Abstract
The execution of a multisite trial frequently includes image collection. The Clinical Trials Processor (CTP) makes removal of protected health information highly reliable. It also provides reliable transfer of images to a central review site. Trials using central review of imaging should consider using CTP for handling image data when a multisite trial is being designed.
Introduction
Imaging plays a critical role in performing clinical trials for most cancers. In multisite trials, independent central review of the images has been shown to produce more consistent results than local review by the individual image acquisition sites themselves [1]. Although central review typically increases the cost per subject of a multisite trial, the improved results, coupled with the opportunity to archive the images for analysis in other contexts, can reduce total study costs and make it the preferred approach.
To preserve the integrity of the data, modern clinical imaging trials and image archive projects in cancer research will typically store, process, and transmit the image data in their original digital form. Although handling data in this way can be very efficient, special tools are required to ensure that the quality and reliability of the data are preserved and the regulatory requirements of Health Insurance Portability and Accountability Act (HIPAA) are met. In this paper, we describe tools developed by the Radiological Society of North America (RSNA; Oak Brook, IL) [the Image Sharing Network (ISN) and Clinical Trials Processor (CTP)] to address these challenges.
Clinical Trial Dataflow
The simplest dataflow topology for a multisite clinical imaging trial is a single principal investigator site surrounded by a constellation of image acquisition sites. The image acquisition sites are responsible for recruiting patients who meet the criteria of the trial, performing the examinations, and transmitting the data to the principal investigator site. The principal investigator site is responsible for validating, organizing, archiving, and analyzing the data and making the data and the analytic results available to the sponsor of the trial. This basic design has been described in [2].
Confidentiality and Deidentification
Almost all image acquisition sites use their standard clinical information systems to register the patient and then order, schedule, and perform the examinations. The data objects produced in this process therefore contain real-world identifiers—identifiers that connect the data objects to the human being who was the trial subject. Such identifiers are known in HIPAA parlance as protected health information (PHI). Descriptions of HIPAA and PHI are outside the scope of this paper. For more information on HIPAA and PHI, we refer you to http://www.hhs.gov/ocr/privacy/hipaa/understanding/summary/.
Because the HIPAA privacy regulations impose strict requirements on anyone who possesses PHI, Institutional Review Board (IRB) tightly restrict usage of PHI in clinical trials and rarely allow PHI to be transmitted to other institutions, so PHI must normally be replaced with research identifiers. A full description of HIPAA requirements for deidentification can be found at http://www.hhs.gov/ocr/privacy/hipaa/understanding/coveredentities/De-identification/guidance.html. Thus, before transmission to the principal investigator site, the data objects must be modified to remove unnecessary PHI and to replace necessary PHI with pseudonyms that preserve the relationships among the data objects while breaking the connection between the data and the human being who was the trial subject. This process is formally termed “deidentification” and is technically “pseudonymization” because there is an identifier for each subject, but in common parlance, it is known as “anonymization,” and systems that implement this process are often called anonymizers.
Most medical images are encoded according to the Digital Imaging and Communications in Medicine (DICOM) standard (http://medical.nema.org/standard.html). DICOM defines a dictionary of several thousand elements, each of which is designed to contain a specific type of information (e.g., patient name, patient ID, image pixels, and other data). A typical image includes fewer than 100 such elements, but there is effectively no limit. Additionally, the standard provides a mechanism for including nonstandard, so-called private elements, with contents that are not constrained by the standard. PHI is intended to appear in many standard elements, and of course, PHI can appear in any private element.
The HIPAA privacy regulations provide the following two methods for deidentifying data objects: the Safe Harbor Method and the Statistical Method. The Safe Harbor Method defines 18 categories of PHI and requires that data in all those categories be removed or replaced with pseudonyms (see Table 1). This approach requires the removal of information (e.g., patient age or sex) that is often necessary for the data to be useful in a trial. The Statistical Method allows some PHI to be preserved if it can be demonstrated that it is insufficient to identify the human being who was the trial subject.
Table 1.
HIPAA PHI Identifiers.
|
HIPAA §164.514(b) specifies 18 categories of data that are considered as potentially identifying (taken from http://www.gpo.gov/fdsys/pkg/CFR-2011-title45-vol1/pdf/CFR-2011-title45-vol1-sec164-514.pdf, page 885).
The deidentification of a DICOM object is an arcane and technical task requiring an understanding of the DICOM standard and sometimes even knowledge of the provenance of the data. Part 15 of the DICOM standard now defines a set of deidentification profiles that specify procedures for deidentifying objects in various circumstances. The most stringent profile, the Basic Profile, generally removes all PHI as required by the Safe Harbor Method, often more than is appropriate for a specific trial. Other profiles preserve certain categories of PHI. In many trials, one or more of these profiles are overlaid on the Basic Profile to retain the necessary data while preserving the anonymity of the trial subjects.
Clinical Trials Processor
The RSNA has developed a specialized tool, CTP, to support the dataflow for clinical trials. CTP organizes related communication and data processing functions into ordered sequences called pipelines.
Data objects are thought of as flowing down a pipeline, encountering each pipeline stage in turn. CTP includes 35 standard pipeline stages in four categories (import services, data processors, storage services, and export services). Using CTP's configuration editor, stages can be arranged in pipelines and configured to implement the desired dataflow for image acquisition sites and principal investigator sites.
CTP includes several stages to deidentify DICOM images:
The DicomAnonymizer stage implements all the deidentification profiles defined in the DICOM standard (ftp://medical.nema.org/medical/dicom/final/sup55_ft.pdf). This stage is highly configurable, allowing a clinical trial administrator to adapt it as required by a specific trial.
The DicomPixelAnonymizer stage blanks regions of images containing PHI burned into the pixels. This stage uses nonpixel information in the data object to identify the manufacturer and model of the image device and blanks the regions defined in a dictionary for that model.
The DicomMammoPixelAnonymizer stage blanks PHI burned into mammography images by examining the pixels directly.
CTP has been employed in numerous multisite trials and other imaging-related research. One of the largest is the National Cancer Institute's Cancer Image Archive [3].
Audit Requirements
The Food and Drug Administration (FDA) imposes regulations on medical software and data management. Title 21 of the Code of Federal Regulations, Part 11 (21CFR11), applies to records in electronic form. In 21CFR11-compliant clinical trials, it is necessary to log changes made to data objects, such as the changes made by an anonymizer [4]. CTP provides an audit logging mechanism that is used by several of its pipeline stages to meet that requirement.
Data Transmission
The transmission of data from image acquisition sites to the principal investigator site is typically accomplished through the Internet, though compact disks were commonly used in the past. Although data are normally deidentified before transmission, most trials nevertheless use encrypted communication and site authentication to minimize the possibility of intercept. The CTP import service and export service pipeline stages support both encrypted and unencrypted communications.
In the modern Information Technology (IT) environment, it is critical that hospital systems be protected from outside attack. This typically requires blocking any unsolicited data being transferred into a hospital computer. In a clinical trial, the receipt of image data at the central site is usually an asynchronous and unsolicited event, and such a restriction would make that impossible. To address this challenge, many central sites place a receiver outside the hospital's restrictive firewall and poll the receiver from the internal network. This, however, requires specialized IT support that is sometimes unavailable from heavily taxed IT departments. Another solution is to use an Internet-based intermediary. An example of such an intermediary is a cloud storage mechanism like Google Drive (Google; Mountain View, CA) or Dropbox (Dropbox, Inc, San Francisco, CA).
The ISN
Although all these transmission alternatives are currently in use in trials employing CTP, the RSNA has developed a more sophisticated approach on the basis of the Cross-Enterprise Document Sharing (XDS) transfer protocol defined by the Integrating the Healthcare Enterprise (Oak Brook, IL; see http://IHE.net) initiative. This approach uses a separate server and special-purpose CTP stages, all developed by the RSNA's ISN project. An Internet-based commercial XDS Repository (the clearinghouse) serves as an intermediary to receive submissions from image acquisition sites and supplies them to principal investigator sites when polled. This is effectively like the model described above where images are collected by a server outside the firewall, but rather than being required for each hospital, it is a central resource supported by the ISN project.
The ISN system uses CTP to provide the anonymization and site-based image transfer capabilities but uses XDS and the clearinghouse to transmit images between sites. As such, the functionality will be similar to that of CTP but eliminates the need for a central site to have a CTP receiver that is visible outside its firewall. From that perspective, any site that has ISN implemented will likely find it preferable to use the ISN for research purposes because it eliminates to purchase and maintain that extra server.
Central Site Management
When the images have been transmitted to the central site, the central site typically validates the data to ensure that the imaging protocol was followed and that artifacts (e.g., motion) do not render the images unusable.
Adherence to the imaging protocol can largely be checked programmatically by comparing values in the DICOM metadata with values required by the protocol. The specific Quality Assurance steps used will vary with each clinical trial. Many will have specified specific acquisition parameters [e.g., slice thickness for computed tomography (CT) and magnetic resonance (MR); peak kilovolts (Kvp) and milliamperes (mA) for CT; and TR, TE, and slice orientation for magnetic resonance imaging (MRI)], and these can be programmatically or manually checked on receipt of a study. CTP provides several pipeline stages that can be used to accomplish these checks. Checking for motion artifacts programmatically is more challenging, but it has been reported to be useful [5].
Once these quality assurance (QA) checks are completed, the measurement and analysis phase can be started. The exact measurements vary with each protocol; many cancer studies will simply want Response Evaluation Criteria in Solid Tumors (RECIST) measurements made, but there is increasing interest in measuring tumor volumes. The FDA requires that a record be maintained of the receipt of, and all changes made to, data that is part of a submission for a drug or device approval. In the context of the processing of image data for clinical trials, this audit trail must document the versions of the software used, including any scripts that control the anonymization that was performed. In the extreme case, the audit trail must include every individual change made in each image. Such audit trails can be created automatically by many electronic tools, including CTP. If the analysis involves more than a few steps, workflow technologies can execute automated steps or provide task lists for human operators. Such a system could also help to guide the user on the correct next steps as well as automatically document the data measurements and manipulations [6].
Using the CTP
The CTP tool is extremely flexible, but with that flexibility comes complexity. Here, we present a common use case of a multisite clinical trial and specifically how to use CTP to enable that clinical trial. The basic assumptions of this use case are that there are N sites participating (“submitter”), each with its own set of research subjects who will be getting one or more imaging examinations that will be transmitted to a central analysis site. We also assume that there is exactly one central analysis site. We assume that each submitter will keep the PHI of their patients at their site and that they have a research ID that will be used to identify each subject to the analysis site. That research ID should be encoded into the image file to maximize the integrity of the process.
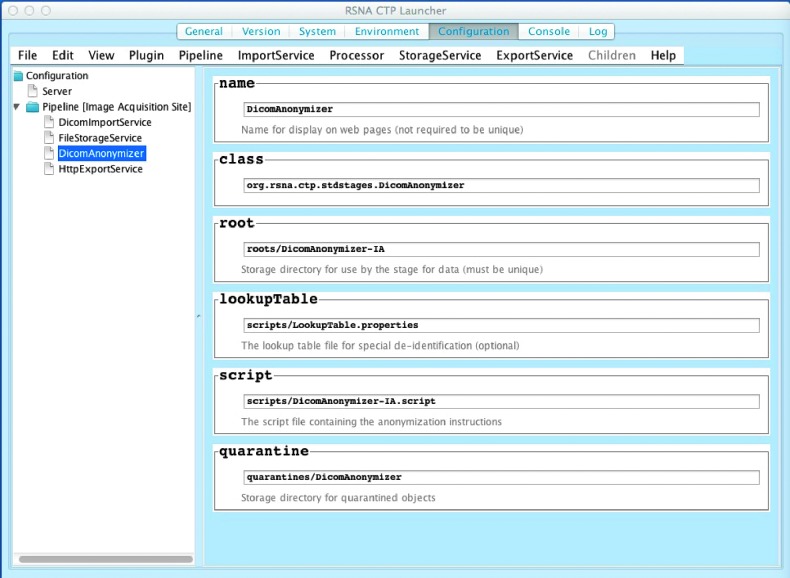
When CTP-installer.jar is downloaded (from http://mirc.rsna.org/query) and executed, it will recognize if it has not been installed before into the selected location. In that case, it will display the “help page” for CTP, which asks the user to select a pipeline. Figure 1 shows the configuration screen for CTP. For a multisite trial where data are sent to another site, the user would select “Image Acquisition Site” (a central site would select “Principal Investigator Site”). Within this pipeline, the user may configure the anonymization options to use, as well as information about how it will receive information from the Picture Archive and Communications System (PACS; DicomImportService).
Figure 1.
Configuration screen for a typical CTP installation for a submitting site. From this screen, one can view and edit the various components of CTP, including the pipeline(s) being used, and the components within a pipeline.
In most cases, the individual sites will have a mapping from their local identifier to the research identifier. The “lookupTable” field in the DicomAnonymizer option of CTP points to the file where that lookup table exists. When a file is transferred to CTP, CTP will look up the medical record number in that file and replace it with the research identifier. It will then also remove other PHI elements according to the instructions specified in the “script” that is a file pointed to by the “scripts” field in the pipeline. An example of such a file is shown in Figure 2 (to comply with HIPAA, there are no real Medical Record Numbers but text that indicates these).
Figure 2.
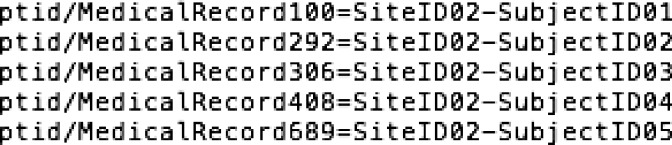
Example of a lookup table file used by CTP for mapping from the local medical record number to the subject ID. In this example, there is a SiteID component as well as a local subject component, but that is not required.
Removing information from the DICOM tags is fairly straightforward, but CTP cannot only remove information, but can also insert calculated values. For instance, a study could add a private but known offset to the date of birth so that only members of the study with that private information would know the correct date of birth. This allows preservation of information without disclosing PHI.
CTP also enables removal of information from portions of an image, depending on the vendor and model of device. The database of devices/models and where they typically “burn in” PHI is supplied with CTP and can easily be included as part of the pipeline. And example of this is shown in Figure 3. In the past, such “burned in” information was common for all images. Now, it is occasionally seen with fluoroscopy and ultrasound images and increasingly with postprocessed images like three-dimensional (3D) renderings.
Figure 3.
Portion of the PixelAnonymizer configuration file for CTP. This section applies to images from a General Electric Advantage Windows © workstation and specific types of images that it can produce. In this case, it looks at tags in the DICOM header to characterize the type of image and then specifies the rectangle on the image that contains PHI.
Other Options for Clinical Trials That Use Images
There are no other free and open-source tools built specifically for multisite trials with anonymization and image transfer. The American College of Radiology (Reston, VA) has developed a robust tool for image transfer called TRIAD, but that is proprietary. There are several open-source tools, as well as free and commercial tools for anonymizing studies. Most of these do a very good job of handling the standard DICOM tags. Few have a robust mechanism for removing all private tags, except those known to be safe. This is critical because many research studies wish to use cutting-edge imaging methods where DICOM standards for storing the scanning parameters may not be defined. Until they are defined, vendors typically store the information in private tags that become known fairly quickly. It is critical that anonymization softwares have a database of “known safe tags” to preserve this information but remove the rest. CTP is implementing this, and it should be available by the time this article is published. Several free and open-source image-viewing softwares can anonymize images, including ImageJ, ClearCanvas (Toronto, CA), and OsiriX (Geneva, Switzerland). Another option focusing on just anonymization is shown in http://www.dclunie.com/pixelmed/software/webstart/DicomCleanerUsage.html.
In some cases, PHI may also be “burned into” the pixels of the image, and proper anonymization also requires removal of this. It is possible to use image editing software to “black out” these pixels, but CTP is the only software we are aware of that has a database of where PHI is placed in images, based on the vendor and model of the imaging device.
Conclusion
Images are important for the conduct of many clinical trials. They are often used for documenting the response of a tumor to therapy or for decision making in a clinical trial—to determine whether a subject is eligible or to determine whether a condition that requires a change in therapy has been reached. Implementing an efficient dataflow in a multisite clinical trial while meeting privacy regulations requires special tools. Here, we have described open-source software that helps researchers to implement such a system.
References
- 1.Ford R, Schwartz L, Dancey J, Dodd LE, Eisenhauer EA, Gwyther S, Rubinstein L, Sargent D, Shankar L, Therasse P, et al. Lessons learned from independent central review. Eur J Cancer. 2009;45:268–274. doi: 10.1016/j.ejca.2008.10.031. [DOI] [PubMed] [Google Scholar]
- 2.Erickson BJ, Pan T, Marcus DS. TSA Imaging Informatics Project Group Whitepapers on imaging infrastructure for research: part 1: general workflow considerations. J Digit Imaging. 2012;25:449–453. doi: 10.1007/s10278-012-9490-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clark K, Vendt B, Smith K, Freymann J, Kirby J, Koppel P, Moore S, Phillips S, Maffitt D, Pringle M, et al. The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. J Digit Imaging. 2013;26:1045–1057. doi: 10.1007/s10278-013-9622-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rhodes C, Moore S, Clark K, Maffitt D, Perry J, Handzel T, Prior F. IEEE Engineering in Medicine and Biology Society. Buenos Aires, Argentina: 2010. Regulatory compliance requirements for an open source electronic image trial management system; pp. 3475–3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mortamet B, Bernstein MA, Jack CR, Jr, Gunter JL, Ward C, Britson PJ, Meuli R, Thiran JP, Krueger G. lzheimer's Disease Neuroimaging Initiative Automatic quality assessment in structural brain magnetic resonance imaging. Magn Reson Med. 2009;62:365–372. doi: 10.1002/mrm.21992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Erickson BJ, Langer SG, Blezek DJ, Ryan WJ, French TL. EWEY: the DICOM-enabled workflow engine system. J Digit Imaging. 2014 doi: 10.1007/s10278-013-9661-0. E-pub ahead of print January 10, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]