Abstract
Tumor recurrence is a leading cause of cancer mortality. Therapies for recurrent disease may fail, at least in part, because the genomic alterations driving the growth of recurrences are distinct from those in the initial tumor. To explore this hypothesis, we sequenced the exomes of 23 initial low-grade gliomas and recurrent tumors resected from the same patients. In 43% of cases, at least half of the mutations in the initial tumor were undetected at recurrence, including driver mutations in TP53, ATRX, SMARCA4, and BRAF, suggesting recurrent tumors are often seeded by cells derived from the initial tumor at a very early stage of their evolution. Notably, tumors from 6 of 10 patients treated with the chemotherapeutic drug temozolomide (TMZ) followed an alternative evolutionary path to high-grade glioma. At recurrence, these tumors were hypermutated and harbored driver mutations in the RB and AKT-mTOR pathways that bore the signature of TMZ-induced mutagenesis.
The genetic landscape of tumors is continually evolving, which can be an impediment to the clinical management of cancer patients with recurrent disease (1, 2). In contrast to the clonal evolution of hematological malignancies (3, 4) and solid tumor metastases (5-7), the local regrowth of solid tumors after surgery occurs under a unique set of evolutionary pressures, which are further impacted by adjuvant therapies. Through acquisition of new mutations, residual tumor cells can progress to a more aggressive state. Grade II astrocytic gliomas are particularly troublesome from this perspective. While surgery is the standard of care, these invasive brain tumors typically recur (8). Many remain grade II at recurrence, while others progress to a higher histological grade with a poor prognosis (9). The incidence and timing of malignant progression is variable and unpredictable (8).
We undertook genome sequence analysis of initial and recurrent human gliomas to address two questions: (i) what is the extent to which mutations in initial tumors differ from their subsequent recurrent tumors?; and (ii) how does chemotherapy with TMZ, a drug commonly used in the treatment of glioma, affect the mutational profile of recurrent tumors? We sequenced the exomes of 23 grade II gliomas at initial diagnosis and their recurrences resected from the same patients up to 11 years later (table S1). We selected initial tumors of predominantly astrocytic histology that capture the full spectrum of glioma progression (histological grade II-IV at recurrence) and adjuvant treatment history. Tumor and matched normal DNA were sequenced to an average 125-fold coverage, enabling the sensitive detection of mutations down to a 10% variant frequency, small insertions/deletions, and DNA copy number alterations (CNAs) (Fig. 1A) (tables S2 and S3) (10).
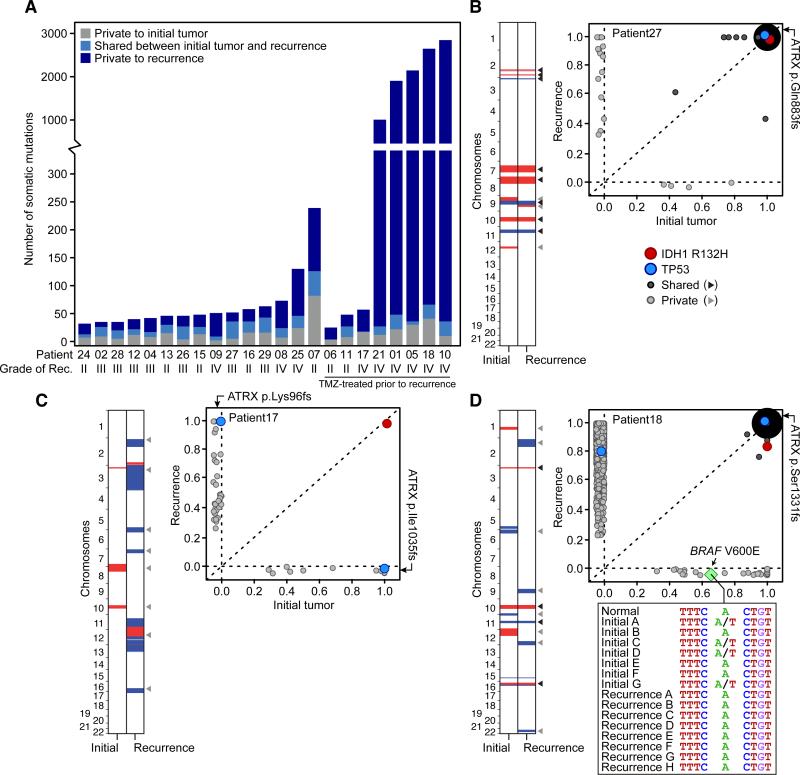
Fig. 1. Genetic landscapes of low-grade gliomas and their patient-matched recurrences.
(A) Total number of mutations private to or shared between the initial and first recurrent glioma of 23 patients. (B to D) Shared and private somatic mutations in paired initial and recurrent tumors (x and y axes respectively) as a function of the estimated fraction of tumor cells carrying the mutant allele. Mutations present in all the cells in both tumors are represented by a single point whose radius is scaled by the log count of such mutations. Shared and private CNAs are indicated (red and blue are gains and losses respectively, white is copy-neutral). In panel C, clonal TP53 and ATRX mutations in the initial tumor were not identified in the recurrent tumor, but different clonal mutations in these two genes were acquired. (D) Inset shows the DNA sequence encompassing BRAF V600E in the normal tissue and in 15 geographically distinct samples of the initial and recurrent tumors.
We identified an average of 33 somatic coding mutations in each initial tumor, of which an average of 54% were also detected at recurrence (shared mutations) (Fig. 1A). The shared mutations included those in IDH1, TP53, and ATRX in most but not all cases (fig. S1) (11-13). All other somatic mutations were identified only in the initial tumor or only in the recurrent tumor from a given patient (private mutations) and thus presumably arose later in tumor evolution. For example, mutations in SMARCA4 were private to the initial or recurrent tumor in six of seven patients and therefore may confer a selective advantage in the context of preexisting early driver events (14, 15). Overall, the initial and recurrent gliomas displayed a broad spectrum of genetic relatedness (fig. S2) (table S4). At one end of this spectrum were four patients whose tumors showed a pattern of linear clonal evolution; we infer that the recurrent tumors in these patients were seeded by cells bearing ≥75% of the mutations detected in the initial tumors (as in patient 27, Fig. 1B). At the other end of the spectrum, tumors from three patients showed branched clonal evolution; we infer that the recurrent tumors in these patients were seeded by cells derived from the initial tumor at an early stage of its evolution, as the recurrent tumors shared ≤25% of mutations detected in the initial tumors. Patient 17 was an extreme example of branched clonal evolution, as the initial and recurrent tumors shared only the IDH1 R132H mutation (Fig. 1C). This further implicates IDH1 mutations as an initiating event in low-grade gliomagenesis (12). Indeed, IDH1 mutation was the only shared mutation in every patient, an observation that supports the current interest in IDH1 as a therapeutic target (16). Paired tumors from the remaining 16 patients formed a continuum between linear and branched clonal evolution. Together, these data illustrate the extent to which genetically similar low-grade gliomas diverge after surgical resection, and that recurrences may emerge from early stages in the evolution of the initial tumor.
Many solid tumors, including glioblastoma (GBM) display intratumoral heterogeneity (17, 18). For example, geographically distinct parts of the tumor may have different mutations. Intratumoral heterogeneity could be a confounding factor in estimates of genetic divergence when only one relatively small fraction of a tumor is sampled. To explore the extent of intratumoral heterogeneity in our cases, we first analyzed the BRAF V600E mutation that was subclonal in the initial tumor of patient 18 and undetectable in the recurrent tumor by either exome sequencing or droplet digital PCR (Fig. 1D) (fig. S3) (10). BRAF V600E was present in three of six additional samples from geographically distinct regions of the initial tumor, while seven additional samples of the recurrence all lacked this mutation. This suggests the BRAF-mutant clone did not expand, despite the proliferative advantage typically conferred by this mutation. This contrasts sharply with the selection and outgrowth of subclonal drivers during the evolution of chronic lymphocytic leukemias (3).
Beyond the actionable BRAF mutation, we sequenced the exomes of additional, geographically distinct samples from three cases to further determine the extent to which apparently private mutations might be misclassified due to intratumoral heterogeneity. For patient 17 in which all mutations except IDH1 were private, intratumoral heterogeneity was observed in the initial and recurrent tumor. From the mutational profiles however, we inferred that three samples of the initial tumor and four samples of the recurrence all derived from a common tumor cell of origin that possessed only an IDH1 R132H mutation (Fig. 2A) (table S5). Moreover, the recurrent tumor contained driver mutations in TP53 and ATRX distinct from those observed in the initial tumor. We found no evidence of these new TP53 or ATRX mutations in the initial tumor at allele frequencies of ~0.1% (fig. S3 and S4), implying convergent phenotypic evolution (5) via a strong ongoing selection for loss of these genes. The initial and recurrent tumors likely did not arise independently, as they also shared three somatic non-coding mutations (fig. S5). Thus, the initial and recurrent tumors were only distantly related and, despite the local and relatively rapid recurrence (fig. S6), exonic mutations other than IDH1 R132H were only transiently present during the course of this patient's disease. Finally, we sequenced the exomes of additional distinct samples of the initial and recurrent tumors from patients 26 and 27, broadening our assessment of the impact of intratumoral heterogeneity on the reported genetic divergence. We found only a small minority of private mutations were actually shared events (7%; table S3) (10). Intratumoral heterogeneity therefore could not explain the majority of the genetic divergence between the initial and recurrent tumors in our cohort, including the driver mutations in initial tumors that were undetected in their recurrence.
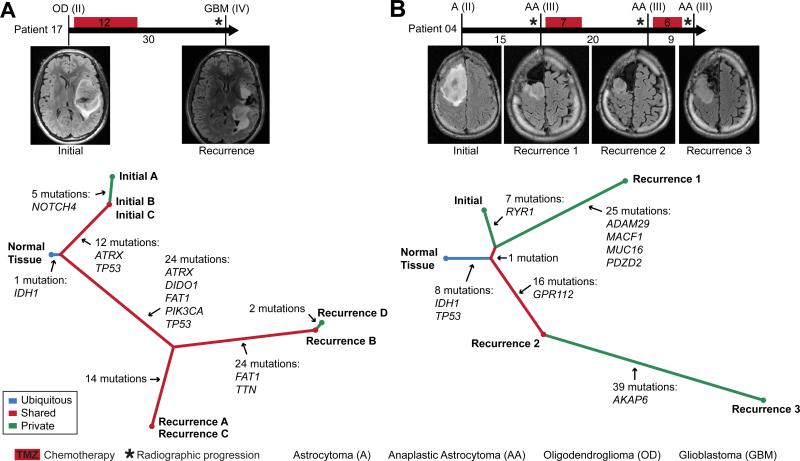
Fig. 2. The temporal and spatial patterns of clonal evolution in the tumors of two glioma patients.
(A and B). A timeline of treatment histories for patient 17 (A) and patient 04 (B) (top, intervals labeled in months). Vertical bars correspond to the time of tumor resection and are labeled with the tumor diagnosis and grade. Representative MRIs are also shown. A phylogenetic tree (bottom) depicts the patterns of clonal evolution of these tumors inferred from the pattern and frequency of somatic mutations, highlighting genes frequently mutated in cancer.
To investigate whether sequential recurrences from a single patient could each be traced to the same evolutionary stage of the initial tumor, we sequenced the exomes of the second and third recurrent tumors from patient 04 and constructed a disease phylogeny by clonal ordering (Fig. 2B) (Fig S7) (table S5) (5, 19). The initial tumor and three sequential local recurrences were clonally related, as indicated by the shared phylogenetic branch containing early driver mutations in IDH1 and TP53. We infer that the tumor cells seeding the second recurrence branched off from the initial tumor at a slightly earlier evolutionary stage than the cells seeding the first recurrence. In contrast, the third recurrent tumor was a direct outgrowth of the second recurrence. These results show that branched and linear patterns of clonal evolution occurred at differing times in the same patient and are therefore not intrinsic properties of the tumor.
Beyond maximal, safe, surgical resection, there is currently no standard of care for patients with low-grade glioma, and options include surveillance, adjuvant radiation alone, TMZ alone, or radiation and TMZ. TMZ is an alkylating agent that induces apoptosis in glioma cells and is sometimes used to defer or delay the use of radiation. However, there is currently no information on whether treatment of grade II astrocytomas with TMZ confers longer overall survival (8). As TMZ is also mutagenic (20), we sought to determine how adjuvant chemotherapy with TMZ affects the mutational profile of recurrent tumors by comparing the initial low-grade gliomas to their post treatment recurrence. While the initial tumors and most of the recurrent tumors in our cohort had 0.2-4.5 mutations per megabase (Mb) (21, 22), six of the ten patients treated with TMZ had recurrent tumors that were hypermutated; that is, they harbored 31.9-90.9 mutations per Mb (table S6). Overall, 97% of these were C>T/G>A transitions predominantly occurring at CpC and CpT dinucleotides, a signature of TMZ-induced mutagenesis distinct from non-hypermutated tumors (fig. S8) (20, 22, 23). We classified C>T/G>A transitions in each hypermutated tumor as TMZ-associated if they were undetected in the matched initial tumor, which was resected before TMZ treatment (Fig. 3A). Although it is difficult to definitively attribute any single mutation to TMZ exposure, comparing the C>T/G>A mutation rates in each tumor pair suggested that >98.7% are due to TMZ-induced mutagenesis (10). To determine whether intratumoral heterogeneity in initial tumors resulted in the misclassification of some mutations as TMZ-associated, we sequenced the exomes of three additional geographically distinct samples of the untreated initial tumor from patient 18. For mutations classified as TMZ-associated, sequencing reads with the mutation were rare in the additional exomes and were found at rates no higher than expected by chance (1.7±0.08%; p-value=0.5, Wilcoxon) (10) further suggesting they are induced by TMZ.
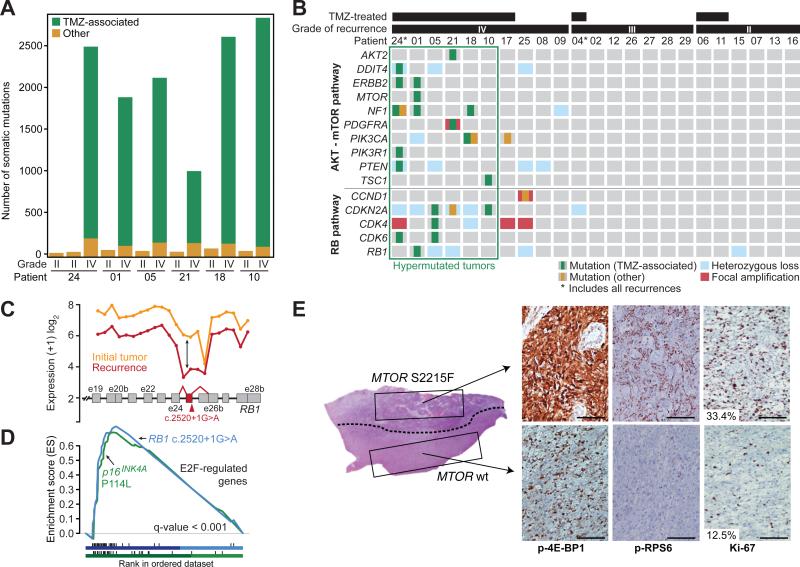
Fig. 3. Recurrent tumors from patients treated with TMZ harbor genetic alterations in the RB and AKT-mTOR signaling pathways.
(A) The number of TMZ-associated mutations and other mutations identified in the six patients with hypermutated recurrent tumors. (B) Somatic mutations and CNAs acquired upon recurrence in key genes of pathways associated with GBM. (C) Expression level of RB1 at each exon and exon-exon junction in the initial and recurrent tumor of patient 01 showing aberrant splicing of the RB1 transcript in the recurrent tumor harboring the RB1 c.2520+1G>A splice-site mutation. The RB1 exon and exon junctions with significant differential usage (red) and the location of the splice-site mutation are shown. (D) Gene set enrichment analysis shows significant enrichment of genes down-regulated by RB1 and up-regulated by E2F in the recurrent tumors of patients 01 (blue) and 10 (green), coincident with the acquisition of TMZ-associated mutations in the RB pathway. (E) Hematoxylin and eosin (H&E)-stained tumor sample from the first recurrent tumor of patient 01. A dotted line separates the two morphologically distinct regions. IHC for phospho-RPS6, phospho-4E-BP1 and Ki-67 show differential activation of mTORC1 targets and proliferation rates in the two adjacent regions. Bars represent 100 microns.
Resistance to TMZ develops in part through the acquisition of mutations that inactivate the DNA mismatch repair (MMR) pathway. MMR pathway dysfunction and continued TMZ exposure can in turn result in hypermutation (22-25). Indeed, we found that hypermutated tumors acquired somatic mutations in MMR genes that were not detected in their initial tumors, as well as aberrant DNA methylation of O6-methylguanine-DNA methyltransferase (MGMT) (fig. S3, S9, and table S1).
The introduction of thousands of de novo mutations may drive the evolution of TMZ-resistant glioma cells to higher states of malignant potential (1, 23). Indeed, all six recurrent tumors that showed evidence of TMZ-induced hypermutation underwent malignant progression to GBM, a high-grade tumor with a worse prognosis (8, 9). To investigate this hypothesis and identify TMZ-associated mutations that may drive the outgrowth of GBM from low-grade glioma, we focused on the RB and AKT-mTOR signaling pathways which are associated with high-grade gliomas (Fig. 3B) (22, 26-28). In each hypermutated recurrence, TMZ-associated mutations affected genes coding for essential signaling molecules in these two pathways. For example, in the RB pathway we identified a TMZ-associated RB1 c.2520+1G>A splice site mutation found previously in the germline of patients with hereditary retinoblastoma (29, 30). Transcriptome sequencing confirmed this mutation triggered aberrant splicing, premature termination, and loss of the RB1 C-terminal domain necessary for growth suppression (Fig. 3C) (31). Recurrent tumors from patient 05 and patient 10 each had a TMZ-associated CDKN2A P114L mutation that prevents it from inhibiting CDK4 or inducing cell cycle arrest (32). The same mutation has been reported in other tumor types (33) and in the germline of patients with familial melanoma (34). Gene set enrichment analysis further confirmed deregulation of RB1-mediated cell cycle control upon tumor recurrence (Fig. 3D), suggesting that TMZ-associated mutations compromise the function of the RB tumor suppressor pathway.
We also investigated TMZ-associated mutations that may activate the AKT-mTOR signaling pathway. We identified a TMZ-associated mutation PIK3CA E542K in the recurrent tumor of patient 18 that drives Akt hyperactivation and induces mTOR-dependent oncogenic transformation (35). Similarly, the TMZ-treated second recurrence of patient 24 had TMZ-associated mutations in PTEN (A121T and G165R) at residues critical to its phosphatase activity (36) that are recurrently mutated in GBM (33). Finally, we validated in vitro that a TMZ-associated MTOR S2215F mutation in the recurrent tumor of patient 01 was constitutively activating (fig. S10), similar to the previously identified MTOR S2215Y (37). Moreover, adjacent regions of this recurrence showed heterogeneous mTORC1 activity (Fig. 3E and fig. S11). Microdissection revealed that while these adjacent regions shared a subset of the mutations found in the initial tumor, MTOR S2215F and other TMZ-associated mutations were present only in the region that stained strongly for mTORC1 activation, which also had higher Ki-67, implying that the TMZ-associated mutations conferred a proliferative advantage. A distal second recurrence harbored the same TMZ-associated mutations and stained strongly and homogeneously for mTORC1 targets (fig. S12). Although both regions of the first recurrence were GBM, the hypermutated subclone underwent in vivo selection, invaded distally, and seeded the second recurrence (fig. S13 and S14). Across our cohort, AKT-mTOR pathway mutations corresponded with elevated phospho-4E-BP1 and RPS6 in vivo, indicating hyper-activated mTORC1 in recurrent GBMs relative to their initial tumors (fig. S12).
There was no evidence that the mutations in the RB and AKT-mTOR signaling pathways preceded TMZ treatment, based on analysis of additional geographically distinct samples of initial tumors from four of the six patients with hypermutated recurrent tumors (table S7). Nonhypermutated recurrent tumors that progressed to GBM also acquired genetic changes in these signaling pathways, but through alternative mechanisms. In contrast, none of the grade II-III recurrences acquired mutations in these pathways. These data suggest a connection between TMZ treatment, driver mutations in oncogenic signaling pathways, and malignant progression.
In summary, through direct comparison of the genomic landscape of gliomas at initial diagnosis and recurrence, we were able to infer the mutational character of the infiltrating tumor cells that give rise to recurrence and that adjuvant therapy with TMZ is intended to eliminate. Recurrences did not typically arise from cells bearing the full set of mutations found in the initial tumor, as would be expected from a local recurrence in the absence of selective pressure from adjuvant chemotherapy. This finding complicates the use of tumor genomics to design precision therapies targeting residual disease. We also demonstrated an alternative evolutionary path of low-grade glioma that is largely determined by adjuvant chemotherapy with TMZ. This extends earlier studies of primary GBMs (23, 25), unpaired recurrent tumors (22), and a cell culture model (20). Future basic and clinical studies must weigh the initial antitumor effects of TMZ against the potential risk of inducing new driver mutations and malignant progression. Ultimately, a better understanding of the invading cells that give rise to recurrent tumors and the effect of adjuvant therapeutics on their evolution will facilitate the development of new strategies to delay or prevent recurrence and malignant progression.
Supplementary Material
Acknowledgments
We thank S. Gonzalez for assistance with the collection of clinical information; R. Kang for assistance with mutation validation; H. van Thuijl for assistance with MGMT methylation; and J. Wiencke and G. Hsuang for assistance with droplet digital PCR. This project was generously supported by Accelerate Brain Cancer Cure; The Grove Foundation; The TDC Foundation; The Anne and Jason Farber Foundation; The Samuel Waxman Cancer Research Foundation; The Alex Lemonade Stand Foundation; The Entertainment Industry Foundation and Anne Feeley; and a generous gift from the Dabbiere family. Additional support by the National Institute Of General Medical Sciences T32GM008568 (T.M.), the National Institutes of Health 1T32CA15102201, R25NS070680 (M.B.), and P50CA097257 (L.E.J., S.J.N, M.S.B., S.M.C., J.F.C. and B.S.T.); the National Cancer Institute R01CA169316-01 (J.F.C.), P01CA81403 (W.C.G, E.C, and W.A.W.), P30CA82103 (A.B.O.), and R01CA163336 (J.S.S.); the National Institute of Neurological Disorders and Stroke K08NS079485 (W.C.G, E.C, and W.A.W); the UCSF Academic Senate and the Sontag Foundation (J.S.S., B.S.T and J.F.C.); the BC Cancer Foundation, Genome BC, and Genome Canada (M.A.M.); and the Goldhirsh Foundation (J.F.C.). This project was supported in part by a research program of the Project for Development of Innovative Research on Cancer Therapeutics (P-Direct) (A.M., N.S., and H.A.), Grant-in-Aid for Scientific Research on Innovative Areas (No. 23134501) (A.M.), and Grant-in-Aid for Scientific Research (S) (No. 24221011) (H.A.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan. C.Y.M was a Damon Runyon Cancer Research Postdoctoral Fellow, M.A.M. is a Canada Research Chair in Genome Science. All exome and transcriptome sequencing data have been deposited in the European Genome-phenome Archive under accession number EGAS00001000579, and data from patients 24 to 29 have also been deposited to the Japanese Genotype-phenotype Archive under accession number JGAS00000000004.
References and Notes
- 1.Gerlinger M, Swanton C. How Darwinian models inform therapeutic failure initiated by clonal heterogeneity in cancer medicine. Br J Cancer. 2010 Oct 12;103:1139. doi: 10.1038/sj.bjc.6605912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012 Jan 19;481:306. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Landau DA, et al. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell. 2013 Feb 14;152:714. doi: 10.1016/j.cell.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ding L, et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 2012 Jan 26;481:506. doi: 10.1038/nature10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerlinger M, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012 Mar 8;366:883. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yachida S, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010 Oct 28;467:1114. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu X, et al. Clonal selection drives genetic divergence of metastatic medulloblastoma. Nature. 2012 Feb 23;482:529. doi: 10.1038/nature10825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanai N, Chang S, Berger MS. Low-grade gliomas in adults. J Neurosurg. 2011 Nov 1;115:948. doi: 10.3171/2011.7.JNS101238. [DOI] [PubMed] [Google Scholar]
- 9.Westphal M, Lamszus K. The neurobiology of gliomas: from cell biology to the development of therapeutic approaches. Nat Rev Neurosci. 2011 Sep 1;12:495. doi: 10.1038/nrn3060. [DOI] [PubMed] [Google Scholar]
- 10.Materials and methods are available as supplementary material on Science Online.
- 11.Jiao Y, et al. Frequent ATRX, CIC, and FUBP1 mutations refine the classification of malignant gliomas. Oncotarget. 2012 Jul 1;3:709. doi: 10.18632/oncotarget.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Watanabe T, Nobusawa S, Kleihues P, Ohgaki H. IDH1 mutations are early events in the development of astrocytomas and oligodendrogliomas. The American journal of pathology. 2009 Apr 1;174:1149. doi: 10.2353/ajpath.2009.080958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Watanabe K, et al. Incidence and timing of p53 mutations during astrocytoma progression in patients with multiple biopsies. Clin Cancer Res. 1997 Apr 1;3:523. [PubMed] [Google Scholar]
- 14.Medina PP, et al. Frequent BRG1/SMARCA4-inactivating mutations in human lung cancer cell lines. Hum Mutat. 2008 May 1;29:617. doi: 10.1002/humu.20730. [DOI] [PubMed] [Google Scholar]
- 15.Glaros S, Cirrincione GM, Palanca A, Metzger D, Reisman D. Targeted knockout of BRG1 potentiates lung cancer development. Cancer research. 2008 May 15;68:3689. doi: 10.1158/0008-5472.CAN-07-6652. [DOI] [PubMed] [Google Scholar]
- 16.Rohle D, et al. An Inhibitor of Mutant IDH1 Delays Growth and Promotes Differentiation of Glioma Cells. Science (New York, NY) Apr 4;:2013. doi: 10.1126/science.1236062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Szerlip NJ, et al. Intratumoral heterogeneity of receptor tyrosine kinases EGFR and PDGFRA amplification in glioblastoma defines subpopulations with distinct growth factor response. Proceedings of the National Academy of Sciences of the United States of America. 2012 Feb 21;109:3041. doi: 10.1073/pnas.1114033109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sottoriva A, et al. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proceedings of the National Academy of Sciences of the United States of America. 2013 Mar 5;110:4009. doi: 10.1073/pnas.1219747110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Merlo LMF, Pepper JW, Reid BJ, Maley CC. Cancer as an evolutionary and ecological process. Nat Rev Cancer. 2006 Dec 1;6:924. doi: 10.1038/nrc2013. [DOI] [PubMed] [Google Scholar]
- 20.Bodell WJ, Gaikwad NW, Miller D, Berger MS. Formation of DNA adducts and induction of lacI mutations in Big Blue Rat-2 cells treated with temozolomide: implications for the treatment of low-grade adult and pediatric brain tumors. Cancer Epidemiol Biomarkers Prev. 2003 Jun 1;12:545. [PubMed] [Google Scholar]
- 21.Greenman C, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007 Mar 8;446:153. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cancer Genome Atlas Research Network, Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008 Oct 23;455:1061. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hunter C, et al. A hypermutation phenotype and somatic MSH6 mutations in recurrent human malignant gliomas after alkylator chemotherapy. Cancer research. 2006 Apr 15;66:3987. doi: 10.1158/0008-5472.CAN-06-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cahill DP, et al. Loss of the mismatch repair protein MSH6 in human glioblastomas is associated with tumor progression during temozolomide treatment. Clin Cancer Res. 2007 Apr 1;13:2038. doi: 10.1158/1078-0432.CCR-06-2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yip S, et al. MSH6 mutations arise in glioblastomas during temozolomide therapy and mediate temozolomide resistance. Clin Cancer Res. 2009 Jul 15;15:4622. doi: 10.1158/1078-0432.CCR-08-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reifenberger G, Reifenberger J, Ichimura K, Meltzer PS, Collins VP. Amplification of multiple genes from chromosomal region 12q13-14 in human malignant gliomas: preliminary mapping of the amplicons shows preferential involvement of CDK4, SAS, and MDM2. Cancer research. 1994 Aug 15;54:4299. [PubMed] [Google Scholar]
- 27.Wang H, et al. Analysis of the activation status of Akt, NFkappaB, and Stat3 in human diffuse gliomas. Lab Invest. 2004 Aug 1;84:941. doi: 10.1038/labinvest.3700123. [DOI] [PubMed] [Google Scholar]
- 28.Louis DN. Molecular pathology of malignant gliomas. Annu Rev Pathol. 2006 Jan 1;1:97. doi: 10.1146/annurev.pathol.1.110304.100043. [DOI] [PubMed] [Google Scholar]
- 29.Tsai T, et al. Rapid identification of germline mutations in retinoblastoma by protein truncation testing. Arch Ophthalmol. 2004 Feb 1;122:239. doi: 10.1001/archopht.122.2.239. [DOI] [PubMed] [Google Scholar]
- 30.Houdayer C, et al. Comprehensive screening for constitutional RB1 mutations by DHPLC and QMPSF. Hum Mutat. 2004 Feb 1;23:193. doi: 10.1002/humu.10303. [DOI] [PubMed] [Google Scholar]
- 31.Qin XQ, Chittenden T, Livingston DM, Kaelin WG. Identification of a growth suppression domain within the retinoblastoma gene product. Genes Dev. 1992 Jun 1;6:953. doi: 10.1101/gad.6.6.953. [DOI] [PubMed] [Google Scholar]
- 32.Koh J, Enders GH, Dynlacht BD, Harlow E. Tumour-derived p16 alleles encoding proteins defective in cell-cycle inhibition. Nature. 1995 Jun 8;375:506. doi: 10.1038/375506a0. [DOI] [PubMed] [Google Scholar]
- 33.Forbes SA, et al. The Catalogue of Somatic Mutations in Cancer (COSMIC). Curr Protoc Hum Genet. Apr 1;:2008. doi: 10.1002/0471142905.hg1011s57. Chapter 10, Unit 10.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fargnoli MC, et al. CDKN2a/p16INK4a mutations and lack of p19ARF involvement in familial melanoma kindreds. J Invest Dermatol. 1998 Dec 1;111:1202. doi: 10.1046/j.1523-1747.1998.00412.x. [DOI] [PubMed] [Google Scholar]
- 35.Kang S, Bader AG, Vogt PK. Phosphatidylinositol 3-kinase mutations identified in human cancer are oncogenic. Proc Natl Acad Sci USA. 2005 Jan 18;102:802. doi: 10.1073/pnas.0408864102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Han SY, et al. Functional evaluation of PTEN missense mutations using in vitro phosphoinositide phosphatase assay. Cancer research. 2000 Jun 15;60:3147. [PubMed] [Google Scholar]
- 37.Sato T, Nakashima A, Guo L, Coffman K, Tamanoi F. Single amino-acid changes that confer constitutive activation of mTOR are discovered in human cancer. Oncogene. 2010 May 6;29:2746. doi: 10.1038/onc.2010.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009 Jul 15;25:1754. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010 Sep 1;20:1297. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Depristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature genetics. Apr 10;:2011. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cibulskis K, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. Feb 10;:2013. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ye K, Schulz MH, Long Q, Apweiler R, Ning Z. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics. 2009 Nov 1;25:2865. doi: 10.1093/bioinformatics/btp394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Research. 2010 Sep 1;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.1000 Genomes Project Consortium, A map of human genome variation from population-scale sequencing. Nature. 2010 Oct 28;467:1061. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wiegand KC, et al. ARID1A mutations in endometriosis-associated ovarian carcinomas. N Engl J Med. 2010 Oct 14;363:1532. doi: 10.1056/NEJMoa1008433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009 May 1;25:1105. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010 May 1;28:511. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Griffith M, et al. Alternative expression analysis by RNA sequencing. Nature Methods. 2010 Oct 1;7:843. doi: 10.1038/nmeth.1503. [DOI] [PubMed] [Google Scholar]
- 49.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005 Oct 25;102:15545. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000 Jan 1;132:365. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 51.Christensen BC, et al. DNA methylation, isocitrate dehydrogenase mutation, and survival in glioma. J Natl Cancer Inst. 2011 Jan 19;103:143. doi: 10.1093/jnci/djq497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Berger MF, et al. The genomic complexity of primary human prostate cancer. Nature. 2011 Feb 10;470:214. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chapman MA, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011 Mar 24;471:467. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Campbell PJ, et al. Subclonal phylogenetic structures in cancer revealed by ultra-deep sequencing. Proceedings of the National Academy of Sciences of the United States of America. 2008 Sep 2;105:13081. doi: 10.1073/pnas.0801523105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Venkatraman ES, Olshen AB. A faster circular binary segmentation algorithm for the analysis of array CGH data. Bioinformatics. 2007 Mar 15;23:657. doi: 10.1093/bioinformatics/btl646. [DOI] [PubMed] [Google Scholar]
- 56.Taylor BS, et al. Functional copy-number alterations in cancer. PLoS ONE. 2008 Jan 1;3:e3179. doi: 10.1371/journal.pone.0003179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Van Loo P, et al. Allele-specific copy number analysis of tumors. Proc Natl Acad Sci USA. 2010 Sep 28;107:16910. doi: 10.1073/pnas.1009843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nik-Zainal S, et al. The life history of 21 breast cancers. Cell. 2012 May 25;149:994. doi: 10.1016/j.cell.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maunakea AK, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010 Jul 8;466:253. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Esteller M, Hamilton SR, Burger PC, Baylin SB, Herman JG. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer research. 1999 Feb 15;59:793. [PubMed] [Google Scholar]
- 61.Rohde C, Zhang Y, Reinhardt R, Jeltsch A. BISMA--fast and accurate bisulfite sequencing data analysis of individual clones from unique and repetitive sequences. BMC Bioinformatics. 2010 Jan 1;11:230. doi: 10.1186/1471-2105-11-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.