Abstract
PURPOSE: To evaluate the ability of various software (SW) tools used for quantitative image analysis to properly account for source-specific image scaling employed by magnetic resonance imaging manufacturers. METHODS: A series of gadoteridol-doped distilled water solutions (0%, 0.5%, 1%, and 2% volume concentrations) was prepared for manual substitution into one (of three) phantom compartments to create “variable signal,” whereas the other two compartments (containing mineral oil and 0.25% gadoteriol) were held unchanged. Pseudodynamic images were acquired over multiple series using four scanners such that the histogram of pixel intensities varied enough to provoke variable image scaling from series to series. Additional diffusion-weighted images were acquired of an ice-water phantom to generate scanner-specific apparent diffusion coefficient (ADC) maps. The resulting pseudodynamic images and ADC maps were analyzed by eight centers of the Quantitative Imaging Network using 16 different SW tools to measure compartment-specific region-of-interest intensity. RESULTS: Images generated by one of the scanners appeared to have additional intensity scaling that was not accounted for by the majority of tested quantitative image analysis SW tools. Incorrect image scaling leads to intensity measurement bias near 100%, compared to nonscaled images. CONCLUSION: Corrective actions for image scaling are suggested for manufacturers and quantitative imaging community.
Introduction
Quantitative imaging techniques are promising research tools both for cancer detection and prediction of therapeutic response [1–5]. Although absolute magnetic resonance imaging (MRI) signal values seldom reflect specific biophysical characteristics, quantitative inference to tissue properties is derived from relative contrast across tissues [6], change in signal with acquisition conditions [2,7], or dynamic signal change over time [5]. The concept of “quantitative imaging” implies a direct or modeled relationship between a measurable image quantity and some targeted biophysical property [1]. Often, the initial quantity of interest is image intensity measured at the pixel or region-of-interest (ROI) level. Well upstream of image creation, image data acquisition is specifically designed to elicit signals sensitive to the desired biophysical tissue/disease property [3,4,8,9]. For example, MRI sequences designed to enhance sensitivity to transient signal changes as an exogenous contrast agent passes through and/or leaks out of vasculature are used to provide quantitative measures of tissue perfusion and endothelial permeability [5]. Typically, MRI scanner hardware settings, such as receiver gain, are automatically adjusted to properly precondition signals for analog-to-digital conversion before image reconstruction. Following reconstruction, images may be stored on disk in manufacturer proprietary format, but eventually, images are networked and archived in Digital Imaging and Communications in Medicine (DICOM) format [10,11].
Analysis of image intensity for quantitative inference to biophysical properties requires knowledge of each step that influences image content. An incorrect assumption on any link in the image acquisition-to-analysis chain can introduce error and potentially invalidate quantitative results. Most quantitative imaging protocols [1,2,5] are designed to account for hardware settings (such as receiver gain) and mitigate, where possible, influence of scanner hardware limitations such as field nonuniformities [12,13]. Unfortunately, source-specific variance in image storage format may be overlooked as an active element in the process. Reliance on manufacturer proprietary formats for accurate deciphering of image signal is undesirable for standardization and generalization of quantitative imaging techniques across vendor platforms.
Adoption of the DICOM standard has effectively removed most barriers to multivendor studies [10,11]. However, the DICOM standard still allows retention of substantial manufacturer-specific information stored in “private tags” of the image header that may confound quantitative analysis. Liberal use of private tags by all vendors is common and necessary given the rapid evolution of imaging technology. Even well-established MR scanning techniques require numerous platform-specific hardware/software (SW) settings retained in private tags. Image scaling relates to numerical multiplication and offset of image pixel values (PVs) to stretch and shift the histogram of intensities to more fully use image storage bit depth (e.g., 16-bit integers), thereby potentially maintaining or extending dynamic range. Image scale parameters are stored in both public and private DICOM tags. For some quantitative imaging applications, “unscaling” intensities is required before subsequent quantitative analysis [5,14]. Additionally, despite being stored in DICOM format, secondary captures, screenshots, and calculated images derived on scanners do not necessarily follow the same rules for image scaling as standard MR images. For example, apparent diffusion coefficient (ADC) maps generated by manufacturer SW have physical units (mm2/s) and an expected intensity range [15].
The degree to which MRI system manufacturers implement image scaling and, equally important, the flexibility of image analysis SW packages to account for source-specific scaling were not thoroughly evaluated before this study. To address this, a cooperative study was designed and conducted within the Image Analysis and Performance Metrics working group of the Quantitative Imaging Network (QIN) [16]. The QIN was established by the National Cancer Institute to improve the role of quantitative imaging for clinical decision making in oncology by the development and validation of data acquisition and analysis methods to tailor treatment to individual patients and to predict or monitor the response to chemical or radiation therapy [16]. A key application where image scaling could affect quantitative results is dynamic contrast-enhanced MRI (DCE-MRI) [5] where signal-inferred T1 change [14] along with the time course of signal change after injection of paramagnetic contrast agent are used to extract quantitative parameters of tissue perfusion and permeability [1,4]. The objective of this study was to use a simple phantom, a well-defined acquisition protocol, and a straightforward analysis procedure to determine whether a given image-source plus analysis SW combination is able to correctly document constant signal in the presence of dynamic signal. If the image-source/analysis package combination fails to properly convey constant signal, its ability to measure dynamic signal is doubtful.
Materials and Methods
Experimental Design
Existence of image-scaling error was directly demonstrated by measurement of signal known to be constant a priori. Because image scaling occurs in response to the histogram of pixel intensities, the test phantom included a variable signal compartment to “provoke” a change in scale factors on scanners that employ image scaling. Image scaling occurs at the source; therefore, images were acquired from three MRI system manufacturers [GE (Piscataway, NJ), Philips (Best, The Netherlands), and Siemens (Erlangen, Germany)]. Flexibility of image analysis SW to appropriately account for source-specific image scaling was assessed by simple ROI measurements performed using 13 SW tools (Table 1). In addition, 3 of these 13 tools were customized to account for source-specific scaling to demonstrate effectiveness of “inverse” scaling. These three modified versions were treated as additional SW packages (SW14–SW16; Table 1). Assessment of a given image-source plus analysis SW combination to address image scaling was binary as “pass” or “fail.” Additionally, to test variable scaling of quantitative parametric maps, DICOM images of ADC maps of an ice-water phantom generated by each MRI system were also provided.
Table 1.
Tested SW Tools.
| Label | SW | Version | Origin |
| SW1 | DynaCAD† | 2.0.0.24083 | Commercial |
| SW2 | DynaSuite† | 2.03.96184 | Commercial |
| SW3 | GE AW | 4.5_02.133_CTT_5.X | Commercial |
| SW4 | K-PACS‡ | 1.6.0 | Public |
| SW5 | MRSC Image§ | 2.19.2012 | In-house |
| SW6 | ImageJ¶ | 1.44p | Public |
| SW7 | MATLAB# | 7.13.0.564 | In-house |
| SW8 | PMOD** | 3.406 | Commercial |
| SW9 | PMOD** | 3.2 | Commercial |
| SW10 | OsiriX†† | 5.5.2, 32-bit | Public |
| SW11 | 3D Slicer‡‡ | 4.2 | Public |
| SW12 | GE FuncTool | 9.4.05 | Commercial |
| SW13 | MATLAB# | 7.11.0.584 | In-house |
| SW14 | MRSC Image§ | 2.19.2013 | Customized |
| SW15 | 3D Slicer‡‡ | 4.2 | Customized* |
| SW16 | MATLAB# | 7.13.0.564 | Customized |
Source code is available at https://github.com/fedorov/DICOMPhilipsRescalePlugin.
Invivo, Gainesville, FL.
Available at http://www.k-pacs.net/.
University of California, San Francisco, CA.
Available at http://imagej.nih.gov/ij/.
MathWorks, Natick, MA.
Zurich, Switzerland.
Available at http://www.osirix-viewer.com/.
Available at http://www.slicer.org/.
Phantoms
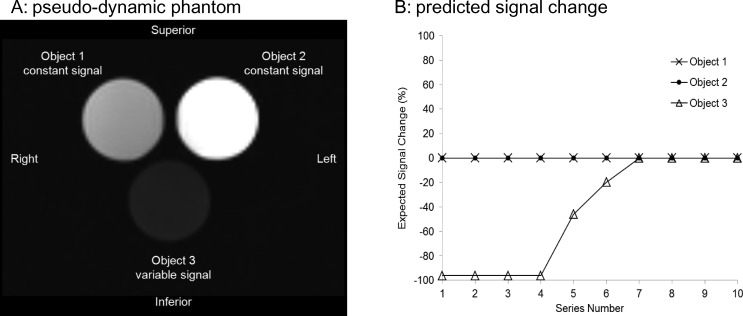
A phantom was designed to provide strong T1 contrast using six 50-mm diameter, 75-ml cylindrical containers filled with: 1) mineral oil, 2) distilled water labeled “0%,” as well as 3) 0.25%, 4) 0.5%, 5) 1.0%, and 6) 2.0% gadoteridol (Prohance; Bracco Diagnostics, Milan, Italy) solution in distilled water by volume. Only three of the six containers were scanned in a single series at a time to mimic a single pseudodynamic time point. Assuming “Head First, Supine” patient entry, phantom samples were positioned on a three-compartment stage in the coronal plane as shown in Figure 1A with the mineral oil container at patient's superior-right (labeled “Object 1”), 0.25% sample at patient's superior-left (labeled “Object 2”), and initially the 0% sample at patient's inferior_center (labeled “Object 3”). Objects 1 and 2 were held fixed in location for all acquisitions and were therefore considered constant signal sources, whereas the 0%, 0.5%, 1%, and 2% samples were alternated in the object 3 compartment from scan to scan. A single six-container phantom and three-compartment stage set was fabricated and shipped for data collection on four MRI platforms at three QIN centers. DICOM images of ADC maps of an ice-water phantom scanned on each of the four MRI systems were also acquired because ice water has a known diffusion coefficient of 1.1 x 10-3 mm2/s [15,17].
Figure 1.
(A) Coronal MR image of pseudodynamic phantom consisting of three 75-ml containers. Object 1 (mineral oil) and object 2 (0.25% gadoteridol) were unaltered to provide constant signal for all acquired series. Object 3 container was manually switched to provide variable signal between series in the following order: 0% gadoteridol for series 1 through 4, 0.5% gadoteridol for series 5, 1.0%gadoteridol for series 6, and 2.0% gadoteridol for series 7 through 10. (B) Expected signal change in objects 1, 2, and 3 relative to last series based on MRI acquisition conditions [18]. Simulated data are depicted with symbols, whereas connecting lines are used as guides.
Image Acquisition
To alter the histogram of pixel intensities across series and provoke differentially scaled images, object 3 was changed from series to series and scanned using an acquisition sequence designed for strong T1 weighting [18], a spoiled gradient recalled echo (SPGR) with the following parameters: repeat time (TR) = 20 ms, echo time (TE) = 5 ms, flip angle = 60°, bandwidth = 244-917 Hz per pixel, field of view = 240 mm, matrix = 128 x 128; single coronal 5-mm-thick slice, number of signal averages = 8, head-coil, image type = magnitude, and series scan time = 20 s. A total of 10 series were acquired on each scanner (i.e., total of 10 images), and scanner hardware settings that affect signal amplitude (e.g., receiver gain) were held constant across series. Object 3 was manually swapped between series as follows: series 1 to 4, object 3 = 0% sample; series 5, object 3 = 0.5% sample; series 6, object 3 = 1% sample; and series 7 to 10, object 3 = 2% sample. These pseudodynamic multiseries scans were performed on four platforms: (scanner 1) Philips 3.0T Ingenia (SW version 4.1.2), (scanner 2) GE 1.5T Signa HDxt (SW version HD16.0_V02_1131.a), (scanner 3) Siemens 3T Tim Trio (SW version VB17A), and (scanner 4) GE 3T Signa (SW version DV22.0_V02_1122.a). For scanner 1, a single-series dynamic acquisition was also performed as a constant scaling control. A manual pause was inserted after each dynamic phase to allow for change of object 3 in the single-series control scan. In addition, a diffusion-weighted scan of an ice-water phantom was acquired using a site-based diffusion-weighted imaging protocol. The central-slice ADC map generated on each of these four scanners and the 10 pseudodynamic series images were provided for image quantification to participating QIN sites.
Image Quantification
Sixteen image view/analysis SW tools used by eight QIN sites were evaluated as listed in Table 1. These tools represent a mixture of commercial (SW1, SW2, SW3, SW8, SW9, and SW12), public-domain free SW (SW4, SW6, SW10, and SW11), and site-customizable in-house SW packages (SW5, SW7, and SW13). SW tools SW14, SW15, and SW16 were modified versions of SW5, SW11, and SW13, respectively, where the modification was to account for vendor-specific image scaling. GE and Siemens platforms did not scale their MR image intensities; however, scaling was detected in Philips images. Relationships between the original pixel floating-point (FP) value, stored PV, rescale intercept (RI), rescale slope (RS), scale slope (SS), scale intercept (SI), and display value (DV) are documented in text headers of Philips PARameter/REConstructed image file formats. These parameters are also available in public tags of Philips MR DICOM: RI in address [0028,1052] and RS in [0028,1053]; and in private tags: in [2005,100D] and SS in [2005,100E]. Relationships between image-scaling parameters, stored, displayed, and FP PVs applicable to Philips MR images are
Each image was quantified using circular ROIs covering approximately 80% of each object along with an ROI in air. An ROI in the center of the ice-water phantom provided an ADC value. For each SW package, 164 ROIs were tabulated {four scanners x [10 series x (three objects + air) + one ADC map]} by mean and SD values of measured pixel intensities, with the following exceptions: SW6 provided only mean values (without SD); SW14, SW15, and SW16 provided measurements only for Philips system (scanner 1); and SW9 provided measurements for scanners 1 to 3 (excluding scanner 4).
Analysis
Measurement of signal change is often the initial step of analysis in quantitative imaging applications. Therefore, in this study, all results are presented as percentage of change relative to the last (i.e., 10th) series (S10),
where Sn is the ROI mean value of the nth series. Normalization by the last series allows for convenient display of all results on the ±100% scale. For each measurement, a deviation of normalized ROI signal change (Equation 3) from that predicted by experimental design (Figure 1B) in excess of twice the ROI measurement error was considered a statistically significant (95% percentile) [19] evidence of (unaccounted) image scaling. For comparative reference, predicted signal change as a function of series number was calculated using the well-established expression for SPGR signal [14], according to:
Contrast agent relaxivity R1gad = 3.7 mmol/s and R2gad = 5.7 mmol/s; and water native T1 = 2 s and T2 = 1.5 s were used along with nominal experimental parameters (TR = 20 ms, TE = 5 ms, flip angle = 60°). Similar to measured signal, the simulated signal was normalized by the signal of the highest (2%) gadoteridol concentration, according to Equation 3.
The normalized ROI measurement error (dispersion) for each object or ADC and scanner was characterized using the average coefficient of variation (%CV) obtained as a series-average ratio of SD to mean ROI value for each scanned object, averaged over all SW tools that provided the corresponding ROI intensity measurements (Table 2):
Table 2.
Average Measurement Variance.
| %CV* | Scanner 1 | Scanner 2 | Scanner 3 | Scanner 4 |
| Object 1 | 3.2 ± 0.6 | 1.9 ± 0.4 | 3.4 ± 0.4 | 5 ± 1 |
| Object 2 | 3.8 ± 0.9 | 2.3 ± 0.3 | 3.7 ± 0.5 | 3.3 ± 0.6 |
| Object 3 | 1.8 ± 0.5 | 3.1 ± 0.7 | 3.6 ± 0.5 | 4.1 ± 0.6 |
| ADCice-water | 1.1 ± 0.1 | 1.1 ± 0.1 | 2.6 ± 0.1 | 2.2 ± 0.4 |
Standard coefficient of variance for an object ROI averaged over image series and software tools as described in text (Equation 5).
This %CV statistics provided a bulk estimate of random error threshold to benchmark measured intensity scaling bias outside of the random variations. Image-scaling bias for each object was estimated as a difference between predicted normalized signal (Figure 1B) and that measured by a specific SW tool for each scanner. The ADC bias was calculated as percent deviation from the known ice-water ADC value of ice water [15,17]. The minimum signal-to-noise ratio (SNR) of a specific scanner was estimated from an average ratio of mean signal for object 3 to the SD of the air ROI for the first four image series (0% gadoteridol). The SNR of the object with the longest T1 provided the sensitivity limit for the scanners that performed pseudodynamic acquisition.
Results
The minimum observed SNR for all scanners was above 85, confirming sufficient imaging sensitivity for the detection of the weakest signal (0% gadoteridol) in this experiment. The average ROI measurement error for all objects, scanners, and SW (Table 2) was 3.5%, ranging from 1.3% (scanner 1, object 3) to 6% (scanner 4, object 1). ADC measurement error was within 2.7% for all scanners. The observed %CV for different scanners reflected mainly SNR limits for the collected images and different sizes and locations of ROIs selected for SW measurements. For example, the data from scanners 1 and 2 had higher average SNR, whereas scanner 4 produced the images with the lowest SNR. Overall, no significant (outside of two SDs) difference was observed for the %CV statistics between the scanners. Results of the noiseless simulation for SPGR intensity change across pseudodynamic series (Equations 3,4) are illustrated in Figure 1B. As with empirical data, expected signal change was expressed relative to the last series (i.e., strongest signal condition, Equation 3). Normalization of simulated signal by the last series reduced sensitivity to specific native relaxation and relaxivity values that are known to depend on field strength (Equation 4) but still allows visual comparison between predicted and empirically observed signal change. The field-dependent change in relaxation parameters for 3T versus 1.5T [e.g., T1 = 2–4 s, R1 = 2.6–4.5 mmol/s would produce negligible (<2%) differences in predicted baseline signal change (Figure 1B)]. By first principles, the correct result for all scanners and SW package pairs should resemble Figure 1B.
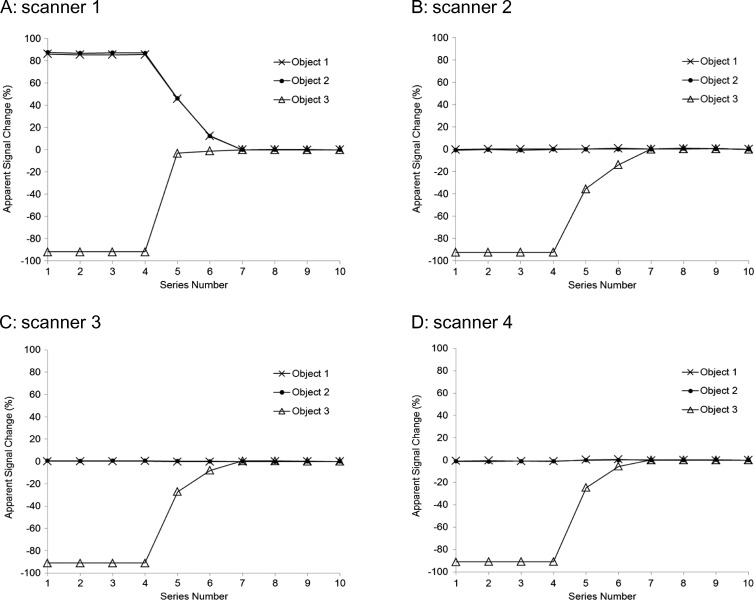
Figure 2 illustrates apparent signal change recorded by SW7 applied to data from all four scanners (A–D). Significant image-scaling bias outside of measurement error (Table 2) is evident for the Philips MR system that is not appropriately accounted for by SW7 (Figure 2A). Not only do objects 1 and 2 appear to falsely exhibit strong signal change (Figure 2A), but curve shape for object 3 is also distorted, suggesting that quantitative analysis of true dynamic change would likewise be in error. Plots in Figure 2, B to D are similar to Figure 1B, suggesting that signal misinterpretation due to image scale factors, if employed, is not an issue for SW7 measurement of GE and Siemens images. However, for these scanners 2 to 4, a small difference from simulation (outside of %CV measurement error) is detected for measured signal change of series 1 to 4, object 3 (Figure 2, B–D) from predicted baseline (Figure 1B). Both proper image scale and baseline close to that predicted in Figure 1B were obtained for Philips data acquired in the single-series dynamic control scan (data not shown).
Figure 2.
Apparent signal change in objects 1, 2, and 3 relative to last series using representative image analysis package SW7 (Table 1) for MR image sources (A) Philips 3.0T (scanner 1), (B) GE 1.5T (scanner 2), (C) Siemens 3T (scanner 3), and (D) GE 3T (scanner 4). Symbols mark the measured data, whereas connecting lines are used as guides. Despite the fact that objects 1 and 2 were unaltered and acquisition conditions were held constant for all 10 series, measured Philips image signal (A) in objects 1 and 2 appears to change by more than 80% relative to the last series. Also note, the shape of object 3 signal change curve is distorted relative to the expected shape shown in Figure 1B. Signal changes measured from the GE and Siemens images (B–D) of objects 1, 2, and 3 are as expected for the experimental conditions (Figure 1B).
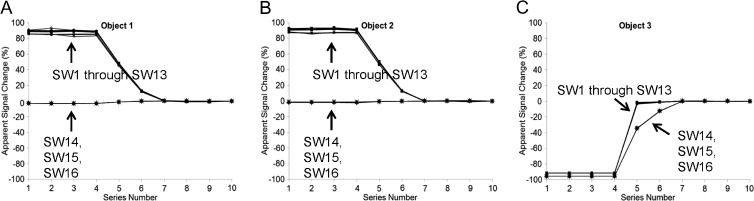
SW7 was representative of SW1 through SW13 packages that all failed to properly account for Philips image scaling as illustrated in Figure 3. Results for SW1 through SW13 applied to GE and Siemens images were similar to Figure 2, B to D, within measurement errors summarized in Table 2 and are not shown. SW packages SW14, SW15, and SW16, modified to return FP PVs according to Equation 2, properly represent signals from objects 1 and 2 as constants that do not change significantly from series to series (Figure 3, A and B) and correctly depict shape of the variable signal from object 3 (Figure 3C). This indicates that quantitative results can be obtained from Philips MR scanners when proper inverse scaling is performed.
Figure 3.
Apparent signal change in object 1 (A), object 2 (B), and object 3 (C) relative to the last series as measured by all SW packages applied to Philips 3.0T MR images. Symbols mark the measured data, whereas connecting lines are used as guides. As exemplified by SW7 in Figure 2, SW packages SW1 through SW13 (Table 1) reveal apparent strong signal changes in (A) object 1 and (B) object 2 even though these objects should exhibit constant signal. Apparent signal change shape by SW1 through SW13 is deviated from expected shape (Figure 1B) for (C) object 3. These observations clearly indicate that SW1 through SW13 do not properly account for image scaling from this image source. SW packages SW14, SW15, and SW16, modified to perform image source-specific scaling, exhibit proper apparent signal changes for all three objects in A, B, and C.
The diffusion coefficient of water at 0°C is known to be 1.1 x 10-3 mm2/s [17]. The ADC measurements provided by all scanners were within 3% from true value (average bias range from 0.1% to 2.5%). Portrayal of scientific notation and physical units in DICOM images is not common, although scaling by powers of 10 is convenient and often performed. Documentation available on Philips MR systems indicates that ADC values are scaled such that an ADC = 1.1 x 10-3 mm2/s would be represented by a DV (in Equation 1) of “1100.” Eleven of 16 SW packages returned a Philips ADC ROI value close to this expected value (mean = 1099.6; range = 1090–1103) with the corresponding %CV statistics summarized in Table 2. Four packages (SW5, SW6, SW7, and SW12) returned Philips ADC values roughly half the expected value (mean = 567; range = 565–570). SW15 returned a value approximately 10-fold too high (ADC = 10436). Direct inspection of DICOM header Philips ADC image scale parameters revealed “RescaleIntercept” (RI) public tag [0028,1052] = 0; “RescaleSlope” (RS) public tag [0028,1053] = 1.95; “ScaleIntercept” (SI) private tag [2005,100D] = 0; and “ScaleSlope” (SS) private tag [2005,100E] = 0.0542. As outlined in Philips documentation, ADC maps are intended to be interpreted in “Display Values” (i.e., DV in Equation 1), and 11 of 16 SW packages appropriately accounted for this scaling, whereas 4 packages did not apply the RS factor and thus were off by a factor of 1.95. Likewise, SW15 was off by factor of 10 because it apparently scaled the ADC map for FP values (Equation 2) instead of DV values (Equation 1).
With only one exception, ROI values derived from GE and Siemens ADC maps were consistently near the “1100” value: for the GE 1.5T scanner, ADC mean = 1127 and range = 1116–1133; for the Siemens 3T, ADC mean = 1099 and range = 1094–1105; and for GE 3T, ADC (excluding one exception) mean = 1119 and range = 1117–1122. DICOM header inspection revealed that GE and Siemens do not use RI and RS public tags [0028,1052] and [0028,1053] for their ADC maps. The single exception was SW8 that returned an ROI value for GE 3T (ADC = 0.001117) that is very close to the correct ADC in mm2/s units. Further inspection of both (1.5T and 3T) GE ADC DICOM header revealed private tag [0051,1002] = “Apparent Diffusion Coefficient (mm2/s)” (i.e., units) and private tag [0051,1004] = 9.9999E-7 would be a suitable scale factor. The differences noted between GE 1.5T and 3T DICOM (e.g., absence of tags [0008,0100] and [0008,0104] for the GE 1.5T ADC map image) may explain why SW8 returned a value of 1126 for the GE 1.5T ADC map. This suggests that SW8 may be designed to use GE-specific scale and units of ADC maps as provided in DICOM.
Discussion
Because objects 1 and 2 were unaltered and hardware settings were held fixed for all pseudodynamic series, measured signal change (Equation 3) for these objects is expected to be close to zero (within measurement %CV) for appropriately scaled images. Our results indicate that the Philips MRI system consistently applies series-specific image scaling. As shown in the results, proper “inverse” scaling of Philips MR images yields appropriate quantitative intensities. Philips has cautioned users to not mathematically combine images acquired across multiple series unless images are properly (inverse) scaled. This fact is known by (some) Philips MRI users, though apparently not accounted for by the image analysis SW developers and users at large. Thus, the majority of SW tools (both public and commercial) for quantitative image analysis need to account for scaling parameters when quantifying multiseries Philips images in DICOM format. We emphasize, this tendency to err only applies when comparing data acquired over multiple series.
Scaling parameters are derived on the basis of the distribution of pixel intensities in a given series. Once determined, scale values are constant for all images acquired in that series for each image type, although multiple image types can be generated in a single series. For example, magnitude-valued (PVs ≥ 0) and real-valued (positive and negative pixels) images produced in a single series will have different image-scaling parameters. Dynamic magnitude images (e.g., standard DCE) acquired in a single series will all have the same scale factor; thus, relative signal change can be accurately quantized independent of image scaling. However, this is not the case when time-course data are acquired in different series, like variable temporal resolution scans and separate precontrast versus postcontrast scans. In addition, acquisition parameters (e.g., flip angle) are changed from series to series for T1 quantification (a prelude to DCE), and signals are expected to change [7,14]. As the physics of true signal change becomes indistinguishable from uncorrected image-scaling effects between series, the level of bias error is, unfortunately, unknown. As shown here, intensity scaling of resulting images may vary between different series of the same experiment depending on relative contrasts of the scanned media. Although different scaling information is recorded in DICOM, the majority of the SW packages used for image analysis and visualization do not account for scaling, thus leading to erroneous quantitative intensity measurements.
Image scaling in MRI is designed to maintain or extend dynamic range of stored images otherwise confined by the bit depth of the stored digital format. Before adoption of the DICOM standard, images were stored in manufacturer proprietary format such as Philips “PAR/REC” binary files. Researchers may continue to use vendor-specific formats, in part, because a method to correct image-scaling effects is fairly explicit in the documentation. However, multicenter clinical trials that incorporate quantitative imaging invariably involve more than one manufacturer platform; thus, management of multiple image formats is necessary [10]. The DICOM standard has essentially removed image network, storage, recall, and display issues. Unfortunately, the fact that an SW package is able to read, display, and derive numbers purported to represent image intensity does not guarantee that it is suitable for quantitative imaging. If Philips MR images are acquired in multiple series, proper (inverse) image scaling must be performed before mathematical combination of the images for quantitative imaging purposes. Even seemingly straightforward comparison of precontrast to postcontrast lesion signal intensity needs to account for signal scaling parameters of Philips images acquired over separate series despite constant acquisition settings, unless the image measurement SW is designed to report FP values and not DVs. In some instances, DVs may be preferred for interpretation of scanner-generated images, such as ADC maps. The majority (11 of 16) of the SW packages tested in this study were designed to return DV values of Philips maps. For GE and Siemens platforms tested, stored PVs of derived ADC maps appear to be uniformly scaled by 106. That is, a true diffusion coefficient of 1 x 10-3 mm2/s is stored and read as “1000.”
Results of this experiment indicate that GE and Siemens MRI systems do not implement image scaling for their standard SPGR images. Public DICOM tags [0028,1052] RescaleIntercept and [0028,1053] RescaleSlope were unused in the GE and Siemens images, and the various SW packages tested in this study essentially returned stored PVs without scaling. This was adequate to reasonably track signal change in the pseudodynamic phantom used in this study. The observed deviation of baseline signal from that predicted by theory was greatest for GE and Siemens scanners, although this may be the result of hardware implementation differences (e.g., true flip angle) or error in the simulation parameters (e.g., native T1).
Some may question the need for image scaling, particularly if it presents a challenge for quantitative analysis SW. However, image scaling is used to preserve or increase dynamic range, which is beneficial to the goals of quantitative imaging. Unfortunately, the fact that the Philips MR employs image scaling is not widely known beyond academic research sites. Expectation to use MRI for quantitative imaging purposes is increasing [1–5], and all MRI manufacturers are encouraged to broadly publicize manipulations that are essential to properly interpret their images. Important scaling
information should be replicated in public tags where possible [10]. Similarly, the use of public DICOM tags is encouraged for quantitative diffusion analysis [2] where b-value and diffusion-direction information is crucial yet is not uniformly provided by manufacturers. Moreover, onus falls on developers of quantitative imaging SW packages to fully understand image characteristics to which their tools are being applied. The pseudodynamic phantom experiment with constant signal described here clearly demonstrated the scope of the problem. The phantom and procedure outlined here are easy to replicate and can be used as an empirical validation for suitability of other image-source/analysis SW combinations for quantitative oncologic imaging applications.
Footnotes
Quantitative Imaging Network and National Institutes of Health funding: U01CA166104, U01CA151235, U01CA154602, U01CA142555, U01CA154601, U01CA140204, U01CA142565, U01CA148131, U01CA172320, U01CA140230, U54EB005149, R01CA136892 and 1S10OD012240-01A1.
References
- 1.Kurland BF, Gerstner ER, Mountz JM, Schwartz LH, Ryan CW, Graham MM, Buatti JM, Fennessy FM, Eikman EA, Kumar V, et al. Promise and pitfalls of quantitative imaging in oncology clinical trials. Magn Reson Imaging. 2012;30:1301–1312. doi: 10.1016/j.mri.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Padhani AR, Liu G, Koh DM, Chenevert TL, Thoeny HC, Takahara T, Dzik-Jurasz A, Ross BD, Van Cauteren M, Collins D, et al. Diffusion-weighted magnetic resonance imaging as a cancer biomarker: consensus and recommendations. Neoplasia. 2009;11:102–125. doi: 10.1593/neo.81328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chenevert TL, Ross BD. Diffusion imaging for therapy response assessment of brain tumor. Neuroimaging Clin N Am. 2009;19:559–571. doi: 10.1016/j.nic.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O'Connor JP, Jackson A, Parker GJ, Roberts C, Jayson GC. Dynamic contrast-enhanced MRI in clinical trials of antivascular therapies. Nat Rev Clin Oncol. 2012;9:167–177. doi: 10.1038/nrclinonc.2012.2. [DOI] [PubMed] [Google Scholar]
- 5.Yankeelov TE, Gore JC. Dynamic contrast enhanced magnetic resonance imaging in oncology: theory, data acquisition, analysis, and examples. Curr Med Imaging Rev. 2009;3:91–107. doi: 10.2174/157340507780619179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Evelhoch JL, LoRusso PM, He Z, DelProposto Z, Polin L, Corbett TH, Langmuir P, Wheeler C, Stone A, Leadbetter J, et al. Magnetic resonance imaging measurements of the response of murine and human tumors to the vascular-targeting agent ZD6126. Clin Cancer Res. 2004;10:3650–3657. doi: 10.1158/1078-0432.CCR-03-0417. [DOI] [PubMed] [Google Scholar]
- 7.Wang J, Qiu M, Kim H, Constable RT. T1 measurements incorporating flip angle calibration and correction in vivo. J Magn Reson. 2006;182:283–292. doi: 10.1016/j.jmr.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 8.Anwar M, Wood J, Manwani D, Taragin B, Oyeku SO, Peng Q. Hepatic iron quantification on 3 Tesla (3 T) magnetic resonance (MR): technical challenges and solutions. Radiol Res Pract. 2013;2013:628150. doi: 10.1155/2013/628150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim H, Taksali SE, Dufour S, Befroy D, Goodman TR, Petersen KF, Shulman GI, Caprio S, Constable RT. Comparative MR study of hepatic fat quantification using single-voxel proton spectroscopy, two-point dixon and three-point IDEAL. Magn Reson Med. 2008;59:521–527. doi: 10.1002/mrm.21561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clunie DA. DICOM structured reporting and cancer clinical trials results. Cancer Inform. 2007;4:33–56. doi: 10.4137/cin.s37032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mildenberger P, Eichelberg M, Martin E. Introduction to the DICOM standard. Eur Radiol. 2002;12:920–927. doi: 10.1007/s003300101100. [DOI] [PubMed] [Google Scholar]
- 12.Belaroussi B, Milles J, Carme S, Zhu YM, Benoit-Cattin H. Intensity non-uniformity correction in MRI: existing methods and their validation. Med Image Anal. 2006;10:234–246. doi: 10.1016/j.media.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 13.Boyes RG, Gunter JL, Frost C, Janke AL, Yeatman T, Hill DL, Bernstein MA, Thompson PM, Weiner MW, Schuff N, et al. Intensity non-uniformity correction using N3 on 3-T scanners with multichannel phased array coils. Neuroimage. 2008;39:1752–1762. doi: 10.1016/j.neuroimage.2007.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schabel MC, Morrell GR. Uncertainty in T 1 mapping using the variable flip angle method with two flip angles. Phys Med Biol. 2009;54:N1–N8. doi: 10.1088/0031-9155/54/1/N01. [DOI] [PubMed] [Google Scholar]
- 15.Chenevert TL, Galbán CJ, Ivancevic MK, Rohrer SE, Londy FJ, Kwee TC, Meyer CR, Johnson TD, Rehemtulla A, Ross BD. Diffusion coefficient measurement using a temperature-controlled fluid for quality control in multicenter studies. J Magn Reson Imaging. 2011;34:983–987. doi: 10.1002/jmri.22363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Quantitative Imaging Network. Available at http://imaging.cancer.gov/programsandresources/specializedinitiatives/qin.
- 17.Holz M, Heil SR, Sacco A. Temperature-dependent self-diffusion coefficients of water and six selected molecular liquids for calibration in accurate H-1 NMR PFG measurements. Phys Chem Chem Phys. 2000;2:4740–4742. [Google Scholar]
- 18.Schabel MC, Parker DL. Uncertainty and bias in contrast concentration measurements using spoiled gradient echo pulse sequences. Phys Med Biol. 2008;53:2345–2373. doi: 10.1088/0031-9155/53/9/010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bland JM, Altman DG. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet. 1986;1:307–310. [PubMed] [Google Scholar]