Abstract
The purpose of this study is to investigate the ability of multivariate analysis of dynamic contrast-enhanced magnetic resonance imaging (DCE-MRI) and diffusion-weighted MRI (DW-MRI) parametric maps, obtained early in the course of therapy, to predict which patients will achieve pathologic complete response (pCR) at the time of surgery. Thirty-three patients underwent DCE-MRI (to estimate Ktrans, ve, kep, and vp) and DW-MRI [to estimate the apparent diffusion coefficient (ADC)] at baseline (t1) and after the first cycle of neoadjuvant chemotherapy (t2). Four analyses were performed and evaluated using receiver-operating characteristic (ROC) analysis to test their ability to predict pCR. First, a region of interest (ROI) level analysis input the mean Ktrans, ve, kep, vp, and ADC into the logistic model. Second, a voxel-based analysis was performed in which a longitudinal registration algorithm aligned serial parameters to a common space for each patient. The voxels with an increase in kep, Ktrans, and vp or a decrease in ADC or ve were then detected and input into the regression model. In the third analysis, both the ROI and voxel level data were included in the regression model. In the fourth analysis, the ROI and voxel level data were combined with selected clinical data in the regression model. The overfitting-corrected area under the ROC curve (AUC) with 95% confidence intervals (CIs) was then calculated to evaluate the performance of the four analyses. The combination of kep, ADC ROI, and voxel level data achieved the best AUC (95% CI) of 0.87 (0.77–0.98).
Introduction
Breast cancer is the second leading cause of cancer death among women in the United States. According to the National Cancer Institute, more than 232,000 new cases in women were diagnosed with breast cancer in 2013, and there were nearly 40,000 breast cancer deaths [1]. On the basis of incidence rates from recent years, it is currently estimated that one in eight women will be diagnosed with breast cancer during their lifetime. Although X-ray mammography and ultrasound imaging are useful in the evaluation of women with breast cancer, they do not provide adequate information for predicting tumor response to therapy to guide clinical decisions. Dynamic contrast-enhanced magnetic resonance imaging (DCE-MRI) and diffusion-weighted MRI (DW-MRI) have matured to the point where they are able to provide quantitative and complimentary information on tumor status [2]. DCE-MRI involves the rapid, serial acquisition of images of a region of interest (ROI) before, during, and after injection of a contrast agent (CA) into a peripheral vein of a patient. As the CA perfuses into the ROI, it changes the tissue's native relaxation times and therefore its measured signal intensity. By fitting the signal intensity time course to an appropriate pharmacokinetic model, physiological parameters that relate to tissue perfusion and permeability (Ktrans), extra-vascular extracellular volume fraction (ve), blood plasma volume (vp), and the efflux rate constant (kep = Ktrans/ve) can be extracted. DW-MRI allows for the in vivo measurement of the motion of water in tissue. By applying two or more diffusion-sensitizing gradients with different amplitudes, the apparent diffusion coefficient (ADC) can be estimated from the resulting data to describe the rate of water diffusion in cellular tissues. In well-controlled studies, it has been shown that the ADC varies inversely with cell density [3].
Parameters derived from DCE-MRI and DW-MRI have been recently evaluated as surrogate biomarkers for assessing and predicting the response of breast tumors to neoadjuvant chemotherapy (NAC). Early DCE-MRI studies focused on semiquantitative analyses through obtaining changes in tumor size, volume, or morphology to evaluate treatment response [4–10]. More recent studies have focused on measuring and tracking changes in Ktrans, ve, vp, and kep [11–19]. There also have been studies investigating the ability of ADC to separate responders from nonresponders after NAC [20–24]. Most of these studies have tracked changes during the course of treatment in parameters obtained from the whole-tumor ROI or histograms describing their distributions. However, these approaches cannot capture the spatial heterogeneity in tissue characteristics and therefore discard all such information. Furthermore, most previous studies reported on either DCE-MRI or DW-MRI and have not evaluated the performance of the combination of these data.
We have previously proposed and validated a longitudinal registration technique to allow registration of MRI and positron emission tomography (PET) data obtained at different time points during therapy [25–27]. In this study, we applied this registration approach to longitudinally align multiparametric MRI maps to perform a novel voxel-by-voxel analysis that incorporates spatial heterogeneity. We investigate whether this voxel-by-voxel approach outperforms the ROI-based analysis when attempting to separate pathologic complete responders (pCR) from nonresponders (non-pCR). Although others have applied multiparameter voxel level analysis [see, e.g., [28]] to predict response in patients with glioma, to the best of our knowledge, this is the first study to integrate multiparametric data analyzed at the voxel level using a multivariate regression model to optimize the prediction of the response of breast cancer to NAC.
Methods
Patient Population
Patients with stage II/III breast cancer were enrolled in a prospective, Institutional Review Board (IRB)-approved study that specified MRI examinations at the following three time points: before initiating NAC (t1), after one cycle of treatment (t2), and after all cycles (t3) of NAC. Forty-one patients completed at least one of the three scans. Data from eight patients were not included for the following reasons: withdrawal from the study after the first scan (n = 6), hardware failure on the scanner (n = 1), and issues with the contrast line (n = 1). All mastectomy specimens were sliced at 5-mm intervals and examined by a breast pathologist for abnormalities. pCR was defined as the absence of any invasive cancer in breast or lymph nodes at the time of surgery following NAC. Patients with any residual invasive cancer in the breast or lymph nodes or patients who progressed before surgery were defined as non-pCR. Determination of pCR and non-pCR status was performed at the time of definitive surgery by a breast pathologist. At completion of NAC, 12 patients achieved pCR, whereas 21 patients were non-pCRs. Table 1 summarizes the salient features of the study population including receptor status, age, neoadjuvant treatment regimens, tumor grade, and the excised tumor size (as measured by a pathologist on the surgical specimen).
Table 1.
Clinical Features of the Study Population.
| Patient No. | Age (yr) | Treatment Regimens | Receptor Status | Tumor Grade | Excised Tumor Size (cm) | Pathologic Response | ||
| ER | PR | HER2 | ||||||
| 1 | 50 | AC → Taxol* | + | + | - | 3 | 0.5 | Residual disease |
| 2 | 52 | Taxotere† | + | - | + | 3 | 1.5 | Residual disease |
| 3 | 60 | AC → Taxol + concurrent trastuzumab | + | + | + | 1 | 2.9 | Residual disease |
| 4 | 36 | Taxol + cisplatin ± everolimus† | - | - | - | 2 | 2.9 | Residual disease |
| 5 | 48 | Dose-dense AC → Taxol | + | + | - | 1 | 1.3 | Residual disease |
| 6 | 43 | Dose-dense AC → Taxol | + | + | - | 2 | 2.6 | Residual disease |
| 7 | 59 | Dose-dense AC → Taxol | + | + | - | 2 | 4.2 | Residual disease |
| 8 | 53 | Taxol + cisplatin ± everolimus | - | . | - | 2 | 1.3 | Residual disease |
| 9 | 35 | Trastuzumab + carboplatin + ixabepilone | + | + | + | 3 | 1.4 | Residual disease |
| 10 | 28 | Taxol + cisplatin ± everolimus | - | - | - | 3 | 0.8 | Residual disease |
| 11 | 33 | AC → Taxol | + | + | - | 3 | 1.2 | Residual disease |
| 12 | 39 | AC → Taxol | + | + | - | 1 | 2.5 | Residual disease |
| 13 | 57 | AC → Taxol | - | - | - | 3 | N/A§ | Residual disease |
| 14 | 67 | Dose-dense AC → Taxol | - | - | + | 3 | 1.8 | Residual disease |
| 15 | 45 | Taxol + cisplatin ± everolimus | - | - | - | 3 | 0.5 | Residual disease |
| 16 | 46 | Taxotere + carboplatin + Herceptin¶ | + | + | + | 3 | 0.3 | Residual disease |
| 17 | 47 | Taxotere → AC | + | + | - | 1 | 8.0 | Residual disease |
| 18 | 36 | AC → Taxol | + | + | + | 2 | 1.0 | Residual disease |
| 19 | 43 | Cisplatin + Taxol ± everolimus | - | - | + | 3 | 0.7 | Residual disease |
| 20 | 55 | AC → Taxol | + | + | - | 2 | 3.5 | Residual disease |
| 21 | 58 | Cisplatin + Taxol ± everolimus | - | + | - | 2 | 1.7 | Residual disease |
| 22 | 53 | AC → concurrent Taxol + trastuzumab | - | - | + | 3 | 0 | pCR |
| 23 | 46 | Taxotere → AC | - | + | - | 3 | 0 | pCR |
| 24 | 46 | AC → concurrent Taxol + trastuzumab | - | - | + | 2 | 0 | pCR |
| 25 | 33 | AC → weekly Taxol | - | - | - | 3 | 0 | pCR |
| 26 | 39 | Trastuzumab and Lapatinib | - | - | + | 2 | 0 | pCR |
| 27 | 46 | AC → Taxol | + | - | - | 3 | 0 | pCR |
| 28 | 42 | Taxol + cisplatin ± everolimus | - | - | - | 3 | 0 | pCR |
| 29 | 34 | Taxotere → AC | - | - | - | 3 | 0 | pCR |
| 30 | 44 | Trastuzumab + Lapatinib | - | - | + | 3 | 0 | pCR |
| 31 | 37 | Taxol + cisplatin ± everolimus | - | - | - | 3 | 0 | pCR |
| 32 | 39 | AC → Taxol | - | - | - | 3 | 0 | pCR |
| 33 | 48 | Taxotere + carboplatin + Herceptin | - | - | + | 3 | 0 | pCR |
AC indicates adriamycin and cyclophosphamide; HER2, human epidermal growth factor receptor 2.
paclitaxel (Taxol; HQ SPCLT PHARMA, Paramus, NJ).
docetaxel (Taxotere; Sanofi-Aventis, Bridgewater, NJ).
The study is ongoing, and we are blinded to the randomization.
This patient was transferred to another hospital, and the tumor size is not available.
trastuzumab (Herceptin; Genentech Inc, San Francisco, CA).
MRI Data Acquisition
DCE-MRI was performed using a Philips 3.0T Achieva MRI scanner (Philips Healthcare, Best, The Netherlands). A 4-channel receive double-breast coil covering both breasts was used for 20 patients (Invivo Inc, Gainesville, FL), whereas a 16-channel double-breast coil was used for 13 patients. Data for constructing a T1 map were acquired with an radio frequency (RF)-spoiled 3-dimensional (3D) gradient echo multiflip angle approach with repetition time/echo time (TR/TE) = 7.9 ms/1.3 ms and 10 flip angles from 2 to 20° in 2° increments. The acquisition matrix was 192 x 192 x 20 (full breast) over a sagittal square field of view (22 cm2) with slice thickness of 5 mm, one signal acquisition, and a sensitivity encoding factor of 2 for an acquisition time of just less than 3 minutes. The dynamic scans used identical parameters and a flip angle of 20°. Each 20-slice set was collected in 16 seconds at 25 time points for just less than 7 minutes of dynamic scanning. For the DCE study, a catheter placed within an antecubital vein delivered 0.1 mmol/kg (9–15 ml, depending on patient weight) of the CA gadopentetate dimeglumine Gadolinium diethylenetriamine pentaacetic acid (Gd-DTPA; Magnevist, Wayne, NJ) at 2 ml/s (followed by a saline flush) through a power injector (Medrad, Warrendale, PA) after the acquisition of three baseline dynamic scans.
DW-MRI was acquired with a single-shot spin-echo echo-planar imaging sequence in three orthogonal diffusion-encoding directions (x, y, and z). For 13 patients, b = 0 and 500 s/mm2, TR/TE = 2500 ms/45 ms, Δ = 21.4 ms, δ = 10.3 ms, and 10 signal acquisitions were acquired. For 20 patients, b = 0 and 600 s/mm2, TR/TE = “shortest” (range, 1800–3083 ms/43–60 ms), Δ = 20.7 to 29 ms, δ = 11.4 to 21 ms, and 10 signal acquisitions were acquired. The total scan time of each DW-MRI data was 4 minutes and 40 seconds. The acquisition matrix was 144 x 144 x 12 over a field of view (19.2 cm2) with a slice thickness of 5 mm.
Quantitative Image Analysis
For each patient at each time point, an ROI was drawn manually to completely surround the enhancing tumor as seen on each slice. The tumor was then defined as the voxels in each ROI displaying a percentage of enhancement larger than 80%, which was the optimal threshold enhancement level determined by a previous study [29]. The Extended Tofts-Kety model [30] was used to estimate physiological parameters from the DCE-MRI data. This pharmacokinetic model assumed a linear relationship between the time-varying longitudinal relaxation time T1 (t), and the concentration of CA in the tissue Ct(t):
| (1) |
where R10 is the R1 value of the tissue before CA administration and r1 is the relaxivity of the CA. The Ct time course was calculated as follows:
| (2) |
where Ktrans is the volume transfer constant, ve is the extravascular extracellular volume fraction, and vp is the plasma volume fraction. The arterial input function (AIF) Cp was a population-averaged AIF constructed from 50 individual AIFs obtained from a total of 24 patients scanned at different time points. All of the 24 patients were included in the 41 patients presented in this study. Each AIF was detected through a semiautomatic AIF-tracking algorithm [31]. The efflux constant kep (≡ Ktrans/ve) was then computed after Ktrans and ve were obtained.
ADC maps were calculated with the following equation: ADC = ln (S1/S2)/(b2 - b1), where S1 and S2 denote the signal acquired with b values of b1 and b2, respectively.
Voxels for which either the Extended Tofts-Kety model or the ADC fitting model did not converge or converged to nonphysical values (i.e., Ktrans > 5.0 min-1, ve > 1.0, vp > 1.0, ADC > 3.0 x 10-3 mm2/s or ADC < 0.01 x 10-3 mm2/s, or any parameter below 0.0) were set equal to zero and not included in subsequent analyses.
Registration Algorithms
A tumor volume-constrained registration algorithm [25,26] was employed for alignment of breast images acquired during therapy. First, a rigid body registration algorithm [32] was used to align the averaged postcontrast images at t1 and t3 to the target images at t2 to obtain a global and rough alignment. This algorithm searched the optimal rotation and translation parameters through maximizing the normalized mutual information. A nonrigid registration method [25], based on the adaptive bases algorithm [33], was then applied to the images to refine the alignment. The cost function of this method was composed of the following two terms: the negative normalized mutual information term and the tumor volume constraint term:
| (3) |
where H(A) and H(B) are the marginal entropy of images A and B, respectively, and H(A,B) is the joint entropy. JT(x) is the Jacobian determinant on the tumor area, and α is the parameter to control the weight of this constraint term; α can be adjusted from 0 to 1 on the basis of individual data sets. For the patient data sets in this study, a range of 0.05 to 0.3 was (empirically) selected for the image sets. The deformation field was modeled by a linear combination of radial basis functions [34]. Through searching the optimal coefficients of radial basis functions, the cost function maximally aligns the breast volumes while minimally distorting the tumor whose volume should be kept true to what is measured at each time point. Details of the method can be found elsewhere [25,26].
As the DW- and DCE-MRI data were acquired within the same imaging session with minimal patient motion, the rigid body registration [35] mentioned above was used to transform the DW-MRI data into the DCE-MRI space at each time point. The transformations obtained from the longitudinal DCE-MRI registration were then applied to register the DW-MRI data at different time points.
Data Analysis
ROI analysis. Our previous studies [35,36] indicated that the accuracy of predicting treatment response was superior using the parameter estimates at t2 rather than those at t1. Thus, in the present study, we focused on the mean values for each parameter (ADC, Ktrans, ve, kep, and vp) in the tumor ROIs at t2 for each patient. The logistic regression (LR) model was then applied to seven cases: kep, ADC, Ktrans, ve, and vp considered individually (five cases), the combination of kep and ADC, and the combination of all parameters, as follows:
| (4) |
where pCR is a binary outcome variable indicating whether or not a subject is a responder and X denotes covariates. For example, for the combination of kep and ADC, βX can be expressed as β0 + β1mkep + β2mADC, where mkep and mADC are the mean values and βi is the coefficient of the input variables.
Voxel-based analysis. As described above, the longitudinal registration algorithm was employed to register the averaged postcontrast images obtained at three time points to a common space. The obtained transformations were then applied to align the serial, multiparametric maps. This step allowed us to detect the subset of voxels on the data at t2 that displayed increases in kep, Ktrans, and vp from t1 to t2 and the voxels that displayed decreases in ve and ADC from t1 to t2. The corresponding percentiles of the subsets of voxels were calculated, and a redundancy analysis [37] was performed to select the most nonpredictable percentiles from the remaining percentiles for the given histograms. As described in [37], after dividing the voxels into 20 equal bins, the most predictable percentiles quantified by the coefficient of determination (equivalent to R2 in linear regression) from the remaining percentiles were removed in a stepwise fashion. Those percentiles selected as the least predictable served as explanatory variables in the logistic ridge regression (LRR) model in each of the seven cases. Instead of regular LR, we employed LRR that has been widely used to handle a case with small sample size and a large number of explanatory variables; i.e., this is an example of “small n and large p” scenario [38]. The difference between the LR and LRR models is that a penalty is added in the cost function in the LRR model. The penalty is calculated as βT β and controls the variance of the coefficients β. The contribution of the penalty to the cost function is regularized by a tuning parameter that can be chosen as described by Cule et al. [39]. For example, the selected percentiles for both kep and ADC were input into the model as follows:
| (5) |
where hkepi and hADCi were the ith percentiles for kep and ADC, respectively. These analyses were performed in R Statistical Software version 3.0.2 (http://CRAN.R-project.org; R Foundation for Statistical Computing, Vienna, Austria).
ROI and voxel analysis. In this approach, for both the single parameter and the parameter combinations, the predictor variables in the LRR model were the mean values and the selected percentiles obtained from the redundancy analysis.
ROI, voxel, and clinical data. In this approach, in addition to the mean parameters and the selected percentiles, clinical data (patient age and tumor grade) were also used as explanatory variables in the LRR model. The deviation of patient age was calculated as the measure of difference between each patient's age and the mean age of the population and input in the LRR model. Each patient's tumor was graded using the modified Nottingham method (shown in Table 1) and recorded as follows: 1 (low grade), 2 (intermediate grade), and 3 (high grade) [40]. Those scores were also input in the LRR model as predictor variables, as well as the age deviation, the mean parameters, and the percentiles.
Statistical Analysis
The regression model returned the probabilities of achieving pCRs for all patients, and receiver-operating characteristic (ROC) analysis was performed to determine the ability of each analysis to predict pCR [41]. The areas under the curve (AUCs) were estimated using the trapezoidal rule. The bootstrap method to correct for overfitting [42,43] was performed to compute the overfitting-corrected AUC and its 95% confidence intervals (CIs) with 500 replicates. At each replicate, a set of bootstrap samples was used to fit an LR (or LRR) model, and then the fitted model was validated against the original data set, which in turn resulted in the overfitting-corrected AUC estimate. This approach is known to be more reliable than the leave-one-out cross-validation that tends to inflate the variance of AUC estimates [44].
Results
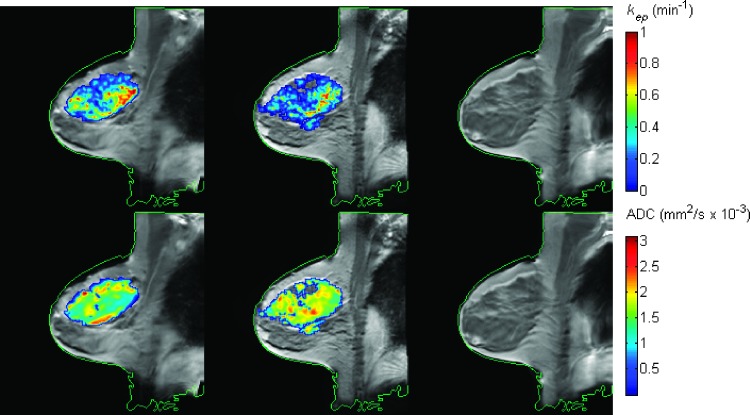
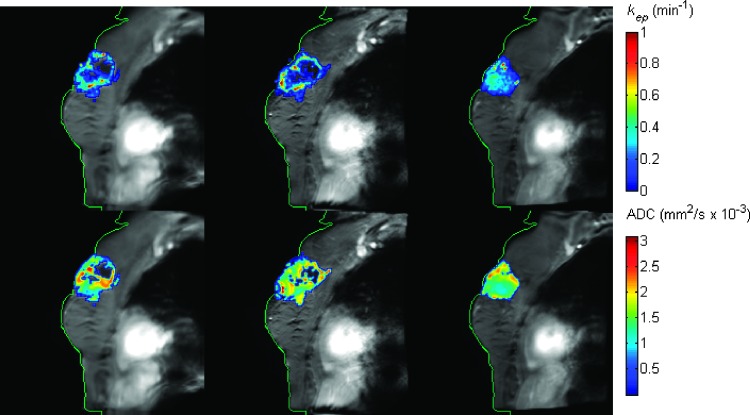
Figure 1 shows the kep (top row) and ADC (bottom row) maps superimposed on the postcontrast DCE-MRI data at the pre-NAC (first column), post-one cycle of NAC (second column), and post-all cycles of NAC (third column) time points for one (representative) patient achieving pCR. The parametric maps obtained at each imaging session were all registered to a common space. The green contour was drawn on the image at t3 and copied to other panels. If the registration is accurate, the green contour at t3 should also match the contours at t1 and t2 after registration. The figure demonstrates the accuracy of the alignment of the images across time (i.e., during NAC). Observe that kep has decreased from 0.42 min-1 at baseline to 0.31 min-1 after one cycle of therapy, whereas the ADC values were 1.47 mm2/s x 10-3 and 1.75 mm2/s x 10-3 at the first two time points, respectively. Figure 2 shows the corresponding data for one patient who is a non-pCR. In this case, the kep values were 0.26 min-1, 0.32 min-1, and 0.23 min-1, respectively, and the ADC values were 1.59 mm2/s x 10-3, 1.60 mm2/s x 10-3, 1.51 mm2/s x 10-3, respectively, at the three time points. The ROI level analysis is focused on how these averaged values changed over time to predict eventual therapeutic response. The next level of analysis is to consider the voxel level data.
Figure 1.
The kep (top row) and ADC (bottom row) maps superimposed on the postcontrast DCE-MRI data pre-NAC (first column), post-one cycle of NAC (second column), and post-all cycles of NAC (third column) for one patient achieving pCR. Multiparametric maps at different time points were all registered to a common space. The green contour was drawn on the images at t3 and copied to the images at the other time points. The well-aligned contours at all three times indicate the accuracy of the temporal alignment of the images. (The area of nonspecific enhancement seen in the medial quadrant of the breast at t3 was biopsied and found to reveal only benign fibrocystic changes.)
Figure 2.
The kep (top row) and ADC (bottom row) maps superimposed on the postcontrast DCE-MRI data pre-NAC (first column), post-one cycle of NAC (second column), and post-all cycles of NAC (third column) for one non-pCR. Multiparametric maps at different time points were all registered to a common space. The green contour was drawn on the images at t3 and copied to the images at the other time points. The well-aligned contours at all three times indicate the accuracy of the temporal alignment of the images.
Using previously published methods [29], there was not a statistical difference in tumor volumes at baseline or post-one cycle of NAC between patients who went on to achieve pCR and those that did not.
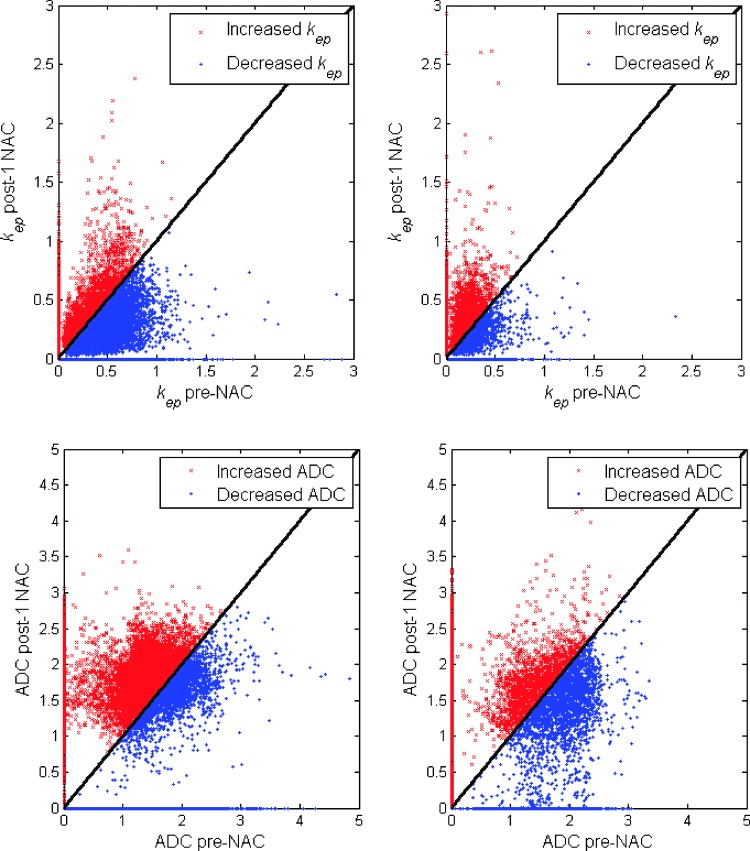
Figure 3 shows the kep (top row) and ADC (bottom row) values at the pre-NAC time point plotted against the kep and ADC values at the post-one cycle of NAC time point for one pCR (left) and one non-pCR (right); red (blue) points indicate those voxels for which there was an increase (decrease) in kep between the two time points. For the pCR (left), 23.7% of tumor voxels are red, and 76.3% of voxels are blue for kep, whereas 63.0% are red, and 37.0% are blue for ADC. For the non-pCR (right), 49.8% of tumor voxels are red, and 50.2% of voxels are blue for kep, whereas 48.6% of tumor voxels are red, and 51.5% of voxels are blue for ADC, respectively.
Figure 3.
The kep (top row) and ADC (bottom row) values at the pre-NAC time point were plotted against the kep and ADC values at the post-one cycle of NAC time point for one pCR (left) and one non-pCR (right); red (blue) points indicate those voxels for which there was an increase (decrease) in kep or ADC between the two time points. For the pCR (left), 23.7% of tumor voxels are red, and 76.3% of voxels are blue for kep, whereas 63.0% are red, and 37.0% are blue for ADC. For the non-pCR (right), 49.8% of tumor voxels are red, and 50.2% of voxels are blue for kep, whereas 48.6% of tumor voxels are red, and 51.5% of voxels are blue for ADC, respectively.
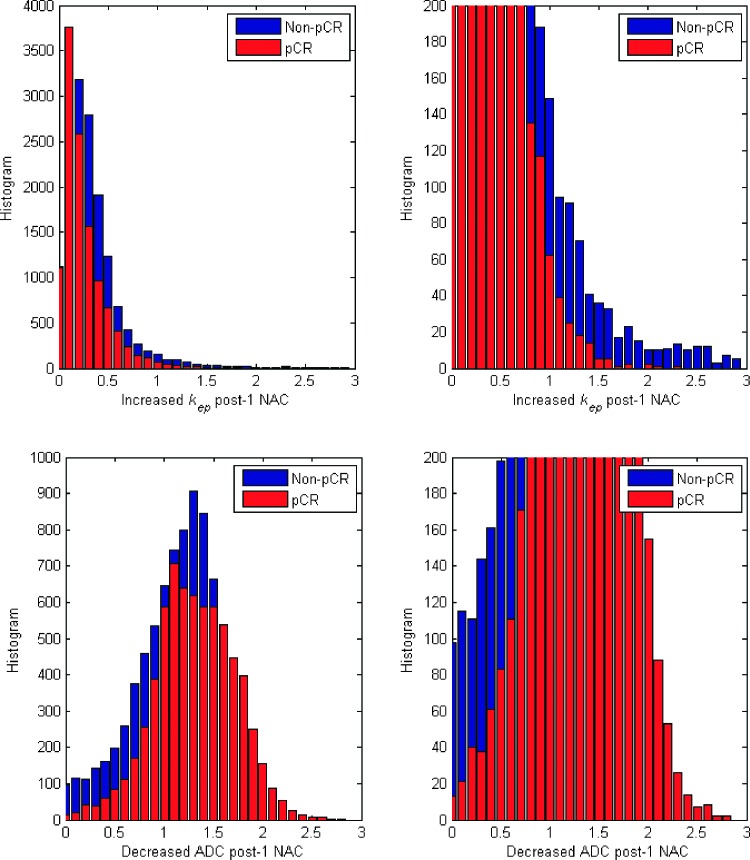
In Figure 4, the left panels show the increased kep histogram (top) and decreased ADC histogram (bottom) of all tumor voxels obtained from 21 patients who are non-pCRs (blue) and 12 patients who achieved pCR (red). The magnified histograms are shown in the right panel, which demonstrates that kep from the non-pCRs yielded a wider distribution and a longer tail than the responders. More specifically, the kurtoses, a measure of the shape of a histogram, of kep were 10.35 and 5.84 for the pCRs and non-pCRs, respectively, and the kurtoses of ADC were 1.92 and 2.35, respectively. The skewnesses, a measure of the extent of asymmetry of a probability distribution, of kep were 2.79 and 2.04 for the pCRs and non-pCRs, whereas the skewnesses of ADC were 0.73 and 0.85, respectively.
Figure 4.
The figure shows the increased kep histogram (top) and decreased ADC histogram (bottom) of all tumor voxels obtained from 21 non-pCRs (blue) and 12 pCRs (red). The right panels show the magnified histograms of kep and ADC.
Table 2 displays the AUCs, as well as the bootstrapped 95% CIs of the AUCs, for seven different parameter combinations (rows) and four different analyses (columns). Considering the first column of the table, kep and ADC outperformed the other individual parameters, though not significantly so as evidenced by the overlapping 95% CIs. When kep and ADC are combined in the regression model, the resulting AUC (0.82) and 95% CIs (0.67–0.98) indicate a substantial improvement over the single measures though, again, not significantly so. Using all five imaging parameters in the regression model (bottom row of the first column) results in a decrease in the AUC (0.73) over that achieved by combining only kep and ADC.
Table 2.
The AUCs of Different Parameters Using Four Different Analyses
| ROI | Voxel | ROI + Voxel | ROI + Voxel + Clinical Data | |||||
| AUC | 95% CI | AUC | 95% CI | AUC | 95% CI | AUC | 95% CI | |
| kep | 0.70 | [0.49–0.96] | 0.72 | [0.53–0.91] | 0.72 | [0.52–0.91] | 0.75 | [0.60–0.90] |
| ADC | 0.77 | [0.40–1.00] | 0.75 | [0.62–0.90] | 0.85 | [0.71–1.00] | 0.84 | [0.55–1.00] |
| Ktrans | 0.68 | [0.47–0.93] | 0.66 | [0.47–0.85] | 0.71 | [0.52–0.90] | 0.77 | [0.62–0.93] |
| ve | 0.49 | [0.29–0.71] | 0.42 | [0.22–0.64] | 0.70 | [0.52–0.89] | 0.74 | [0.56–0.93] |
| vp | 0.68 | [0.39–0.96] | 0.62 | [0.33–0.96] | 0.71 | [0.52–0.90] | 0.77 | [0.64–0.91] |
| kep and ADC | 0.82 | [0.67–0.98] | 0.75 | [0.60–0.92] | 0.87 | [0.77–0.98] | 0.81 | [0.67–0.93] |
| kep, ADC, Ktrans, ve, and vp | 0.73 | [0.49–0.95] | 0.72 | [0.51–0.96] | 0.77 | [0.58–0.98] | 0.81 | [0.65–0.97] |
The second and third columns display the AUC values when the voxel level data are used and when both the ROI and voxel level data are used to populate the logistic model, respectively. Finally, the rightmost column summarizes the results of combing the ROI level data, voxel level data, and select (available) clinical data into the regression model. Note that the combined ROI and voxel analysis yielded better AUCs for all parameters than either ROI- or voxel-based analyses. The best AUC of 0.87 was obtained when both the ROI and voxel level data were used for the combination of kep and ADC. In general, there is a substantial improvement in predictive ability of individual parameters as the ROI, voxel, and clinical data are used. However, as indicated by the overlapping 95% CIs, there are only a small number of entries that are statistically different. The combination of ROI, voxel level kep, and ADC data provides an optimal balance of model complexity and ROC performance. In addition, by further including Ktrans, ve, and vp in the regression model, the AUC values decrease with the increased number of parameters involved in the model.
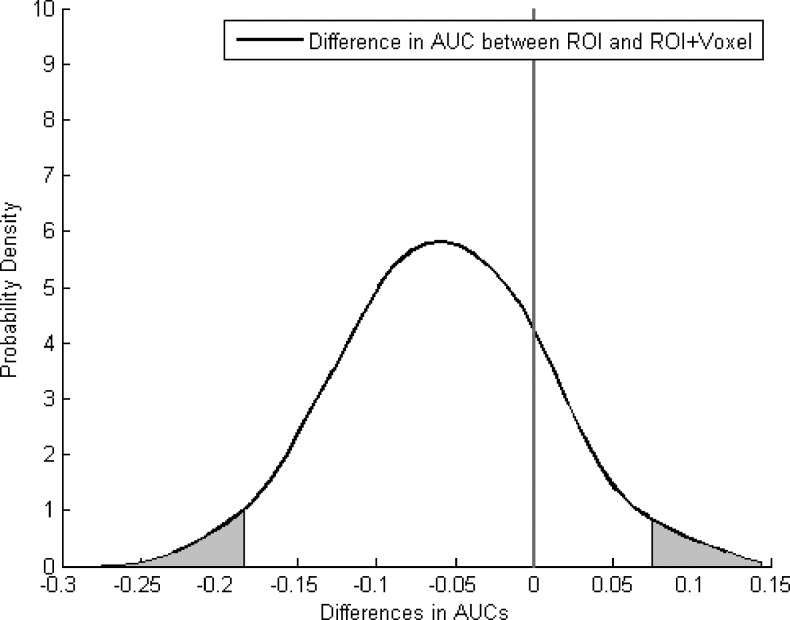
Figure 5 displays the density distributions of the AUC differences between the ROI and the combined ROI and voxel analyses for the combination of kep and ADC. Specifically, 500 AUCs were generated through the boot strap method for the analysis, and the AUC differences between the two analyses were calculated; the figure displays the probability density functions, which described the density distributions of the AUC differences. The 95% CIs of the AUC differences were (-0.18–0.075). For clarity, the areas outside the 95% CIs are shadowed for the distribution. Note that the shadowed areas on the right side are close to zero (the reference line), indicating a moderate trend approaching significance between the ROI and the combined ROI and voxel data analyses. [CIs (95%) that lie completely to the left or right of the zero mark would indicate a significant difference between two methods at the P < .05 level.] When these data are combined with those in Table 2, they support the selection of the logistic model that includes kep and ADC as the most preferable, as they combine the desirable features of being the most parsimonious approach with the greatest ROC performance.
Figure 5.
The figure displays the density distribution of the AUC differences between the ROI analysis and the combined ROI and voxel analysis for the combination of kep and ADC. The areas outside the 95% CIs are shadowed for the distribution. The 95% CIs of the AUC differences were (-0.18–0.075), indicating a moderate trend approaching significance.
Discussion
To the best of our knowledge, this is the first report to indicate that integrating spatial information characterizing intratumoral heterogeneity improves the ability of quantitative MRI to predict the response of breast tumors to NAC. A longitudinal registration technique was employed to spatially align parametric maps obtained at the pre-one cycle and post-one cycle of NAC time points, thereby enabling subsets of voxels to be selected on the basis of their changes in parameter values. By incorporating the ROI and voxel level data into the LRR model, the overall predictive ability was quite strong (AUC = 0.87). It is important to note that the methodology presented here is readily generalizable to include other data types including, for example, other imaging modalities or genomic data.
There are some limitations in the present study. First, although these initial results are encouraging, future work needs to investigate the proposed approach on a larger cohort of patients. Second, the temporal resolution of 16 seconds is not optimal for characterizing the AIF, and this can confound a quantitative DCE-MRI analysis. (This temporal resolution was chosen as a compromise between temporal and spatial resolution and field-of-view coverage; details on this technical point are available in [29].) Third, the NAC regimens used in this study were left to the discretion of the treating physicians and were therefore quite varied. It is certainly possible that the imaging biomarkers could vary both by the biology of the disease and by the agents used. A final limitation in the approach outlined here concerns the registration method itself. It is important to note that, although previous studies [25–27,45] have shown that longitudinal registration of the breast MRI data can be done accurately and robustly, it remains quite challenging. The improved AUCs also indicated the importance of longitudinal registration and thus warrant further investigation. One difficulty of the algorithm is that, for some data sets, it is not straightforward to find an optimal registration parameter setting (e.g., the number of radial basis functions, the number of resolution levels, or the weight of the constraint term) to deform the healthy tissues while simultaneously constraining the tumor to minimize its distortion. Once the registration is complete, the optimal approach for analyzing the aligned parametric maps has also yet to be established; in particular, the method proposed in the present study may simply not be the best. Hence, future work includes the investigation of different approaches to analyze the aligned parameter maps.
In conclusion, our study shows that tumor heterogeneity contains important information related to treatment response and should be included with ROI level data to maximize the ability of quantitative MRI to predict the response of breast tumors to NAC.
Acknowledgments
A special thank you is offered to the patients and their families for participating in our studies. We thank the Kleberg Foundation for generous support of the imaging program at our Institution.
Footnotes
We thank the National Institutes of Health for funding through NCI 1R01CA129961, NCI 1U01CA142565, NCI 1P50 098131, NCI P30 CA68485, U01CA174706, and NCRR/NIH UL1 RR024975-01 (Vanderbilt Clinical and Translational Science Award grant).
References
- 1.American Cancer Society, author. How many women get breast cancer? 2013. Available at: http://www.cancer.org/cancer/breastcancer/overviewguide/breastcancer-overview-key-statistics.
- 2.Yankeelov TE, Arlinghaus LR, Li X, Gore JC. The role of magnetic resonance imaging biomarkers in clinical trials of treatment response in cancer. Semin Oncol. 2011;38:16–25. doi: 10.1053/j.seminoncol.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson AW, Xie J, Pizzonia J, Bronen RA, Spencer DD, Gore JC. Effects of cell volume fraction changes on apparent diffusion in human cells. Magn Reson Imaging. 2000;18:689–695. doi: 10.1016/s0730-725x(00)00147-8. [DOI] [PubMed] [Google Scholar]
- 4.Cheung YC, Chen SC, Su MY, See LC, Hsueh S, Chang HK, Lin YC, Tsai CS. Monitoring the size and response of locally advanced breast cancers to neoadjuvant chemotherapy (weekly paclitaxel and epirubicin) with serial enhanced MRI. Breast Cancer Res Treat. 2003;78:51–58. doi: 10.1023/a:1022153327339. [DOI] [PubMed] [Google Scholar]
- 5.Chou CP, Wu MT, Chang HT, Lo YS, Pan HB, Degani H, Furman-Haran E. Monitoring breast cancer response to neoadjuvant systemic chemotherapy using parametric contrast-enhanced MRI: a pilot study. Acad Radiol. 2007;14:561–573. doi: 10.1016/j.acra.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 6.Martincich L, Montemurro F, De Rosa G, Marra V, Ponzone R, Cirillo S, Gatti M, Biglia N, Sarotto I, Sismondi P, et al. Monitoring response to primary chemotherapy in breast cancer using dynamic contrast-enhanced magnetic resonance imaging. Breast Cancer Res Treat. 2004;83:67–76. doi: 10.1023/B:BREA.0000010700.11092.f4. [DOI] [PubMed] [Google Scholar]
- 7.Wasser K, Klein SK, Fink C, Junkermann H, Sinn HP, Zuna I, Knopp MV, Delorme S. Evaluation of neoadjuvant chemotherapeutic response of breast cancer using dynamic MRI with high temporal resolution. Eur Radiol. 2003;13:80–87. doi: 10.1007/s00330-002-1654-1. [DOI] [PubMed] [Google Scholar]
- 8.Drew PJ, Kerin MJ, Mahapatra T, Malone C, Monson JR, Turnbull LW, Fox JN. Evaluation of response to neoadjuvant chemoradiotherapy for locally advanced breast cancer with dynamic contrast-enhanced MRI of the breast. Eur J Surg Oncol. 2001;27:617–620. doi: 10.1053/ejso.2001.1194. [DOI] [PubMed] [Google Scholar]
- 9.Abraham DC, Jones RC, Jones SE, Cheek JH, Peters GN, Knox SM, Grant MD, Hampe DW, Savino DA, Harms SE. Evaluation of neoadjuvant chemotherapeutic response of locally advanced breast cancer by magnetic resonance imaging. Cancer. 1996;78:91–100. doi: 10.1002/(SICI)1097-0142(19960701)78:1<91::AID-CNCR14>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 10.Gilles R, Guinebretière JM, Toussaint C, Spielman M, Rietjens M, Petit JY, Contesso G, Masselot J, Vanel D. Locally advanced breast cancer: contrast-enhanced subtraction MR imaging of response to preoperative chemotherapy. Radiology. 1994;191:633–638. doi: 10.1148/radiology.191.3.8184039. [DOI] [PubMed] [Google Scholar]
- 11.Yankeelov TE, Lepage M, Chakravarthy A, Broome EE, Niermann KJ, Kelley MC, Meszoely I, Mayer IA, Herman CR, McManus K, et al. Integration of quantitative DCE-MRI and ADC mapping to monitor treatment response in human breast cancer: initial results. Magn Reson Imaging. 2007;25:1–13. doi: 10.1016/j.mri.2006.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Padhani AR, Hayes C, Assersohn L, Powles T, Makris A, Suckling J, Leach MO, Husband JE. Prediction of clinicopathologic response of breast cancer to primary chemotherapy at contrast-enhanced MR imaging: initial clinical results. Radiology. 2006;239:361–374. doi: 10.1148/radiol.2392021099. [DOI] [PubMed] [Google Scholar]
- 13.Ah-See ML, Makris A, Taylor NJ, Harrison M, Richman PI, Burcombe RJ, Stirling JJ, d'Arcy JA, Collins DJ, Pittam MR, et al. Early changes in functional dynamic magnetic resonance imaging predict for pathologic response to neoadjuvant chemotherapy in primary breast cancer. Clin Cancer Res. 2008;14:6580–6589. doi: 10.1158/1078-0432.CCR-07-4310. [DOI] [PubMed] [Google Scholar]
- 14.Pickles MD, Manton DJ, Lowry M, Turnbull LW. Prognostic value of pre-treatment DCE-MRI parameters in predicting disease free and overall survival for breast cancer patients undergoing neoadjuvant chemotherapy. Eur J Radiol. 2009;71:498–505. doi: 10.1016/j.ejrad.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 15.Heldahl MG, Bathen TF, Rydland J, Kvistad KA, Lundgren S, Gribbestad IS, Goa PE. Prognostic value of pretreatment dynamic contrast-enhanced MR imaging in breast cancer patients receiving neoadjuvant chemotherapy: overall survival predicted from combined time course and volume analysis. Acta Radiol. 2010;51:604–612. doi: 10.3109/02841851003782059. [DOI] [PubMed] [Google Scholar]
- 16.Johansen R, Jensen LR, Rydland J, Goa PE, Kvistad KA, Bathen TF, Axelson DE, Lundgren S, Gribbestad IS. Predicting survival and early clinical response to primary chemotherapy for patients with locally advanced breast cancer using DCE-MRI. J Magn Reson Imaging. 2009;29:1300–1307. doi: 10.1002/jmri.21778. [DOI] [PubMed] [Google Scholar]
- 17.Newitt DC, Partridge SC, Chang B, Joe BN, Hylton N. Optimization of the percent enhancement threshold for breast MRI tumor volume measurement during neoadjuvant treatment of breast cancer for predicting recurrence free survival time. Proc Intl Soc Mag Reson Med (ISMRM) 2011;19:508. [Google Scholar]
- 18.Li SP, Makris A, Beresford MJ, Taylor NJ, Ah-See ML, Stirling JJ, d'Arcy JA, Collins DJ, Kozarski R, Padhani AR. Use of dynamic contrast-enhanced MR imaging to predict survival in patients with primary breast cancer undergoing neoadjuvant chemotherapy. Radiology. 2011;260:68–78. doi: 10.1148/radiol.11102493. [DOI] [PubMed] [Google Scholar]
- 19.Yu HJ, Chen JH, Mehta RS, Nalcioglu O, Su MY. MRI measurements of tumor size and pharmacokinetic parameters as early predictors of response in breast cancer patients undergoing neoadjuvant anthracycline chemotherapy. J Magn Reson Imaging. 2007;26:615–623. doi: 10.1002/jmri.21060. [DOI] [PubMed] [Google Scholar]
- 20.Pickles MD, Gibbs P, Lowry M, Turnbull LW. Diffusion changes precede size reduction in neoadjuvant treatment of breast cancer. Magn Reson Imaging. 2006;24:843–847. doi: 10.1016/j.mri.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 21.Sharma U, Danishad KK, Seenu V, Jagannathan NR. Longitudinal study of the assessment by MRI and diffusion-weighted imaging of tumor response in patients with locally advanced breast cancer undergoing neoadjuvant chemotherapy. NMR Biomed. 2009;22:104–113. doi: 10.1002/nbm.1245. [DOI] [PubMed] [Google Scholar]
- 22.Belli P, Costantini M, Ierardi C, Bufi E, Amato D, Mule' A, Nardone L, Terribile D, Bonomo L. Diffusion-weighted imaging in evaluating the response to neoadjuvant breast cancer treatment. Breast J. 2011;17:610–619. doi: 10.1111/j.1524-4741.2011.01160.x. [DOI] [PubMed] [Google Scholar]
- 23.Woodhams R, Kakita S, Hata H, Iwabuchi K, Kuranami M, Gautam S, Hatabu H, Kan S, Mountford C. Identification of residual breast carcinoma following neoadjuvant chemotherapy: diffusion-weighted imaging—comparison with contrast-enhanced MR imaging and pathologic findings. Radiology. 2010;254:357–366. doi: 10.1148/radiol.2542090405. [DOI] [PubMed] [Google Scholar]
- 24.Nilsen L, Fangberget A, Geier O, Olsen DR, Seierstad T. Diffusion-weighted magnetic resonance imaging for pretreatment prediction and monitoring of treatment response of patients with locally advanced breast cancer undergoing neoadjuvant chemotherapy. Acta Oncol. 2010;49:354–360. doi: 10.3109/02841861003610184. [DOI] [PubMed] [Google Scholar]
- 25.Li X, Dawant BM, Welch EB, Chakravarthy AB, Freehardt D, Mayer I, Kelley M, Meszoely I, Gore JC, Yankeelov TE. A nonrigid registration algorithm for longitudinal breast MR images and the analysis of breast tumor response. Magn Reson Imaging. 2009;27:1258–1270. doi: 10.1016/j.mri.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li X, Dawant BM, Welch EB, Chakravarthy AB, Xu L, Mayer I, Kelley M, Meszoely I, Means-Powell J, Gore JC, et al. Validation of an algorithm for the nonrigid registration of longitudinal breast MR images using realistic phantoms. Med Phys. 2010;37:2541–2552. doi: 10.1118/1.3414035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li X, Abramson RG, Arlinghaus LR, Chakravarthy AB, Abramson V, Mayer I, Farley J, Delbeke D, Yankeelov TE. An algorithm for longitudinal registration of PET/CT images acquired during neoadjuvant chemotherapy in breast cancer: preliminary results. EJNMMI Res. 2012;2:62. doi: 10.1186/2191-219X-2-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Galbán CJ, Chenevert TL, Meyer CR, Tsien C, Lawrence TS, Hamstra DA, Junck L, Sundgren PC, Johnson TD, Galbán S, et al. Prospective analysis of parametric response map-derived MRI biomarkers: identification of early and distinct glioma response patterns not predicted by standard radiographic assessment. Clin Cancer Res. 2011;17:4751–4760. doi: 10.1158/1078-0432.CCR-10-2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li X, Arlinghaus LR, Ayers GD, Chakravarthy AB, Abramson RG, Abramson VG, Atuegwu N, Farley J, Mayer IA, Kelley MC, et al. DCE-MRI analysis methods for predicting the response of breast cancer to neoadjuvant chemotherapy: pilot study findings. Magn Reson Med. 2013 doi: 10.1002/mrm.24782. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Daldrup H, Shames DM, Wendland M, Okuhata Y, Link TM, Rosenau W, Lu Y, Brasch RC. Correlation of dynamic contrast-enhanced MR imaging with histologic tumor grade: comparison of macromolecular and small-molecular contrast media. AJR Am J Roentgenol. 1998;171:941–949. doi: 10.2214/ajr.171.4.9762973. [DOI] [PubMed] [Google Scholar]
- 31.Li X, Welch EB, Chakravarthy AB, Lei X, Arlinghaus LR, Farley J, Loveless ME, Mayer I, Kelley M, Meszoely I, et al. A novel AIF detection method and a comparison of DCE-MRI parameters using individual and population based AIFs in human breast cancer. Phys Med Biol. 2011;56:5753–5769. doi: 10.1088/0031-9155/56/17/018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maes F, Collignon A, Vandermeulen D, Marchal G, Suetens P. Multimodality image registration by maximization of mutual information. IEEE Trans Med Imaging. 1997;16:187–198. doi: 10.1109/42.563664. [DOI] [PubMed] [Google Scholar]
- 33.Rohde GK, Aldroubi A, Dawant BM. The adaptive bases algorithm for intensity-based nonrigid image registration. IEEE Trans Med Imaging. 2003;22:1470–1479. doi: 10.1109/TMI.2003.819299. [DOI] [PubMed] [Google Scholar]
- 34.Wu Z. Multivariate compactly supported positive definite radial functions. Adv Comput Math. 1995;4:283–292. [Google Scholar]
- 35.Li X, Arlinghaus LR, Chakravarthy AB, Welch EB, Farley J, Mayer I, Abramson V, Abramson R, Kelley M, Meszoely I, et al. Towards optimization of DCE-MRI analysis for early prediction of the response of breast cancer patients to neoadjuvant chemotherapy. Proc Intl Soc Mag Reson Med (ISMRM) 2012;20:1464. [Google Scholar]
- 36.Li X, Arlinghaus LR, Chakravarthy AB, Abramson RG, Abramson V, Farley J, Ayers GD, Mayer I, Kelley M, Meszoely I, et al. Quantitative DCE-MRI to predict the response of primary breast cancer to neoadjuvant therapy. Cancer Res. 2012;72(24) Suppl 3 SABCS12-P4-01-03. [Google Scholar]
- 37.Van Den Wollenberg AL. Redundancy analysis an alternative for canonical correlation analysis. Psychometrika. 1977;42:207–219. [Google Scholar]
- 38.Le Cessie S, Van Houwelingen JC. Ridge estimators in logistic regression. J R Stat Soc. 1992;41:191–201. [Google Scholar]
- 39.Cule E, De Iorio M. Ridge regression in prediction problems: automatic choice of the ridge parameter. Genet Epidemiol. 2013;37:704–714. doi: 10.1002/gepi.21750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Elston CW, Ellis IO. Assessment of histological grade. In: Elston CW, Ellis IO, editors. The Breast. Vol. 13. Edinburgh, NY: Churchill Livingstone; 1998. pp. 356–384. [Google Scholar]
- 41.Hanley JA, McNeil BJ. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology. 1982;143:29–36. doi: 10.1148/radiology.143.1.7063747. [DOI] [PubMed] [Google Scholar]
- 42.Efron B. Estimating the error rate of a prediction rule: improvement on cross-validation. J Am Stat Assoc. 1983;78:316–331. [Google Scholar]
- 43.Harrell F., Jr . Regression Modeling Strategies: With Applications to Linear Models, Logistic Regression, and Survival Analysis. New York, NY: Springer; 2001. Resampling, validating, describing, and simplifying the model. [Google Scholar]
- 44.Breiman L, Spector P. Submodel selection and evaluation in regression: the X-random case. International Statistical Review. 1992;60:291–319. [Google Scholar]
- 45.Ma B, Meyer CR, Pickles MD, Chenevert TL, Bland PH, Galbán CJ, Rehemtulla A, Turnbull LW, Ross BD. Voxel-by-voxel functional diffusion mapping for early evaluation of breast cancer treatment. Inf Process Med Imaging. 2009;21:276–287. doi: 10.1007/978-3-642-02498-6_23. [DOI] [PMC free article] [PubMed] [Google Scholar]