites are covered by at least one extended read. We define a methylation score for any region in the genome as number of extended reads per kb. A CpG island is defined as unmethylated if its methylation score is less than 20 reads/kb, as partially methylated if its methylation score is between 20 and 50 reads/kb, and as completely methylated if its methylation score is greater than 50 reads/kb. See Supplementary Fig. S3 for distribution of MeDIP-score across CpG sites and Fig S8 for MeDIP-score across CpG islands.
MRE-seq and MRE-score for the human
We sequenced the same sample with Illumina GAI and GAII with a total number of reads about 20 million. We mapped these reads to the human genome assembly (hg18) with MAQ with an additional constraint that the 5′ end of a read must map to the CpG site within a MRE site. This resulted in about 11 million mapped MRE-reads. About 1.5 million CpG sites have at least one mapped MRE-read. We define MRE-score for each CpG site as the number of MRE-reads that map to the site, regardless of the orientation. We define MRE-score for each CpG island as the average MRE-score for all CpG sites that have a score within the island. See Supplementary Fig. S2 for a distribution of MRE-score across CpG sites and Fig S7 for MRE-score across CpG islands.
NIC (Normalized Internal Coverage) score
For any genome-wide data presented in wiggle format, NIC for any given region is defined as the total area of the data profile within the region normalized by the length of the region. See Supplementary Fig. S13 for distribution of NIC scores of CpG islands with respect to H3K4me3.
CAGE association
We used published CAGE data from mouse and human. Tissue-specific CAGE data is available as “wiggle” tracks. For each CpG island, we extend the island boundary by 200bp in both upstream and downstream directions. If the extended island overlaps with any wiggle signal from the CAGE dataset, we calculate NIC score for the island.
Identifying conserved CpG islands between human and mouse
We first syntenically mapped all human CpG islands to the mouse genome assembly (mm8) and filtered out those that don’t map. We further filtered out ones that when mapped to the mouse, they do not overlap annotated CpG islands. Next, we compared classification of these islands (promoter, intragenic, 3′ of transcript or intergenic) and filtered out those pairs whose classifications do not match. This results in 2400 pairs of conserved CpG islands between human and mouse, 500 of which are intragenic.
RNA-seq; Identification of putative transcription start sites; Gene expression measurements
100 ng of total RNA was used to synthesize full-length single-stranded cDNAs using the SMART PCR cDNA Synthesis Kit (Clontech, Mountain View, CA, USA) following the protocol as described by Morin et al. The resulting double-stranded cDNAs was assessed using an Agilent DNA 1000 series II assay (Agilent, Mississauga ON, Canada) and Nanodrop 7500 spectrophotometer (Nanodrop, Wilmington, DE, USA). Sonication was performed for a total of 50 minutes using Bioruptor UCD-200 (Diagenode Inc. Sparta, NJ, USA). The sheared cDNA was size separated by 8% PAGE and the 200–250bp DNA fraction excised and eluted from the gel slice overnight at 4 °C in 300 μl of elution buffer (5:1, LoTE buffer (3 mM Tris-HCl, pH 7.5, 0.2 mM EDTA)-7.5 M ammonium acetate), and purified using a QIAquick purification kit (Qiagen, Mississauga, ON, Canada). The library was constructed following the Illumina genomic DNA paired end library protocol with 10 cycles of PCR (Illumina Inc., Hayward CA, USA). The resulting PCR product was purified using 8% PAGE to remove small products including adapter dimers, and the DNA quality was assessed using an Agilent DNA 1000 series II assay and quantified by Qubit fluorometer (Invitrogen, Burlington, ON, Canada) and then diluted to 10nM. The final concentration was double checked and determined by Quant-iT dsDNA HS Assay Kit using Qubit fluorometer (Invitrogen). Cluster generation and paired-end sequencing was performed on the Illumina cluster station and Genome Analyzer following manufacturer’s instructions (Illumina Inc., Hayward CA, USA).
In total, 93 million paired-end reads (186 million reads) were generated for the frontal cortex WTSS-lite library. Custom scripts were used to identify 56.4 million reads that contained the SMART oligo sequence and a variable G stretch (added by the RT terminal transferase activity) on the 5′ end. Putative TSS were found by identifying WTSS reads containing sequence corresponding to SMART oligo tags, clipping these tags informatically, and aligning the resulting sequence tag (representing the 5′ end of a full-length mRNA) using Maq. In detail: paired end reads were split into forward (read1) and reverse (read2) reads. Read 1s were parsed for those which contained reads starting with the SMART tag followed by a variable number of Gs and clipped after the terminal G. These variable length sequence strings were written to the SMART file (56.4 million reads). All Read2s and those Read1s that did not contain the SMART sequence tag were written to a NOSMART file (129.6 million reads). The SMART file was split into 14 subfiles based on read length and Maq (0.7.1) alignments were run and the resulting .map files merged. The NOSMART file was split into 2 subfiles (for the 75 and 50bp read lengths), and Maq aligned and the resulting .map files merged. The .map files were used to generate SMART and NOSMART wig tracks using FindPeaks 2 (xset5; no threshold). For gene expression analysis, the clipped and non-clipped reads were pooled (SMART and NOSMART .map files merged), and read counts generated at the exon and gene level using custom scripts.
To assess promoter activities of individual CpG islands, we first extended each island boundary by 200bp in both upstream and downstream directions and looked for evidence of TSS based on RNA-seq data in these regions. We tallied number of SMART and NOSMART RNA-seq reads overlapping with each island, and defined TSS activity as (1) having at least 5 SMART tagged reads, and (2) at least 70% of total RNA-seq reads are SMART tagged reads.
Normal tissues and cultured primary cells
For the SHANK3 experiments normal human brain samples were provided from the Neurosurgery Tissue Bank at the University of California San Francisco (UCSF) and we collected adult peripheral blood lymphocytes (PBL) from healthy volunteers. All samples were obtained with informed consent, and their use was approved by the Committee on Human Research at UCSF. Normal human primary adult keratinocytes and normal human fetal astrocytes were purchased from Cambrex and were cultured for fewer than three passages. Normal human ES cells (HSF6) were kindly provided by Mary Firpo while at UCSF. Mouse whole brain, cerebella, hippocampi, lung, pancreas, heart, PBL, and sperm were isolated from normal 8-week old C57BL/6J mice. Keratinocytes from the skin of normal newborn NIH/Ola pups were isolated by physical separation of the epidermal layer from whole skin. In addition to adult stages, brain and lung tissues were derived from mice at pre- and post-natal developmental time points where indicated in the text. Astrocyte monolayers were derived from the postmortem cerebral cortex and hippocampus of postnatal day 7 C57BL/6J mice. The cerebral cortex dissection was preformed in such a way as to exclude all cells of the ventricular or subependymal region. Primary cultures were generated by mincing the tissue and incubating it with papain enzyme, after which cells were filtered through a 70 μm cell strainer. The resulting cell suspensions were seeded on laminin coated plates in DMEM/F12 medium containing 10% (vol/vol) FCS supplemented with 2 mM glutamine and allowed to grow to confluence. The cells were confirmed to be astrocytes based on morphology and expression of the astrocyte-specific glial fibrillary acidic protein. Mouse ES cells (from C57BL/6J blastocysts) were kindly provided by Miguel Ramalho-Santos (UCSF). All tissue samples were homogenized for isolation of nucleic acids. All cultured cells were collected by trypsinization using 0.25% trypsin-EDTA and washed before cell lysis.
Demethylation and deacetylation experiments
Primary mouse astrocytes were seeded at 1 × 105 cells per well of a six-well plate, incubated for 24 hours in Dulbecco’s Modified Eagle’s Medium (DMEM) high glucose with 10% serum, and then supplemented with fresh media containing 5-aza-2′-deoxycytidine (5azadC) (1 or 5 μM; Sigma-Aldrich) for 72 hours or trichostatin A (TSA) (100 ng/ml; Sigma-Aldrich) for 12 hours. For the combination treatment, 1 or 5 μM 5azadC was present for 72 hours and TSA was added for the last 12 hours. The media containing drugs were changed every 24 hours.
Bisulfite treatment, PCR and sequencing
We treated total genomic DNA with sodium bisulfite for 16 hours and carried out PCR using primers listed in Supplementary Table 2, and cloned products into pCR2.1/TOPO (Invitrogen). We selected a specified number of individual colonies and sequenced inserts using the ABI 3700 automated DNA sequencer. DNA methylation patterns and levels were determined only from highly (>95%) converted sequences.
5′-Rapid amplification of cDNA ends
Total RNA from brain and lung of normal 8-week old C57BL/6J mice were used to amplify the 5′ end of SHANK3 mRNA with the Gene Racer kit (Invitrogen) based on the protocol supplied by the manufacturer. The mRNA was ligated to the Gene Racer oligo, reverse-transcribed, and amplified using SHANK3-specific reverse primers R1 or R2 (Supplementary Table 2) with PfuUltra high-fidelity DNA polymerase (Stratagene) under the following 3-step ‘touch-down’ cycling parameters: (1) 5 cycles of 94°C for 30 sec, 72°C for 1 min, (2) 5 cycles of 94°C for 30 sec, 70°C for 1 min, (3) 30 cycles of 94°C for 30 sec, 62°C for 30 sec, and 72°C for 1 min, followed by 72°C for 10 min. The amplification products were gel purified, cloned into pCR4-TOPO (Invitrogen), and inserts were sequenced. The sequence data for the novel SHANK3 transcripts, 22t and 32t, have been deposited into the dbEST database and correspond to accession numbers GD253656 and GD253657, respectively. The unique first exon sequences of 22t and 32t correspond to chr15:89,354,730–89,355,012 and chr15:89,363,250–89,363,804, respectively (Mouse July 2007 assembly; http://genome.ucsc.edu). Another transcript with a transcription start site downstream of 32t and lacking the full-length SHANK3 exon 18 was also identified by 5′-RACE (accession number: GD253658).
Reverse transcription, standard and real-time reverse transcription-PCR
Reverse transcription reactions were performed essentially as previously described. From mouse samples, we measured the expression of full-length SHANK3 and an internal control GusB with probe/primer assays Mm00498775_m1 and Mm00446953_m1 (Applied Biosystems), respectively, by real-time RT-PCR using the Opticon2 Continuous Fluorescence Detector (MJ Research) and calculated relative expression levels using the deltaCt-method. Expression levels of 22t and 32t were measured by RT-PCR using 18S and β-actin as internal controls for mouse and human samples, respectively. Primers and their corresponding PCR conditions are listed in Supplementary Table 1.
Integration of promoter-associated features at SHANK3
For the SHANK3 locus (chr15:89,328,288–89,388,754; Mouse July 2007 assembly), we combined three distinct ‘features’ associated with promoters described in the text. We identified ECRs throughout SHANK3 using ‘ECR Browser’: http://ecrbrowser.dcode.org. CAGE tag sequences along SHANK3 were obtained from: http://fantom31p.gsc.riken.jp/cage_analysis. ECRs with 4 or more CAGE tags are shown with arrows in Fig 3a. ChIP-Seq data of H3K4me3 and H3K27me3 marks across SHANK3 in ES cells were obtained from: http://www.broad.mit.edu/seq_platform/chip. Because all of these features are sequence-based, we were able to precisely align them in relationship to the corresponding SHANK3 genomic sequence.
Cloning of ECRs, transfection, and promoter-reporter assays
From mouse or human genomic DNA, selected ECR sequences were PCR amplified with PfuUltra high-fidelity DNA polymerase (Stratagene) using primers designed to contain specific restriction sites (Supplementary Table 2). We subcloned each PCR product into the TOPO-TA cloning vector, selected and sequenced positive colonies, and isolated plasmid DNA containing correct insert sequences. We digested the plasmids, gel-purified the inserts, and re-ligated them into a similarly digested pGL3-Basic vector (Promega). We screened for and confirmed positive colonies by restriction digestion and sequencing, respectively, and isolated plasmid DNA. Using the FuGENE6 reagent (Roche) and according to the manufacturer’s instructions, 1 μg of each construct and 10 ng of an internal control vector (pRL-hTK; Promega) were co-transfected into HEK-293 cells that were cultured in six-well plates containing DMEM media with 10% serum. The pGL3-Basic vector without insert and the pGL3 vector containing an SV40 promoter served as negative and positive controls, respectively. Firefly luciferase and Renilla luciferase activities were each measured 48 hours after transfection by the Dual-Luciferase Reporter Assay System (Promega). As a measure of ‘promoter’ strength, luciferase activities were calculated from the intensity of light produced as a consequence of beetle luciferin oxidation by Firefly luciferase expressed from each ECR construct relative to that of the promoter-less pGL3-basic vector after normalizing for transfection efficiency as measured by the intensity of light produced as a consequence of coelenterazine oxidation by Renilla luciferase expressed from a co-transfected plasmid. Sequences containing promoter activity within ECR5, ECR22, and ECR32 have been deposited into the GenBank database and correspond to accession numbers FJ215690, FJ215689, FJ215688, respectively.
In vitro DNA methylation assay
Each pGL3-ECR promoter construct was treated with 2 mM S-adenosylmethionine (New England Biolabs) in the presence (methylated) or absence (‘mock’-methylated) of 6 units of M.SssI (CpG) methylase per μg of DNA for 4 hours at 37°C. Aliquots of purified constructs were digested with HpaII to confirm the methylation status (data not shown).
Supplementary Material
Acknowledgments
We thank Scott Vandenberg for technical assistance and The Pleiades Promoter Project and their funders Genome Canada, Genome British Columbia, GlaxoSmithKline R&D Ltd., BC Mental Health and Addiction Services, Child & Family Research Institute, UBC Institute of Mental Health, and UBC Office of the Vice President Research. This work was supported in part by an NIH NRSA-F31 fellowship to A.M. and an NIH NRSA-F32 fellowship to R.P.N., a grant from the National Brain Tumor Society and Goldhirsh Foundation to J.F.C., and by the British Columbia Cancer Foundation. T.W. was a Helen Hay Whitney Fellow and M.A.M. is a Terry Fox Young Investigator and a Michael Smith Senior Research Scholar.
Footnotes
Supplementary Information is linked to the online version of this paper at www.nature.com/nature.
Author Contributions
A.K.M. conceived and performed SHANK3 experiments; R.P.N. designed and performed MeDIP-seq and MRE-seq and qRT-PCR; M.B., C.D., C.N., Y.Z., G.T. and S.S.J.J. performed and analysed brain ChIP-seq; M.A.M., M.H., Y.J. supervised and analysed IGAII sequencing, and participated in project coordination; S.D.F. performed bisulfite sequencing. C.H. performed bisulfite sequencing and luciferase assay experiments; B.E.J. helped perform MRE-seq and bisulfite sequencing. A.D. wrote the script to parse the SMART and non-SMART containing tags from RNA-seq data. R.V. performed the iterative alignments from RNA-seq and N.T. generated the gene expression measures from the alignments. K.S., V.M.H., and D.H.R. performed mouse brain dissections and isolated astrocytes, neurons and neuronal precursors; T.W., T.J.B., X.X., C.F., M.M performed bioinformatics analyses. D.H. participated in project coordination and SHANK3 genomic conservation analysis. A.K.M., R.P.N., T.W. and J.F.C. coordinated the project, wrote the manuscript and incorporated revisions from co-authors.
Sequencing reads are available through the NCBI SRA, accession number SRP002318 (http://www.ncbi.nlm.nih.gov/sra/?term=SRP002318). Browser tracks (hg18 assembly) are available at http://genome.ucsc.edu/. The sequence data for the novel SHANK3 transcripts, 22t and 32t, have been deposited into the dbEST database (accession numbers GD253656 and GD253657, respectively).
Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
References
- 1.Cokus SJ, et al. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452 (7184):215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Flanagan JM, Wild L. An epigenetic role for noncoding RNAs and intragenic DNA methylation. Genome Biol. 2007;8 (6):307. doi: 10.1186/gb-2007-8-6-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lorincz MC, Dickerson DR, Schmitt M, Groudine M. Intragenic DNA methylation alters chromatin structure and elongation efficiency in mammalian cells. Nat Struct Mol Biol. 2004 doi: 10.1038/nsmb840. [DOI] [PubMed] [Google Scholar]
- 4.Ball MP, et al. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat Biotechnol. 2009;27 (4):361–368. doi: 10.1038/nbt.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rauch TA, et al. A human B cell methylome at 100-base pair resolution. Proc Natl Acad Sci U S A. 2009;106 (3):671–678. doi: 10.1073/pnas.0812399106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eckhardt F, et al. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat Genet. 2006;38 (12):1378–1385. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ching TT, et al. Epigenome analyses using BAC microarrays identify evolutionary conservation of tissue-specific methylation of SHANK3. Nat Genet. 2005;37 (6):645–651. doi: 10.1038/ng1563. [DOI] [PubMed] [Google Scholar]
- 8.Illingworth R, et al. A novel CpG island set identifies tissue-specific methylation at developmental gene loci. PLoS Biol. 2008;6 (1):e22. doi: 10.1371/journal.pbio.0060022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Song F, et al. Association of tissue-specific differentially methylated regions (TDMs) with differential gene expression. Proc Natl Acad Sci U S A. 2005;102 (9):3336–3341. doi: 10.1073/pnas.0408436102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weber M, et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37 (8):853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 11.Carninci P, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309 (5740):1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 12.Carninci P, et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat Genet. 2006;38 (6):626–635. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- 13.Kim TH, et al. A high-resolution map of active promoters in the human genome. Nature. 2005;436 (7052):876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kapranov P, et al. Examples of the complex architecture of the human transcriptome revealed by RACE and high-density tiling arrays. Genome Res. 2005;15 (7):987–997. doi: 10.1101/gr.3455305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kimura K, et al. Diversification of transcriptional modulation: large-scale identification and characterization of putative alternative promoters of human genes. Genome Res. 2006;16 (1):55–65. doi: 10.1101/gr.4039406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meissner A, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454 (7205):766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Durand CM, et al. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat Genet. 2007;39 (1):25–27. doi: 10.1038/ng1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilson HL, et al. Molecular characterisation of the 22q13 deletion syndrome supports the role of haploinsufficiency of SHANK3/PROSAP2 in the major neurological symptoms. J Med Genet. 2003;40 (8):575–584. doi: 10.1136/jmg.40.8.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Appanah R, et al. An unmethylated 3′ promoter-proximal region is required for efficient transcription initiation. PLoS genetics. 2007;3 (2):e27. doi: 10.1371/journal.pgen.0030027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang X, et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in arabidopsis. Cell. 2006;126 (6):1189–1201. doi: 10.1016/j.cell.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 21.Lister R, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462 (7271):315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Irizarry RA, et al. The human colon cancer methylome shows similar hypo-and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41 (2):178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Valen E, et al. Genome-wide detection and analysis of hippocampus core promoters using DeepCAGE. Genome Res. 2009;19 (2):255–265. doi: 10.1101/gr.084541.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carninci P. Tagging mammalian transcription complexity. Trends Genet. 2006;22 (9):501–510. doi: 10.1016/j.tig.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 25.Post-transcriptional processing generates a diversity of 5′-modified long and short RNAs. Nature. 2009;457 (7232):1028–1032. doi: 10.1038/nature07759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Birol I, et al. De novo transcriptome assembly with ABySS. Bioinformatics (Oxford, England) 2009;25 (21):2872–2877. doi: 10.1093/bioinformatics/btp367. [DOI] [PubMed] [Google Scholar]
- 27.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125 (2):315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 28.Walsh CP, Bestor TH. Cytosine methylation and mammalian development. Genes Dev. 1999;13 (1):26–34. doi: 10.1101/gad.13.1.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baylin S, Bestor TH. Altered methylation patterns in cancer cell genomes: cause or consequence? Cancer Cell. 2002;1 (4):299–305. doi: 10.1016/s1535-6108(02)00061-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.