Abstract
Pharmacokinetic analysis of dynamic contrast-enhanced magnetic resonance imaging (DCE-MRI) time-course data allows estimation of quantitative parameters such as Ktrans (rate constant for plasma/interstitium contrast agent transfer), ve (extravascular extracellular volume fraction), and vp (plasma volume fraction). A plethora of factors in DCE-MRI data acquisition and analysis can affect accuracy and precision of these parameters and, consequently, the utility of quantitative DCE-MRI for assessing therapy response. In this multicenter data analysis challenge, DCE-MRI data acquired at one center from 10 patients with breast cancer before and after the first cycle of neoadjuvant chemotherapy were shared and processed with 12 software tools based on the Tofts model (TM), extended TM, and Shutter-Speed model. Inputs of tumor region of interest definition, pre-contrast T1, and arterial input function were controlled to focus on the variations in parameter value and response prediction capability caused by differences in models and associated algorithms. Considerable parameter variations were observed with the within-subject coefficient of variation (wCV) values for Ktrans and vp being as high as 0.59 and 0.82, respectively. Parameter agreement improved when only algorithms based on the same model were compared, e.g., the Ktrans intraclass correlation coefficient increased to as high as 0.84. Agreement in parameter percentage change was much better than that in absolute parameter value, e.g., the pairwise concordance correlation coefficient improved from 0.047 (for Ktrans) to 0.92 (for Ktrans percentage change) in comparing two TM algorithms. Nearly all algorithms provided good to excellent (univariate logistic regression c-statistic value ranging from 0.8 to 1.0) early prediction of therapy response using the metrics of mean tumor Ktrans and kep (=Ktrans/ve, intravasation rate constant) after the first therapy cycle and the corresponding percentage changes. The results suggest that the interalgorithm parameter variations are largely systematic, which are not likely to significantly affect the utility of DCE-MRI for assessment of therapy response.
Introduction
With advances in targeted molecular therapy for cancer treatment, change in tumor size in response to therapy, which is routinely used in standard care for therapeutic monitoring, is often found to manifest later than changes in underlying tumor characteristics [1–8], such as vascularization and vascular permeability, cellularity, metabolism, and biochemistry. Thus, imaging modalities that can quantify tumor functions are becoming increasingly important for evaluation and prediction of therapy response. Dynamic contrast-enhanced magnetic resonance imaging (DCE-MRI) is a minimally invasive imaging method that measures changes in tissue microvascular properties and has been widely used in research or early phase clinical trial settings to provide assessment of tumor therapeutic response [1–8], as many cancer drugs affect tumor vasculature directly or indirectly [9]. The quantitative approach for DCE-MRI data analysis using pharmacokinetic models allows extraction and mapping of quantitative parameters of tumor biology in vivo. These parameters are usually variants of Ktrans, a rate constant for contrast agent (CA) molecule plasma/interstitium transfer, ve, the volume fraction of interstitial space (extracellular and extravascular, the putative CA distribution volume), and vp, the plasma volume fraction. The CA intravasation rate constant, kep, can be calculated as Ktrans/ve. Recent workshops on both sides of the Atlantic have generated guidelines and recommendations on acquisition and analysis of DCE-MRI data for the purpose of assessing tumor therapy response [9–11].
Unlike qualitative (such as description of curve shape) or semi-quantitative (such as calculation of maximum signal change) analysis, the parameters derived from pharmacokinetic modeling of DCE-MRI time-course data should, in principle, be independent of MRI scanner platform (vendor and field strength), data acquisition details (pulse sequence and parameters), CA dose and/or injection rate, personnel skills, and so on, which makes them desirable imaging end points in multicenter clinical trial studies. However, the accuracy and precision of these parameters can be affected by a plethora of factors, including errors in quantification of pre-contrast T1 [12–15] and determination of arterial input function (AIF) [4,9,16,17], inadequate temporal resolution [9,18,19] or signal-to-noise ratio [15], as well as selection of models to fit the data [9,20,21]. A recent study [22] comparing four commercial software packages for quantitative DCE-MRI data analysis has revealed considerable variability in pharmacokinetic parameter quantification from data sets of 15 subjects, with up to 74% within-subject coefficient of variation (wCV) among the tools, even though all four software packages were presumably based on the same Tofts model (TM) [23]. Commercialization of software tools for kinetic modeling of DCE-MRI data represents a necessary step for wide dissemination of DCE-MRI as a quantitative imaging biomarker in clinical trials and general practice. However, the poor reproducibility shown by this study among the available commercial solutions is one of the major obstacles in integration of quantitative DCE-MRI into standard care. Thorough comparison and validation of algorithms/software tools for DCE-MRI data analysis are necessary within the context of monitoring tumor response to therapy.
Recognizing the importance of quantitative imaging for assessment of cancer response to therapy and rapid evaluation of the efficacy of new anticancer drugs, the National Cancer Institute has recently established the Quantitative Imaging Network (QIN) to provide resources for developing and validating quantitative imaging tools. The main mission of the QIN Image Analysis and Performance Metrics Working Group is to provide guidance and reach consensus on quantitative image analysis methods through comparison and validation of analysis algorithms. The available QIN infrastructure facilitates collaborative challenge projects involving multiple QIN centers. Here, we report the results from a DCE-MRI data analysis challenge project, in which several QIN centers performed analyses of DCE-MRI data from a digital reference object (DRO) [24] and human breast tumors using site-specific employment of algorithms/software tools. The overall goal of the project was to compare and validate DCE-MRI data analysis tools available within the QIN. Because ultimately the utility of a quantitative imaging method for assessing cancer therapy response is judged by its robustness in evaluation/prediction of clinical and/or pathologic end points of response, the DCE-MRI pharmacokinetic parameters and their changes following therapy were correlated with pathologic response status of the patients with breast cancer to compare the capabilities of the algorithms/tools in predicting complete response versus non-complete response.
Materials and Methods
DCE-MRI Challenge Participating QIN Centers
The QIN centers that participated in this DCE-MRI data analysis challenge project are Oregon Health and Science University (OHSU), Vanderbilt University (VU), University of Pittsburgh (UP), Brigham and Women's Hospital (BWH) and in collaboration with General Electric Research and Development (BWH-GE), University of Michigan (UM), University of Washington (UW), and Icahn School of Medicine at Mount Sinai (ISM).
Simulated DRO DCE-MRI Data Sharing and Analysis
A software phantom or DRO with known pharmacokinetic parameter values can be an effective means to compare and validate different DCE-MRI data analysis packages by providing a reference. The Quantitative Imaging Biomarker Alliance (QIBA) and Duke University have generated a variety of simulated DCE-MRI data sets using a matrix of parameter values [24]. For this challenge project, the DRO data of the QIBA_v6_Tofts version [25] were made available to the participating QIN centers. The DRO data (Figure 1) were constructed with 30 combinations of six Ktrans (0.01, 0.02, 0.05, 0.1, 0.2, and 0.35 min-1) and five ve (0.01, 0.05, 0.1, 0.2, and 0.5) values according to the TM [23] and consisted of a single image slice of simulated DCE-MRI signal intensity time-course data (with 0.5-second temporal resolution) without noise in DICOM format with headers for both Siemens and GE scanner platforms. Each Ktrans and ve combination occupied a 10 x 10 pixel square area of the slice, allowing evaluation of reproducibility in fitting pixel data with the same Ktrans and ve combination. Details of simulated data acquisition parameters and fixed parameters used for the TM can be found in references [24] and [25].
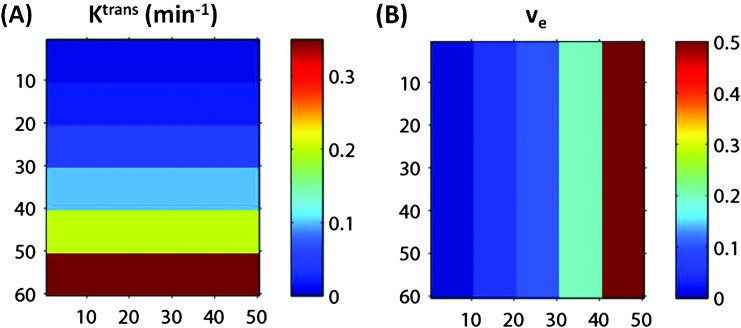
Figure 1.
Two-dimensional image view of the arrays of Ktrans (A) and ve (B) values used to construct the simulated DRO DCE-MRI data without noise. Each color stripe represents one of six Ktrans (0.01, 0.02, 0.05, 0.1, 0.2, and 0.35 min-1) (A) or one of five ve (0.01, 0.05, 0.1, 0.2, and 0.5) (B) values. The numbers along the x- and y-axes represent pixel numbers in both panels. Combination of the two panels results in 30 squares with 10 x 10 pixels each, representing the possible 30 combinations of Ktrans and ve values for simulated DRO data.
The DRO data were downloaded from the Duke University websites [24] and, with permission of the creators, stored at a secure website (with help from the QIN Bioinformatics Working Group) for download by the participating QIN sites. Before proceeding to analyze the human breast DCE-MRI data, the participating sites were asked to process the simulated DRO DCE-MRI data using their TM-based algorithms/software tools with the purpose of validating the mathematical formulations implemented in the TM algorithms. The results of DRO data analyses were submitted as parametric maps in Matlab, Nifti, or Nrrd formats.
Human Breast DCE-MRI Data Acquisition and Sharing
As part of the quantitative imaging studies at OHSU as an individual QIN center, breast DCE-MRI data were acquired with consent from patients with locally advanced breast cancer who underwent preoperative neoadjuvant chemotherapy (NACT). Four DCE-MRI sessions were undertaken during the treatment course: pre-NACT (visit 1, V1), after the first cycle of NACT (visit 2, V2), at a NACT midpoint (usually after the third NACT cycle; visit 3), and after the completion of NACT (visit 4). The study was conducted under the approval of the local Institutional Review Board (IRB). For this challenge project, the V1 and V2 data sets from 10 of 16 consecutive patients were shared among the participating QIN sites. The data from the other six patients were not selected because either the V2 data were not acquired or severe motion degraded data quality at V1 and/or V2. All breast MRI studies were performed using a Siemens 3T system with a body coil and a four-channel bilateral phased-array breast coil as the transmitter and receiver, respectively. Following pilot scans and pre-CA T2-weighted MRI with fat saturation and T1-weighted MRI without fat saturation, axial bilateral DCE-MRI images with fat saturation and full breast coverage were acquired with a three-dimensional (3D) gradient echo-based Time-resolved angiography WIth Stochastic Trajectories (TWIST) sequence, which employs the strategy of k-space under-sampling during acquisition and data sharing during reconstruction [26,27]. DCE-MRI acquisition parameters included 10° flip angle, 2.9/6.2 millisecond echo time/repetition time (TE/TR), a parallel imaging acceleration factor of two, 30 to 34 cm field of view (FOV), 320 x 320 in-plane matrix size, and 1.4-mm slice thickness. The total acquisition time was ∼10 minutes for 32 to 34 image volume sets of 112 to 120 slices each with 18- to 20-second temporal resolution. The CA Gd (HP-DO3A) [ProHance] IV injection (0.1 mmol/kg at 2 ml/s) by a programmable power injector was timed to commence after acquisition of two baseline image volumes, followed by a 20-ml saline flush. Breast tumor regions of interest (ROIs) were drawn by experienced OHSU breast radiologists on post-CA multislice images covering the contrast-enhanced tumor.
For DCE-MRI data sharing, the V1 and V2 raw data sets in Matlab, Nifti, and DICOM formats together with the registered predrawn tumor ROIs from the 10 patients—a total of 20 data sets—were uploaded to the same secure website that housed the DRO data and downloaded by the participating sites. Since the human MRI data were acquired under strictly coded names (e.g., “Breast Chemo1” as the last name and “Visit 1” as the first name) and without any entries of identifiable patient health information, no patient health information was ever shared among the QIN sites. The sharing of the anonymous human breast MRI data within the QIN was approved by the OHSU IRB.
Human Breast DCE-MRI Data Analysis
The pharmacokinetic models used by the participating sites for breast tumor DCE-MRI data analysis generally fell into the following three categories: the TM [23], the extended TM (ETM) [28,29], and the fast exchange regime (FXR)-allowed Shutter-Speed Model (SSM) [17,30]. Equations 1, 2, and 3 below represent the basic formulations for the TM, ETM, and FXR-SSM, respectively:
| (1) |
| (2) |
| (3) |
where Ct(t) represents tissue CA concentration at time t; Cp(t′) is the arterial plasma CA concentration time course, or AIF; R1*(t) is the tissue apparent longitudinal relaxation rate constant; R1i is the intracellular longitudinal relaxation rate constant; R10 is the pre-CA tissue R1; r1 is the tissue CA relaxivity; and τi is the mean intracellular water molecule lifetime. Both the TM and ETM neglect the finite intercompartmental water exchange kinetics, assuming the system is in the fast exchange limit condition. As a result, R1(t) [or R1*(t)] is linearly related to Ct(t):
| (4) |
The SSM accounts for the finite water exchange kinetics during the CA passage through the tissue of interest, and consequently, R1(t) is not linearly related to Ct(t). The FXR version is a two-compartment three-parameter (extracellular and intracellular compartments; Ktrans, ve, and τi) SSM, taking into account transcytolemmal water exchange kinetics but assuming single exponential longitudinal MR signal decay [17,30].
Each QIN site had the option of using one or more models for DCE-MRI time-course data fitting. This challenge project focuses on comparisons of different pharmacokinetic models and associated software packages for therapy response assessment. Therefore, to minimize variations in derived kinetic parameters caused by factors other than differences in models and algorithms, such as uncertainties in AIF determination and R10 quantification, a population averaged AIF was provided to each site for analysis of all 20 data sets and the R10 value was given for each of the 20 studies. The population-averaged AIF was obtained by averaging individually measured AIFs from an axillary artery in a previous sagittal breast DCE-MRI study [31–33], which employed the same CA injection protocol, including dose, injection rate, and injection site (antecubital vein). The average R10 value for each tumor was determined by comparing signal intensities of pre-CA baseline DCE images with those of the spatially registered proton density images acquired just before DCE-MRI [27] and was used for kinetic modeling of tumor ROI or pixel (within the ROI) DCE data by each site. In addition, the tumor ROI definitions on multiple image slices covering the tumor were also provided for each data set (see above).
The data analysis tools used by the participating sites included software packages built in-house, obtained as free open source, or purchased commercially, as well as prototype research software under development by GE. For results reporting, each site was required to provide mean tumor ROI pharmacokinetic parameter values, the corresponding percentage changes (V2 relative to V1, V21), and parametric maps. Some sites calculated the mean tumor ROI parameters by fitting ROI time-course data from each slice and then averaging the derived slice ROI parameter values, while the others computed those by averaging pixel parameter values obtained from fittings of all pixel time-course data. The details of pharmacokinetic analyses performed by each site from model selection to mean parameter calculation are summarized in Table 1.
Table 1.
Details of Pharmacokinetic Analysis of Human Breast DCE-MRI Data at Participating Sites.
| Characteristics | OHSU | VU | UP | UM | UW | ISM | BWH-GE | BWH-3D Slicer |
| Selected models | TM, FXR-SSM | TM, ETM, FXR-SSM | TM | ETM | ETM | TM | TM, ETM | TM |
| Returned parameters | Ktrans, ve, kep, τi (SSM only) | Ktrans, ve, kep, vp (ETM only), τi (SSM only) | Ktrans, ve, kep | Ktrans, ve, kep, vp | Ktrans, ve, kep, vp | Ktrans, ve, kep | Ktrans, ve, kep, vp (ETM only) | Ktrans, ve, kep |
| Software origin and platform | In-house Matlab | In-house Matlab, IDL | In-house Matlab | In-house Matlab | In-house and commercial* | In-house Matlab | Research prototype for commercialization | Open source 3D Slicer† |
| Tumor ROI definition | Provided | Provided | Provided | Provided | Provided | Provided | Provided | Provided |
| 〈AIF〉 | Provided | Provided | Provided | Provided | Provided | Provided | Provided | Provided |
| 〈R10〉 | Provided | Provided | Provided | Provided | Provided | Provided | Provided | Provided |
| Motion correction | NA | NA | NA | NA | NA | NA | NA | NA |
| CA arrival time estimation | Site-specific | Site-specific | Site-specific | Site-specific | Site-specific | Site-specific | Site-specific | Site-specific |
| Model goodness of fit | Site-specific | Site-specific | Site-specific | Site-specific | Site-specific | Site-specific | Site-specific | Site-specific |
| Mean tumor ROI parameter calculation‡ | Averaging slice ROI values | Averaging pixel values | Averaging slice ROI values | Averaging pixel values | Averaging slice ROI values | Averaging slice ROI values | Averaging pixel values | Averaging pixel values |
〈AIF〉, population-averaged AIF; 〈R10〉, average pre-contrast longitudinal relaxation rate constant.
In-house Matlab script was generated to load image data and covert ROI signal intensity time course to CA concentration time course, which was then fit with the ETM using a commercial software package, PMOD (PMOD Technologies Ltd., Zurich, Switzerland).
Free open source extension for 3D Slicer, http://www.slicer.org/slicerWiki/index.php/Documentation/4.3/Modules/PkModeling.
“Averaging slice ROI values”: Slice ROI parameter value was obtained by fitting tumor ROI DCE-MRI data from the slice, and these values from all slices covering the targeted tumor were then averaged; “averaging pixel values”: Pixel parameter value was obtained by fitting pixel DCE-MRI data, and these values from all pixels within all tumor ROIs were then averaged.
Determination of Pathologic Response Status
The pathologic response (to NACT) status of each patient with breast cancer was determined by pathologic analyses and comparisons of the pre-NACT core biopsy specimen with the post-NACT resection specimen. The two previously published and validated indices [34,35] were measured: Relative Change in Tumor Density (RCTD) and Residual Cancer Burden. A pathologic complete response (pCR) is defined as the absence of residual tumor (RCTD = -1.0; Residual Cancer Burden = 0). A pathologic nonresponse (pNR) is defined as tumor cell density in resection specimen equal to or greater than that in core biopsy specimen (RCTD ≥ 0). Pathologic partial response is defined as findings intermediate between pCR and pNR. For this study, the binary classification of pCR and non-pCR (pNR and pathologic partial response) was used for the 10-patient cohort.
Statistical Analysis
For DRODCE-MRI data analysis, the estimated Ktrans and ve values from data fitting using site-specific TM algorithms were compared to the simulated “true” Ktrans and ve values that were used to construct the DRO data.
For human breast DCE-MRI data analysis, descriptive statistical analysis was conducted to summarize the pharmacokinetic parameter values returned by each algorithm/software tool. To assess the reproducibility of each parameter across all algorithms and within each model (i.e., TM, ETM, or SSM), linear mixed models were fit to obtain the maximum likelihood estimates of the intraclass correlation coefficient (ICC) and wCV, and their corresponding 95% confidence intervals (CIs). The concordance correlation coefficients (CCCs) and the corresponding 95% CIs were estimated to represent the level of pairwise linear agreement to a 45° line of which the intercept is forced to be zero. Linear mixed models were fit to determine whether the parameter mean differs for pairwise comparisons of the three kinetic models. Univariate logistic regression (ULR) models were fit to compute the c-statistic, a measure equivalent to the area under the Receiver Operating Characteristic curve, to reflect the predictive ability of each parameter for tumor therapy response. While logarithm transformation will help address the skewed distribution of some parameters, the analysis results are rather robust to using values with or without transformation. Therefore, the original parameter values returned from all algorithms were used for statistical analysis throughout, which was conducted using SAS 9.3 (Cary, NY). SAS macro %ICC9 and %mccc were used for estimations of ICC, wCV, and CCC.
Results
DRO DCE-MRI Data Analysis
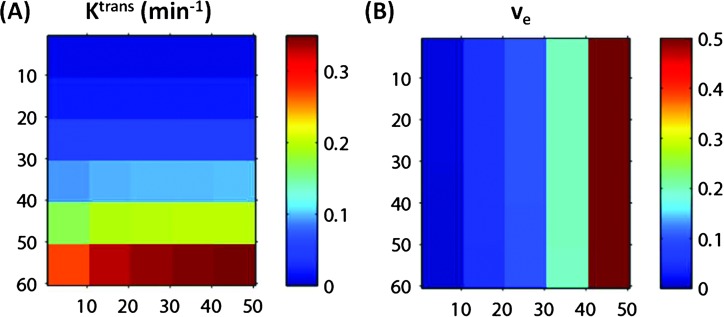
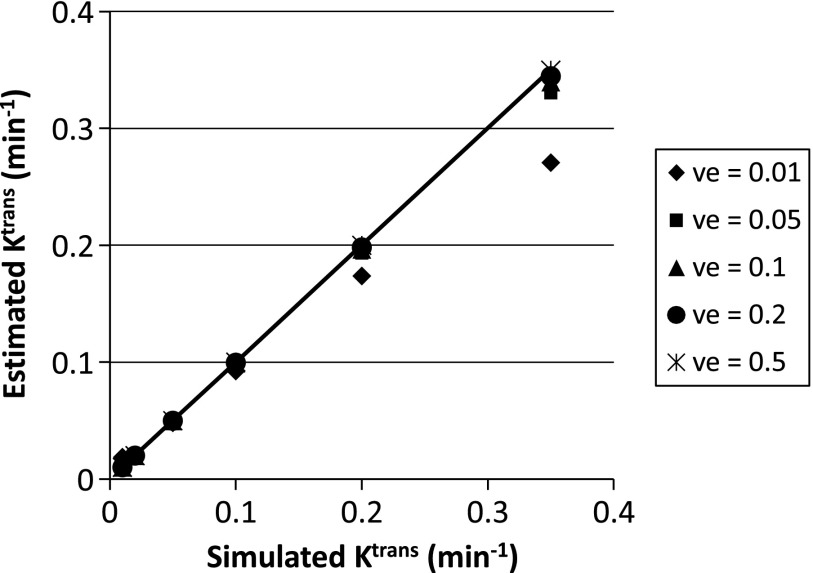
DRO data analyses by five of six TM algorithms/software packages used in this challenge generally returned fairly accurate Ktrans and ve values with <10% errors for both parameters compared to the simulated values. The exceptions occurred in the lower left area of the simulated Ktrans and ve arrays (Figure 1), where high Ktrans and low ve values are combined—the errors for the estimated parameters were the highest for the combination of Ktrans = 0.35 min-1 and ve = 0.01. As an example, Figure 2 shows the 2D arrays of the estimated Ktrans (Figure 2A) and ve (Figure 2B) values obtained with the OHSU TM algorithm. The highest error can be clearly seen in the lower left square of the Ktrans array (Figure 2A), which represents an estimated Ktrans value of 0.27 min-1, 23% smaller than the simulated value of 0.35 min-1. The estimated ve value in the same square is 11% smaller than the simulated value of 0.01. However, the error is not clearly identifiable in the ve array (Figure 2B) due to the small absolute value of the error (0.01 x 11%= 0.0011). The pattern of uniform color in each square of 10 x 10 pixels indicates high reproducibility of the OHSU TM algorithm in analyzing pixel DCE-MRI time-course data constructed with the same Ktrans and ve combination. Figure 3 shows a scatter plot of estimated Ktransversus simulated Ktrans values based on results obtained with the ISM TM algorithm, illustrating nearly the same pattern of estimated Ktrans values as that generated by the OHSU TM algorithm (Figure 2A): good agreement between estimated and simulated Ktrans values except for where ve = 0.01 and Ktrans ≥ 0.2 min-1, and the error increases with Ktrans value. All five TM software packages generated consistent maximum errors at the simulated combination of Ktrans = 0.35 min-1 and ve = 0.01: -23% for Ktrans and -11% for ve.
Figure 2.
Two-dimensional image view of the arrays of estimated Ktrans (A) and ve (B) values obtained by fitting the DRO data with the TM algorithm implemented at OHSU. Good agreements are seen between the estimated Ktrans (A) and simulated “true” Ktrans (Figure 1A) values except for in the lower left areas of the panel with high Ktrans and low ve combinations. There are no visible differences seen between the estimated ve (B) and simulated “true” ve (Figure 1B) values.
Figure 3.
The estimated Ktrans values obtained by fitting the DRO data with the TM algorithm implemented at ISM are plotted against the simulated “true” Ktrans values at five simulated ve values. The straight line is the line of unity, representing perfect agreement between the estimated and simulated Ktrans values.
Human Breast DCE-MRI Analysis
As summarized in Table 1, the V1 and V2 DCE-MRI data from the 10 patients were processed by seven institutions using 12 pharmacokinetic analysis algorithms: 6 TM (OHSU, VU, UP, ISM, BWH-GE, and BWH-3D Slicer [36]), 4 ETM (VU, UM, UW, and BWH-GE), and 2 SSM (OHSU and VU).
Variance analysis.
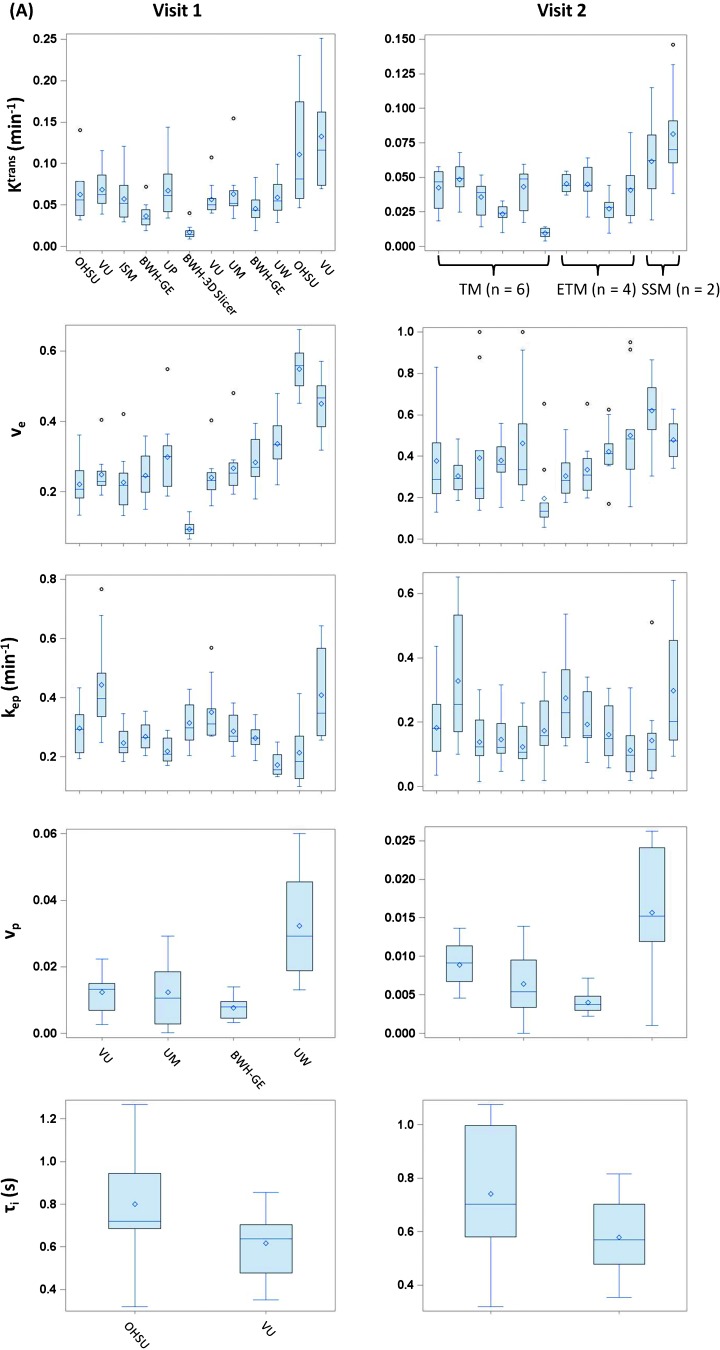
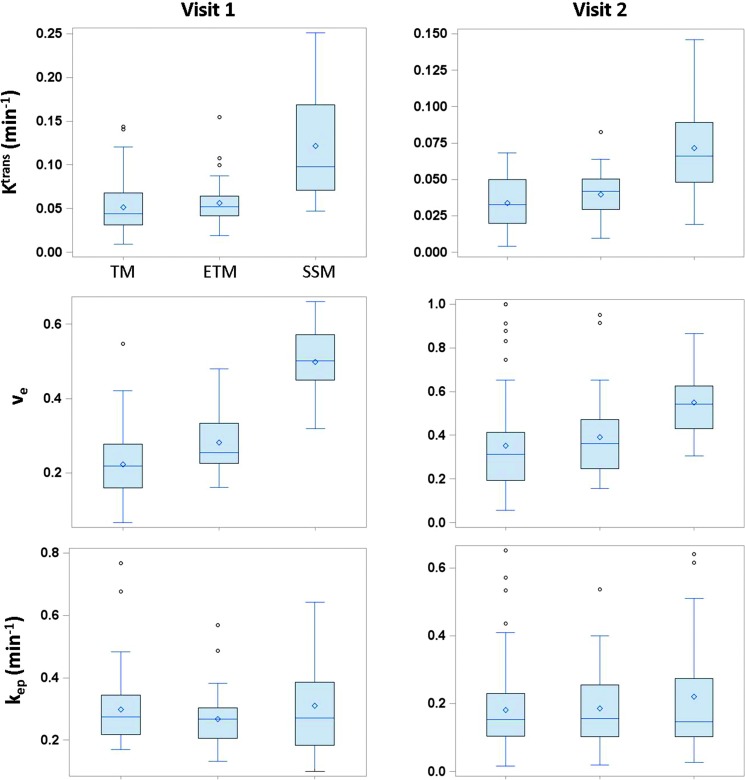
Figure 4A shows the box plots of mean tumor Ktrans, ve, and kep values at V1 and V2 obtained using all 12 algorithms, vp values from the 4 ETM algorithms, and τi values from the 2 SSM algorithms only, whereas Figure 4B displays the box plots of percentage changes (V21) of the same parameters. Considerable variations are observed for all returned parameters at V1 or V2 across different algorithms with the Ktrans and ve values returned by the SSM algorithms (e.g., range at V1: Ktrans, 0.047–0.25 min-1; ve, 0.32–0.66) generally larger than those returned by either the TM (range at V1: Ktrans, 0.0090–0.14 min-1; ve, 0.066–0.55) or the ETM (range at V1: Ktrans, 0.019–0.15 min-1; ve, 0.16–0.48) algorithms. Interestingly, the percentage changes of the DCE-MRI parameters (Figure 4B) show substantial improvement in stability across all the software tools, e.g., the range of the mean values of Ktrans percentage change obtained from the 12 algorithms is relatively small: -13% to -28%.
Figure 4.
(A) Box plots of V1 (left column) and V2 (right column) mean tumor Ktrans, ve, kep, vp, and Ti values from the 10 patients with breast cancer. (B) Box plots of percentage changes (V2 relative to V1) of the five parameters. The central bar and diamond symbols represent the median and mean values, respectively. Ktrans, ve, and kep are obtainable with all three pharmacokinetic models: TM, ETM, and SSM. The box plots associated with the same model are grouped together: institution abbreviations are labeled in the V1 Ktrans panel, while the model abbreviations are labeled in the V2 Ktrans panel. The same labeling orders also apply to the V1 and V2 ve and kep panels in A and Ktrans, ve, and kep percentage change panels in B. With the models used in this study, the vp and Ti parameters can be derived only with the ETM and SSM, respectively. The institution labeling orders in V1 vp and Ti panels (A) apply to the V2 vp and Ti panels in A and vp and Ti percentage change panels in B, respectively.
Figure 5 shows the column graphs of wCV for all five parameters at V1 and V2. wCV cannot be calculated for percentage changes because negative values are not allowed. A smaller wCV value indicates less variation in measurements on the same subject by different approaches. In this study, the wCV values range from 0.25 for τi at V2 to 0.82 for vp at V1. Among the five DCE-MRI parameters, the vp parameter obtained with the ETM shows the largest variations.
Figure 5.
Column graph of mean wCV for Ktrans, ve, kep, vp, and Ti parameters at V1 (blue) and V2 (red) obtained with all 12 algorithms.
Table 2 lists the Ktrans ICC and the corresponding 95% CI values for comparisons of all 12 algorithms, TM only, ETM only, and SSM only. ICC represents the proportion of total variation contributed by between-subject difference, with high ICC indicating good agreement among different measurement approaches. The ICC values are generally lower when all 12 software tools are compared as opposed to only algorithms within the same model are compared. It is notable in Table 2 that percentage change of Ktrans generally has higher ICC values than absolute Ktrans value at either V1 or V2. The same patterns are observed for the other parameters (results not shown here). Among all the parameters, vp has the lowest ICC values of 0.09, 0.04, and 0.11 for V1, V2, and percentage change, respectively.
Table 2.
Ktrans ICC.
| Algorithm Comparison | V1 | V2 | % Change (V21) |
| All algorithms (n = 12) | 0.27 (0.11, 0.53) | 0.18 (0.06, 0.44) | 0.69 (0.46, 0.85) |
| TM (n = 6) | 0.44 (0.20, 0.71) | 0.17 (0.04, 0.54) | 0.91 (0.79, 0.96) |
| ETM (n = 4) | 0.62 (0.34, 0.84) | 0.36 (0.11, 0.71) | 0.63 (0.34, 0.84) |
| SSM (n = 2) | 0.84 (0.58, 0.95) | 0.72 (0.36, 0.92) | 0.89 (0.69, 0.97) |
% Change (V21), percentage change (V2 relative to V1).
The two numbers in parenthesis represent 95% CIs.
Concordance analysis.
Concordance correlation analysis was conducted to assess parameter agreement between any two algorithms within the same pharmacokinetic model (TM, ETM, or SSM). Table 3 tabulates the Ktrans CCC and 95% CI values for any combination of two algorithms belonging to the same pharmacokinetic model. Though the CCC values can be as low as 0.047 for V2 Ktrans between two TM software tools, those for Ktrans percentage change are consistently high—many are greater than 0.9. Similar patterns are also observed for kep, but not for ve, vp, and τi. Table 4 lists the ranges of the CCC values for ve, kep, vp, and τi. The ve, kep, and vp parameters all have large CCC ranges at V1 and V2, but only the kep percentage change has a much tighter CCC range for either the TM or ETM. Note that comparison of the two SSM algorithms generated only one CCC value.
Table 3.
CCC for Ktrans within the TM, ETM, or SSM.
| V1 | V2 | % Change (V21) | |
| TM | |||
| OHSU_TM and VU_TM | 0.870 (0.597, 0.962) | 0.734 (0.318, 0.913) | 0.886 (0.634, 0.968) |
| OHSU_TM and ISM_TM | 0.968 (0.896, 0.991) | 0.807 (0.509, 0.932) | 0.868 (0.600, 0.961) |
| OHSU_TM and BWH-GE_TM | 0.512 (0.127, 0.763) | 0.295 (0.012, 0.534) | 0.911 (0.723, 0.974) |
| OHSU_TM and UP_TM | 0.981 (0.940, 0.994) | 0.911 (0.705, 0.975) | 0.899 (0.683, 0.971) |
| OHSU_TM and BWH-3D Slicer_TM | 0.179 (-.065, 0.404) | 0.058 (-.060, 0.175) | 0.781 (0.406, 0.931) |
| VU_TM and ISM_TM | 0.850 (0.592, 0.950) | 0.538 (0.161, 0.778) | 0.972 (0.912, 0.991) |
| VU_TM and BWH-GE_TM | 0.403 (0.121, 0.625) | 0.212 (0.025, 0.384) | 0.961 (0.873, 0.989) |
| VU_TM and UP_TM | 0.883 (0.624, 0.967) | 0.683 (0.206, 0.898) | 0.959 (0.860, 0.988) |
| VU_TM and BWH-3D Slicer_TM | 0.125 (-.023, 0.268) | 0.047 (-.028, 0.121) | 0.922 (0.776, 0.974) |
| ISM_TM and BWH-GE_TM | 0.598 (0.229, 0.817) | 0.438 (0.087, 0.692) | 0.978 (0.922, 0.994) |
| ISM_TM and UP_TM | 0.934 (0.820, 0.976) | 0.840 (0.620, 0.937) | 0.985 (0.947, 0.996) |
| ISM_TM and BWH-3D Slicer_TM | 0.198 (-.044, 0.417) | 0.081 (-.053, 0.213) | 0.953 (0.839, 0.987) |
| BWH-GE_TM and UP_TM | 0.447 (0.088, 0.703) | 0.269 (-.009, 0.508) | 0.980 (0.926, 0.994) |
| BWH-GE_TM and BWH-3D Slicer_TM | 0.386 (0.102, 0.612) | 0.186 (-.001, 0.361) | 0.938 (0.792, 0.983) |
| UP_TM and BWH-3D Slicer_TM | 0.156 (-.063, 0.361) | 0.056 (-.060, 0.172) | 0.922 (0.743, 0.978) |
| ETM | |||
| VU_ETM and UM_ETM | 0.822 (0.479, 0.947) | 0.715 (0.229, 0.916) | 0.795 (0.416, 0.939) |
| VU_ETM and BWH-GE_ETM | 0.710 (0.310, 0.897) | 0.247 (0.007, 0.460) | 0.650 (0.151, 0.885) |
| VU_ETM and UW_ETM | 0.591 (0.012, 0.874) | 0.428 (-.192, 0.803) | 0.424 (-.224, 0.812) |
| UM_ETM and BWH-GE_TM | 0.570 (0.090, 0.835) | 0.417 (0.114, 0.649) | 0.787 (0.392, 0.937) |
| UM_ETM and UW_ETM | 0.597 (0.024, 0.875) | 0.546 (-.044, 0.853) | 0.546 (-.062, 0.858) |
| BWH-GE_ETM and UW_ETM | 0.616 (0.161, 0.855) | 0.391 (-.098, 0.728) | 0.804 (0.429, 0.943) |
| SSM | |||
| OHSU_SSM and VU_SSM | 0.859 (0.592, 0.956) | 0.761 (0.445, 0.908) | 0.902 (0.679, 0.972) |
% Change (V21), percentage change (V2 relative to V1).
The two numbers in parenthesis represent 95% CIs.
Note: The institution abbreviation is listed ahead of the model abbreviation.
Table 4.
Range of Pairwise CCC for ve, kep, vp, and τi.
| Parameter | Algorithm Comparison | V1 | V2 | % Change (V21) |
| ve | TM (n = 6) | 0.020–0.885 | 0.414–0.965 | 0.062–0.914 |
| ETM (n = 4) | 0.246–0.891 | 0.407–0.931 | 0.132–0.969 | |
| SSM (n = 2) | 0.175–0.175 | 0.456–0.456 | 0.432–0.432 | |
| kep | TM (n = 6) | 0.030–0.782 | 0.267–0.973 | 0.754–0.992 |
| ETM (n = 4) | 0.069–0.792 | 0.183–0.882 | 0.571–0.913 | |
| SSM (n = 2) | 0.134–0.134 | 0.530–0.530 | 0.511–0.511 | |
| vp | ETM (n = 4) | 0.108–0.764 | 0.077–0.407 | 0.087–0.909 |
| τi | SSM (n = 2) | 0.437–0.437 | 0.461–0.461 | 0.067–0.067 |
% Change (V21), percentage change (V2 relative to V1).
Comparison of model parameter mean values.
Table 5 shows the P values for comparisons of the mean DCE-MRI parameters (at V1 and V2, and percentage change) between the three kinetic models using the mixed-models method. Only the means of Ktrans, ve, and kep were compared because each model used in this study generates these three parameters. Figure 6 shows the box plots of these three parameters stratified by the models at V1 and V2. Clearly, the Ktrans and ve values returned by the SSM are substantially greater than those returned by either the TM or ETM, and the differences are statistically significant with very small P values (<.0001 for either TM vs SSM or ETM vs SSM at V1 or V2). It is important to note that despite the significant differences in absolute Ktrans values at V1 and V2, there are no statistically significant differences in Ktrans percentage change between the SSM and TM or ETM. For kep, except for the TM versus SSM comparison of its percentage change, there are no statistically significant differences in its value or percentage change between the models.
Table 5.
P Values for Pairwise Comparison of the Mean DCE-MRI Parameter across Three Kinetic Models.
| Parameter | V1 | V2 | % Change (V21) | ||||||
| TM versus ETM | TM versus SSM | ETM versus SSM | TM versus ETM | TM versus SSM | ETM versus SSM | TM versus ETM | TM versus SSM | ETM versus SSM | |
| Ktrans | .2479 | <.0001 | <.0001 | .0447 | <.0001 | <.0001 | .1016 | .5382 | .697 |
| ve | <.0001 | <.0001 | <.0001 | .1659 | <.0001 | <.0001 | .0658 | .0049 | .0067 |
| kep | .1197 | .6602 | .1704 | .8428 | .1465 | .2289 | .0985 | .0345 | .2594 |
% Change (V21), percentage change (V2 relative to V1).
Figure 6.
Box plots of mean tumor Ktrans, ve, and kep values obtained with algorithms based on the TM (six algorithms), ETM (four algorithms), and SSM (two algorithms) at V1 (left column) and V2 (right column). The central bar and diamond symbols represent the median and mean values, respectively. The TM, ETM, and SSM labeling orders in the V1 Ktrans plot apply to all the other plots.
Early prediction of pCR.
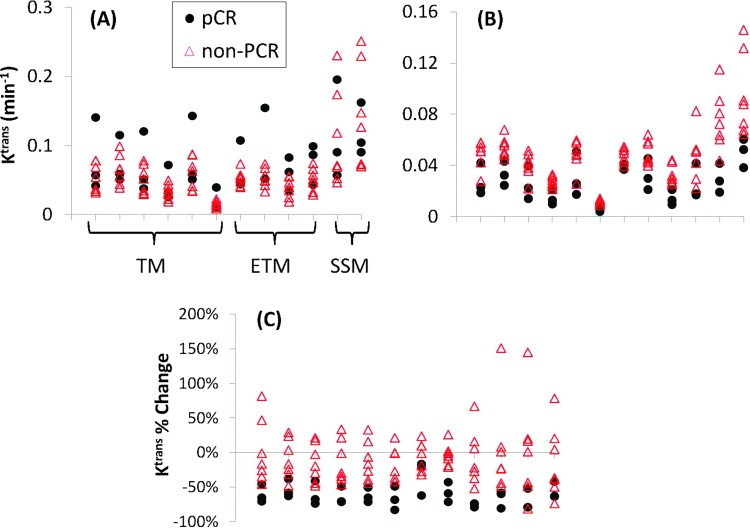
The pathologic reviews of the patients' resection and pre-NACT biopsy specimens revealed that in this cohort, three patients achieved pCR following NACT, while the other seven were non-pCRs (all pPRs). The ULR c-statistic values for early discrimination of pCR and non-pCR are listed in Table 6 for each DCE-MRI parameter (obtained from multiple algorithms) at V1 and V2, as well as its percentage change. None of the parameters at V1 (pre-NACT) provides good (0.8 ≤ c < 0.9) to excellent (0.9 ≤ c ≤ 1.0) prediction of response, except for a very few cases: ve of OHSU_TM (c = 0.81), kep of BWH-GE_TM and BWH-GE_ETM (both c = 0.81), vp of UM_ETM (c = 0.857), and τi of OHSU_SSM (c = 0.857). However, nearly all algorithms from participating QIN centers achieve good to excellent early discriminations of pCR and non-pCR using the Ktrans and kep parameters at V2 and their percentage changes (V21), with majority having c values equal to 1 (indicating complete separation of pCR and non-pCR) or greater than 0.9. Both ve and vp parameters, at V2 or their percentage changes, are not reliable predictors of response, with large variations in the c value across the algorithms. Many of them have no (c < 0.6), poor (0.6 ≤ c < 0.7), or fair (0.7 ≤ c < 0.8) predictive capabilities. As an example of excellent predictive abilities of Ktrans at V2 and its percentage change (V21) across all algorithms, Figure 7 presents the scatter plots of mean tumor Ktrans values at V1 (Figure 7A), V2 (Figure 7B), and their percentage changes (Figure 7C) obtained by all algorithms from the 10 patients. While there are clear overlaps in Ktrans values at baseline (V1) between pCRs and non-pCRs, the separations of the two groups are quite remarkable using V2 Ktrans and Ktrans percentage change values obtained by almost all the algorithms. The same patterns are observed for the kep parameter.
Table 6.
DCE-MRI Parameter ULR c-Statistic Values for Early Prediction of Pathologic Response.
| Parameter | Algorithm | V1 | V2 | % Change (V21) |
| Ktrans | OHSU_TM | 0.667 | 0.952 | 0.952 |
| VU_TM | 0.571 | 1 | 0.952 | |
| ISM_TM | 0.619 | 0.81 | 0.905 | |
| BWH-GE_TM | 0.595 | 1 | 1 | |
| UP_TM | 0.619 | 0.786 | 1 | |
| BWH-3D Slicer_TM | 0.524 | 1 | 1 | |
| VU_ETM | 0.524 | 0.952 | 0.762 | |
| UM_ETM | 0.714 | 0.857 | 1 | |
| BWH-GE_ETM | 0.786 | 1 | 1 | |
| UW_ETM | 0.762 | 0.905 | 1 | |
| OHSU_SSM | 0.571 | 1 | 0.857 | |
| VU_SSM | 0.476 | 1 | 0.81 | |
| ve | OHSU_TM | 0.81 | 0.762 | 0.667 |
| VU_TM | 0.571 | 0.762 | 0.595 | |
| ISM_TM | 0.667 | 0.81 | 0.81 | |
| BWH-GE_TM | 0.595 | 0.857 | 0.905 | |
| UP_TM | 0.643 | 0.81 | 0.857 | |
| BWH-3D Slicer_TM | 0.762 | 0.81 | 0.952 | |
| VU_ETM | 0.667 | 0.667 | 0.452 | |
| UM_ETM | 0.619 | 0.643 | 0.5 | |
| BWH-GE_ETM | 0.548 | 1 | 0.905 | |
| UW_ETM | 0.667 | 1 | 0.857 | |
| OHSU_SSM | 0.667 | 0.81 | 0.714 | |
| VU_SSM | 0.714 | 0.714 | 0.5 | |
| kep | OHSU_TM | 0.5 | 1 | 0.952 |
| VU_TM | 0.762 | 0.81 | 1 | |
| ISM_TM | 0.5 | 0.952 | 0.905 | |
| BWH-GE_TM | 0.81 | 1 | 1 | |
| UP_TM | 0.667 | 1 | 0.952 | |
| BWH-3D Slicer_TM | 0.667 | 0.952 | 1 | |
| VU_ETM | 0.619 | 0.714 | 0.81 | |
| UM_ETM | 0.738 | 0.857 | 1 | |
| BWH-GE_ETM | 0.81 | 1 | 1 | |
| UW_ETM | 0.762 | 1 | 1 | |
| OHSU_SSM | 0.476 | 1 | 0.905 | |
| VU_SSM | 0.571 | 0.81 | 0.857 | |
| vp | VU_ETM | 0.571 | 0.619 | 0.571 |
| UM_ETM | 0.857 | 0.905 | 0.405 | |
| BWH-GE_ETM | 0.476 | 0.857 | 0.643 | |
| UW_ETM | 0.667 | 0.714 | 0.667 | |
| τi | OHSU_SSM | 0.857 | 0.571 | 0.952 |
| VU_SSM | 0.452 | 0.548 | 0.548 |
% Change (V21), percentage change (V2 relative to V1).
Note: The institution abbreviation is listed ahead of the model abbreviation.
Figure 7.
Scatter plots of mean tumor Ktrans at V1 (A), V2 (B), and its percentage change (V21) (C) for three pCRs (black circles) and seven non-pCRs (red triangles). Each column represents results returned by one data analysis algorithm. The columns associated with the same kinetic model are grouped together (labeled in A), as explained in Figure 4.
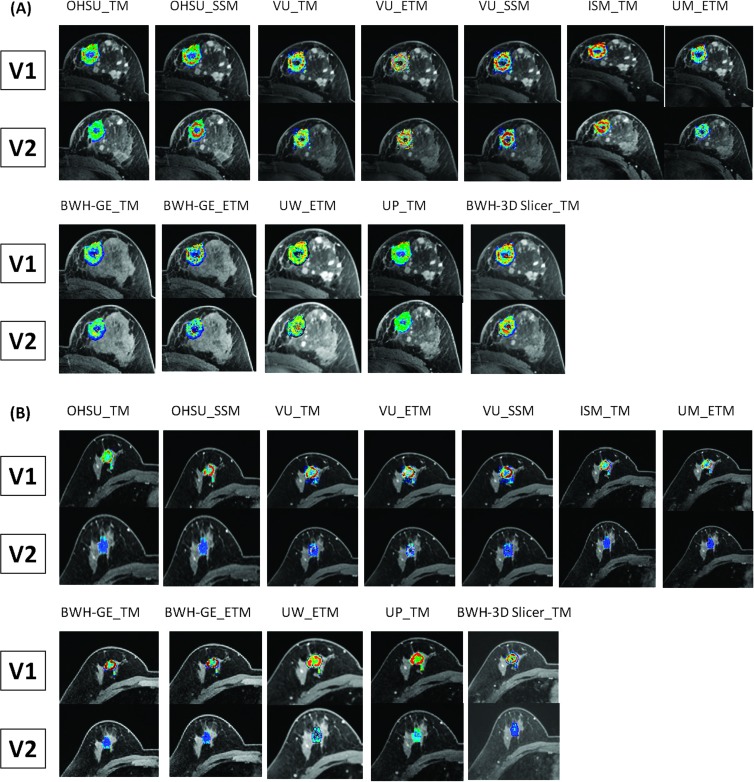
Figure 8 shows the color tumor Ktrans maps of a non-pCR (Figure 8A) and a pCR (Figure 8B) at V1 and V2, computed and generated by all 12 algorithms. For each DCE-MRI data set, the parametric maps are generated for all tumor ROIs on multiple image slices. The Ktrans maps presented in Figure 8 are from the same image slice (for each subject at each visit) through the center of the tumor. Note that except for the algorithms of BWH-GE_TM and BWH-GE_ETM, all Ktrans maps are overlaid on post-contrast images. Despite the differences in software algorithms/tools and display color scales, one observation is apparent and common on all the Ktrans maps: the minimal change in the tumor Ktrans map from V1 to V2 for the non-pCR is in stark contrast with the dramatic “cooling” of the Ktrans map color—red to blue (decrease in value)—from V1 to V2 for the pCR. The color kep maps (not shown here) show nearly identical patterns for the same two patients.
Figure 8.
V1 and V2 single slice tumor Ktrans parametric maps generated by all 12 algorithms for a non-pCR (A) and a pCR (B). The primary tumor was in the left breast of the non-pCR and the right breast of the pCR. The color Ktrans maps are overlaid on post-contrast or pre-contrast DCE images. Although the color scales for these maps are different between subjects and among algorithms, they are kept the same for V1 and V2 maps generated by the same algorithm for the same subject.
Discussion
Though DCE-MRI is becoming one of the imaging modalities of choice for assessment of cancer therapy response in early phase clinical trials, systematic comparison and validation of methods for pharmacokinetic analysis of DCE-MRI time-course data are needed to determine sources of parameter errors and variations, examine the effects of these variations on evaluation of tumor therapy response, and reach consensus on the best practice for DCE-MRI in a multicenter clinical trial. To the best of our knowledge, this is the first study in which shared DCE-MRI data from a longitudinal therapy monitoring study are analyzed by multiple centers using site-specific pharmacokinetic models and associated algorithms. The overall purpose of this multicenter DCE-MRI data analysis challenge is to evaluate variations in DCE-MRI prediction of breast cancer therapy response that are caused by differences in pharmacokinetic models and associated software algorithms only.
With the exception of the 2 SSM algorithms, the other 10 algorithms employed in this challenge are the commonly used TM and ETM. The reasonable results from fittings of the TM-simulated DRO data with five of the six involved TM algorithms confirm the accuracy of mathematical formulation and software coding in these TM software packages. The large errors (>20%) in estimated Ktrans were mostly resulted from fittings of DRO data with high Ktrans and low ve combinations, which were generally not the cases for the studied breast tumors. Though the reasons for large errors from these particular Ktrans and ve combinations warrant further investigations, a complete and in-depth analysis of the DRO data-fitting results is beyond the scope of this manuscript. Despite the fixed inputs of tumor ROI definition, AIF, and T10 in analyzing the shared human data for all algorithms, there are substantial variations in returned DCE-MRI parameters. When comparing all 12 algorithms, the main contributions to the variations of the two common parameters, Ktrans and ve, are the large values returned by the two SSM algorithms and small values returned by the BWH-3D Slicer TM software tool, with the latter returning Ktrans values two to three times and ve value three to four times smaller than the other algorithms (Figure 4A). It is not surprising that Ktrans and ve parameters derived from SSM analysis are significantly greater than their TM or ETM counterparts at either V1 or V2 (Figure 6 and Table 5), as the same patterns have been observed in DCE-MRI studies of tumors in different organs [17,27,31–33,37–39]. The differences between the SSM and TM in derived Ktrans and ve parameter values are systematic ones directly resulted from inclusion or exclusion of the water exchange effects by the SSM and TM, respectively. The exact reason for the out-of-range small Ktrans and ve values returned by the BWH-3D Slicer TM algorithm is unclear. DRO data fitting by the same algorithm consistently returned Ktrans and ve values with >10% errors for all combinations of simulated Ktrans and ve values with maximum error of 71%for Ktrans estimation. Heye et al. observed up to 10-fold to 100-fold difference in Ktrans and ve values obtained with different commercial software packages [22]. They attributed these massive discrepancies to possible differences in scaling factors used by commercial vendors for reporting or displaying estimated pharmacokinetic parameters. The same reason may be the cause of the two-fold to four-fold parameter value difference observed in this study between the BWH-3D Slicer TM software tool and most other algorithms. Most of the DCE-MRI parametric maps generated from the shared human data were spatially heterogeneous (Figure 8) as a result of breast tumor heterogeneity. The difference in computing tumor mean parameter values by each algorithm—averaging pixel parameter values or averaging slice ROI parameter values (Table 1)—is another cause of parameter variance.
It is reasonable that the agreement in the three common DCE-MRI parameters (Ktrans, ve, and kep) that are obtainable by all three kinetic models is generally improved when only algorithms within the same kinetic model are compared, as opposed to comparisons across all 12 algorithms (Table 2). However, there are still substantial variations in returned parameter values by different software tools based on the same model. Besides the scaling factor variation that may have contributed to the markedly small Ktrans and ve values obtained with the BWH-3D Slicer TM algorithm, other contributions to parameter variations among algorithms based on the same model may come from differences in fixed physiological and MR parameters (such as hematocrit, pre-contrast blood T1, and so on), contrast arrival time (or AIF shift), AIF curve resampling strategy, and goodness of fitting criteria employed in each algorithm. These factors were not controlled for any algorithm used in this data analysis challenge. It should be noted that the BWH-3D Slicer tool is the only one of the tested software packages that is available as free open source software. Availability of the software source code can facilitate reproducibility of results [40] and aid further investigation of sources of variability and inconsistency in comparison with other algorithm implementations.
One important observation that is not available in the study by Heye et al. [22] but is afforded by the longitudinal nature of the shared DCE-MRI data in this study is the agreement in parameter percentage change (V2 relative to V1) among the algorithms. It is interesting and important to note that agreements in parameter percentage changes are generally better than those of the absolute parameter values at either V1 or V2 regardless whether the comparisons are made across all algorithms or within each kinetic model (Figure 4 and Tables 2, 3, and 4). The CCC values of Ktrans at V1 and V2 are quite small for any pairwise comparison of TM algorithms that includes the BWH-3D Slicer TM algorithm, but those of the corresponding Ktrans percentage changes are drastically increased (Table 3), indicating greater agreement between the pair of algorithms. The reason for such improvement in agreement among the algorithms is probably partial cancellation of systematic differences (or variations) among different algorithms in percentage change calculations. For example, if the application of a scaling factor in the BWH-3D Slicer TM algorithm caused unusually small Ktrans and ve values, the effects of the scaling factor were canceled in calculating percentage changes of these parameters, resulting in percentage change values comparable to those obtained with the other algorithms.
Among the 12 algorithms used in this challenge, only the 4 based on the ETM generate the vp parameter through data fitting. Compared to the Ktrans and ve parameters estimated with these four ETM algorithms, the variance in vp is relatively high, reflected by its large wCV (Figure 5), small ICC (see above), and small CCC (Table 4) values. This is most likely due to the relatively low temporal resolution (∼20 seconds) of the raw breast DCE-MRI data. vp is a parameter of the vascular compartment that contributes a significant fraction of the DCE-MRI signal only during the early phase of rapid signal rise. The inability of the data acquisition protocol used in this study to accurately capture the DCE curve shape during this time window leads to low accuracy and precision in vp quantification [20,21] and hence its high variance. A simulation study [41] shows that at least 4-second temporal resolution is needed for accurate vp estimation from breast DCE-MRI data. Given clinical preference for full breast coverage and high spatial resolution in breast MRI, such high temporal resolution is difficult to achieve even with combined use of k-space undersampling scheme and parallel imaging acceleration, as is the case for the shared breast data in this study. The low accuracy and precision of the vp parameter, and possibly the resultant reduced capability in prediction of therapy response found (see below) in this study, demonstrate the importance of proper model selection for pharmacokinetic analysis of DCE-MRI data. Using a model that includes the vascular compartment to fit low temporal resolution DCE-MRI data will introduce more errors and uncertainties in the derived parameters [20,21] and thus diminish the effectiveness of DCE-MRI for evaluation of therapy response.
Despite the considerable variances in parameter values obtained with different algorithms, it is rather encouraging within the context of therapy response assessment, however, that nearly all 12 algorithms provided good to excellent (ULR c ≥ 0.8) early prediction of pathologic response using V2 Ktrans and kep or their corresponding percentage changes (V2 relative to V1) as predictive markers (Table 6). For V2 Ktrans and kep, there are 9 and 8 (of 12) algorithms providing excellent predictions with ULR c ≥ 0.9, respectively, while for Ktrans and kep percentage changes, the numbers of algorithms with ULR c ≥ 0.9 are 9 and 10 (of 12), respectively. The predictive abilities of the V2 Ktrans and kep parameters across all algorithms may be due to 1) the substantial decreases in perfusion and permeability of the three pCR tumors in comparison with the seven non-pCR tumors outweighed parameter variations introduced by algorithm differences, and/or 2) the interalgorithm variations are systematic, i.e., error signs in parameter misestimations are the same for both pCR and non-pCR tumors: overestimation or underestimation for both groups. The second, potentially the most important, reason is clearly illustrated in Figure 7B for V2 Ktrans, where the black (pCR) and red (non-pCR) data points more or less move in the same up or down direction going from one algorithm to another, maintaining similar pCR versus non-pCR data point gaps among the algorithms and thus similar discriminative capabilities. The predictive capabilities of Ktrans and kep percentage changes across all algorithms are most likely due to cancellation of the interalgorithm systematic errors (or variations) in percentage change calculation. Figure 7C shows relatively stable distribution of Ktrans percentage changes for both pCRs and non-pCRs across the 12 algorithms in contrast to absolute V1 (Figure 7A) and V2 (Figure 7B) Ktrans values that are more variable across algorithms. Similar patterns can also be seen in Figure 4. With less systematic errors (variations), the more stable Ktrans and kep percentage change values allowed the tested algorithms to estimate the intrinsic differences between the pCRs and non-pCRs and gave fairly uniform predictions of therapy response.
In this multicenter data analysis challenge, the three major aspects in DCE-MRI data acquisition and analysis that can cause significant parameter variations (i.e., tumor ROI definition, T10 measurement, and AIF determination) were controlled to focus on comparisons of pharmacokinetic models and associated algorithms in assessment of breast cancer response to NACT. The results suggest that variations in DCE-MRI parameters caused by differences in models/algorithms only are mostly systematic. As a result, all models/algorithms performed fairly consistently in prediction of therapy response, especially using the percentage change metrics in which the interalgorithm systematic variations are significantly reduced. In this particular study setting, Ktrans and kep percentage changes computed with most of the algorithms provided excellent early prediction of breast cancer response to NACT. The introduction of variations in tumor ROI definition and errors in T10 and AIF determinations in a multicenter clinical trial setting where DCE-MRI data are acquired and analyzed at each individual site will add random errors and variations in derived DCE-MRI parameters. This will not only cause more severe parameter variance but also affect DCE-MRI performance in evaluation of therapy response. Therefore, it is of paramount importance in a multicenter clinical trial to strictly standardize data acquisition protocol (such as temporal resolution) and perform frequent scanner quality assurance/quality control (QA/QC) [9–11] to minimize interscanner platform or interacquisition protocol random errors in quantification of T10 and AIF in a longitudinal DCE-MRI study of tumor therapy response. Random errors due to variations in manual drawing of tumor ROI are difficult to avoid. Use of automatic or semiautomatic algorithms for tumor ROI definition may help mitigate such errors. One possible approach to reduce random errors and variations in a multicenter trial and improve performance consistency in response assessment is centralized DCE-MRI data analysis in which fixed inputs for pharmacokinetic modeling, such as single observer-defined tumor ROIs and population-averaged AIF, could be used.
In conclusion, considerable parameter variations were observed when shared breast DCE-MRI data sets were analyzed with different algorithms based on the TM, ETM, and SSM in a multicenter data analysis challenge. However, nearly all algorithms provided good to excellent early prediction of breast cancer response to therapy using the Ktrans and kep parameters after the first cycle of NACT and their percentage changes, suggesting that the utility of DCE-MRI for assessment of therapy response is not diminished by interalgorithm systematic variations.
Acknowledgments
The authors thank William Woodward for assistance in breast DCE-MRI data acquisition, Aneela Afzal for preparation of the raw breast DCE-MRI data for sharing by participating QIN centers, and Karen Oh and Nicole Roy for drawing tumor ROIs. The authors acknowledge and appreciate the Radiological Society of North America and NIH/National Institute of Biomedical Imaging and Bioengineering (NIBIB) Contract No. HHSN268201000050C for supporting development and evaluation of the DCE-MRI DROs.
Footnotes
This study was supported by National Institutes of Health (NIH) grants U01-CA154602, U01-CA142565, U01-CA148131, U01-CA166104, U01-CA172320, U01-CA151261, U01-CA140230, U01-CA154601, and U54-EB005149.
References
- 1.O'Connor JPB, Jackson A, Parker GJM, Roberts C, Jayson GC. Dynamic contrast-enhanced MRI in clinical trials of antivascular therapies. Nat Rev Clin Oncol. 2012;9:167–177. doi: 10.1038/nrclinonc.2012.2. [DOI] [PubMed] [Google Scholar]
- 2.Padhani AR, Miles KA. Multiparametric imaging of tumor response to therapy. Radiology. 2010;256:348–364. doi: 10.1148/radiol.10091760. [DOI] [PubMed] [Google Scholar]
- 3.Harry VN, Semple SI, Parkin DE, Gilbert FJ. Use of new imaging techniques to predict tumour response to therapy. Lancet Oncol. 2010;11:92–102. doi: 10.1016/S1470-2045(09)70190-1. [DOI] [PubMed] [Google Scholar]
- 4.Yankeelov TE, Gore JC. Dynamic contrast enhanced magnetic resonance imaging in oncology: theory, data acquisition, analysis, and examples. Curr Med Imaging Rev. 2009;3:91–107. doi: 10.2174/157340507780619179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marcus CD, Ladam-Marcus V, Cucu C, Bouche O, Lucas L, Hoeffel C. Imaging techniques to evaluate the response to treatment in oncology: current standards and perspectives. Crit Rev Oncol Hematol. 2009;72:217–238. doi: 10.1016/j.critrevonc.2008.07.012. [DOI] [PubMed] [Google Scholar]
- 6.Turkbey B, Kobayashi H, Ogawa M, Bernardo M, Choyke PL. Imaging of tumor angiogenesis: functional or targeted? AJR. 2009;193:304–313. doi: 10.2214/AJR.09.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Desar IME, van Herpen CML, van Laarhoven HWM, Barentsz JO, Oyen WJG, van der Graaf WTA. Beyond RECIST: molecular and functional imaging techniques for evaluation of response to targeted therapy. Cancer Treat Rev. 2009;35:309–321. doi: 10.1016/j.ctrv.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 8.Brindle K. New approaches for imaging tumour responses to treatment. Nat Rev Cancer. 2008;8:94–107. doi: 10.1038/nrc2289. [DOI] [PubMed] [Google Scholar]
- 9.Leach MO, Morgan B, Tofts PS, Buckley DL, Huang W, Horsfield MA, Chenevert TL, Collins DJ, Jackson A, Lomas D, et al. Imaging vascular function for early stage clinical trials using dynamic contrast-enhanced magnetic resonance imaging. Eur Radiol. 2012;22:1451–1464. doi: 10.1007/s00330-012-2446-x. [DOI] [PubMed] [Google Scholar]
- 10.Evelhoch J, Garwood M, Vigneron D, Knopp M, Sullivan D, Menkens A, Clarke L, Liu G. Expanding the use of magnetic resonance in the assessment of tumor response to therapy: workshop report. Cancer Res. 2005;65:7041–7044. doi: 10.1158/0008-5472.CAN-05-0674. [DOI] [PubMed] [Google Scholar]
- 11.Leach MO, Brindle KM, Evelhoch JL, Griffiths JR, Horsman MR, Jackson A, Jayson GC, Judson IR, Knopp MV, Maxwell RJ, et al. The assessment of antiangiogenic and antivascular therapies in early-stage clinical trials using magnetic resonance imaging: issues and recommendations. Br J Cancer. 2005;92:1599–1610. doi: 10.1038/sj.bjc.6602550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tofts PS, Berkowitz B, Schnall MD. Quantitative analysis of dynamic Gd-DTPA enhancement in breast tumors using a permeability model. Magn Reson Med. 1995;33:564–568. doi: 10.1002/mrm.1910330416. [DOI] [PubMed] [Google Scholar]
- 13.Ashton E. Quantitative MR in multi-center clinical trials. J Magn Reson Imaging. 2010;31:279–288. doi: 10.1002/jmri.22022. [DOI] [PubMed] [Google Scholar]
- 14.Schabel MC, Morrell GR. Uncertainty in T1 mapping using the variable flip angle method with two flip angles. Phys Med Biol. 2009;54:N1–N8. doi: 10.1088/0031-9155/54/1/N01. [DOI] [PubMed] [Google Scholar]
- 15.Schabel MC, Parker DL. Uncertainty and bias in contrast concentration measurements using spoiled gradient echo pulse sequences. Phys Med Biol. 2008;53:2345–2373. doi: 10.1088/0031-9155/53/9/010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang C, Karczmar GS, Medved M, Oto A, Zamora M, Stadler WM. Reproducibility assessment of a multiple reference tissue method for quantitative dynamic contrast enhanced-MRI analysis. Magn Reson Med. 2009;61:851–859. doi: 10.1002/mrm.21912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yankeelov TE, Rooney WD, Li X, Springer CS. Variation of the relaxographic “shutter-speed” for transcytolemmal water exchange affects the CR bolus-tracking curve shape. Magn Reson Med. 2003;50:1151–1169. doi: 10.1002/mrm.10624. [DOI] [PubMed] [Google Scholar]
- 18.Heisen M, Fan X, Buurman J, van Riel NAW, Karczmar GS, ter Haar Romeny BM. The influence of temporal resolution in determining pharmacokinetic parameters from DCE-MRI data. Magn Reson Med. 2010;63:811–816. doi: 10.1002/mrm.22171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Di Giovanni P, Azlan CA, Ahearn TS, Semple SI, Gilbert FJ, Redpath TW. The accuracy of pharmacokinetic parameter measurement in DCE-MRI of the breast at 3 T. Phys Med Biol. 2010;55:121–132. doi: 10.1088/0031-9155/55/1/008. [DOI] [PubMed] [Google Scholar]
- 20.Sourbron SP, Buckley DL. On the scope and interpretation of the Tofts models for DCE-MRI. Magn Reson Med. 2011;66:735–745. doi: 10.1002/mrm.22861. [DOI] [PubMed] [Google Scholar]
- 21.Sourbron SP, Buckley DL. Tracer kinetic modelling in MRI: estimating perfusion and capillary permeability. Phys Med Biol. 2012;57:R1–R33. doi: 10.1088/0031-9155/57/2/R1. [DOI] [PubMed] [Google Scholar]
- 22.Heye T, Davenport MS, Horvath JJ, Feuerlein S, Breault SR, Bashir MR, Merkle EM, Boll DT. Reproducibility of dynamic contrast-enhanced MR imaging. Part I. Perfusion characteristics in the female pelvis by using multiple computer-aided diagnosis perfusion analysis solutions. Radiology. 2013;266:801–811. doi: 10.1148/radiol.12120278. [DOI] [PubMed] [Google Scholar]
- 23.Tofts PS, Kermode AG. Measurement of the blood-brain barrier permeability and leakage space using dynamic MR imaging. Magn Reson Med. 1991;17:357–367. doi: 10.1002/mrm.1910170208. [DOI] [PubMed] [Google Scholar]
- 24. https://dblab.duhs.duke.edu/modules/QIBAcontent/index.php?id=1.
- 25. https://dblab.duhs.duke.edu/wysiwyg/downloads/Dynamic_v6_beta1_description.txt.
- 26.Song T, Laine AF, Chen Q, Rusinek H, Bokacheva L, Lim RP, Laub G, Kroeker R, Lee VS. Optimal k-space sampling for dynamic contrast-enhanced MRI with an application to MR renography. Magn Reson Med. 2009;61:1242–1248. doi: 10.1002/mrm.21901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tudorica LA, Oh KY, Roy N, Kettler MD, Chen Y, Hemmingson SL, Afzal A, Grinstead JW, Laub G, Li X, et al. A feasible high spatiotemporal resolution breast DCE-MRI protocol for clinical settings. Magn Reson Imaging. 2012;30:1257–1267. doi: 10.1016/j.mri.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tofts PS. Modeling tracer kinetics in dynamic Gd-DTPA MR imaging. J Magn Reson Imaging. 1997;7:91–101. doi: 10.1002/jmri.1880070113. [DOI] [PubMed] [Google Scholar]
- 29.Tofts PS, Brix G, Buckley DL, Evelhoch JL, Henderson E, Knopp MV, Larsson HBW, Lee TY, Mayr NA, Parker GJM, et al. Estimating kinetic parameters from dynamic contrast-enhanced T1-weighted MRI of a diffusable tracer: standardized quantities and symbols. J Magn Reson Imaging. 1999;10:223–232. doi: 10.1002/(sici)1522-2586(199909)10:3<223::aid-jmri2>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 30.Li X, Rooney WD, Springer CS. A unified pharmacokinetic theory for intravascular and extracellular contrast agents. Magn Reson Med. 2005;54:1351–1359. doi: 10.1002/mrm.20684. [Erratum. Magn Reson Med 2006;55:1217.] [DOI] [PubMed] [Google Scholar]
- 31.Huang W, Li X, Morris EA, Tudorica LA, Seshan VE, Rooney WD, Tagge I, Wang Y, Xu J, Springer CS. The magnetic resonance shutter speed discriminates vascular properties of malignant and benign breast tumors in vivo. Proc Natl Acad Sci USA. 2008;105:17943–17948. doi: 10.1073/pnas.0711226105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li X, Huang W, Morris EA, Tudorica LA, Seshan VE, Rooney WD, Tagge I, Wang Y, Xu J, Springer CS. Dynamic NMR effects in breast cancer dynamic-contrast-enhanced MRI. Proc Natl Acad Sci USA. 2008;105:17937–17942. doi: 10.1073/pnas.0804224105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang W, Tudorica LA, Li X, Thakur SB, Chen Y, Morris EA, Tagge IJ, Korenblit M, Rooney WD, Koutcher JA, et al. Discrimination of benign and malignant breast lesions by using shutter-speed dynamic contrast-enhanced MR imaging. Radiology. 2011;261:394–403. doi: 10.1148/radiol.11102413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Symmans WF, Peintinger F, Hatzis C, Rajan R, Kuerer H, Valero V, Assad L, Poniecka A, Hennessy B, Green M, et al. Measurement of residual breast cancer burden to predict survival after neoadjuvant chemotherapy. J Clin Oncol. 2007;25:4414–4422. doi: 10.1200/JCO.2007.10.6823. [DOI] [PubMed] [Google Scholar]
- 35.Rajan R, Poniecka A, Smith TL, Yang Y, Frye D, Pusztai L, Fiterman DJ, Gal-Gombos E, Whitman G, Rouzier R, et al. Change in tumor cellularity of breast carcinoma after neoadjuvant chemotherapy as a variable in the pathologic assessment of response. Cancer. 2004;100:1365–1373. doi: 10.1002/cncr.20134. [DOI] [PubMed] [Google Scholar]
- 36.Fedorov A, Beichel R, Kalpathy-Cramer J, Finet J, Fillion-Robin J-C, Pujol S, Bauer C, Jennings D, Fennessy F, Sonka M, et al. 3D slicer as an image computing platform for the Quantitative Imaging Network. Magn Reson Imaging. 2012;30:1323–1341. doi: 10.1016/j.mri.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li X, Priest RA, Woodward WJ, Tagge IJ, Siddiqui F, Huang W, Rooney WD, Beer TM, Garzotto MG, Springer CS. Feasibility of shutter-speed DCE-MRI for improved prostate cancer detection. Magn Reson Med. 2013;69:171–178. doi: 10.1002/mrm.24211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chang EY, Li X, Jerosch-Herold M, Priest RA, Enestvedt CK, Xu J, Springer CS, Jobe BA. The evaluation of esophageal adenocarcinoma using dynamic contrast-enhanced magnetic resonance imaging. J Gastrointest Surg. 2008;11:166–175. doi: 10.1007/s11605-007-0253-5. [DOI] [PubMed] [Google Scholar]
- 39.Kim S, Quon H, Loevner LA, Rosen MA, Dougherty L, Kilger AM, Glickson JD, Poptani H. Transcytolemmal water exchange in pharmacokinetic analysis of dynamic contrast-enhanced MRI data in squamous cell carcinoma of the head and neck. J Magn Reson Imaging. 2007;26:1607–1617. doi: 10.1002/jmri.21207. [DOI] [PubMed] [Google Scholar]
- 40.Ince DC, Hatton L, Graham-Cumming J. The case for open computer programs. Nature. 2012;482:485–488. doi: 10.1038/nature10836. [DOI] [PubMed] [Google Scholar]
- 41.Henderson E, Rutt BK, Lee TY. Temporal sampling requirements for the tracer kinetics modeling of breast disease. Magn Reson Imaging. 1998;16:1057–1073. doi: 10.1016/s0730-725x(98)00130-1. [DOI] [PubMed] [Google Scholar]