Abstract
Concomitant inhibition of multiple cancer-driving kinases is an established strategy to improve the durability of clinical responses to targeted therapies. The difficulty of discovering kinase inhibitors with an appropriate multi-target profile has, however, necessitated the application of combination therapies, which can pose significant clinical development challenges. Epigenetic reader domains of the bromodomain family have recently emerged as novel targets for cancer therapy. Here we report that several clinical kinase inhibitors also inhibit bromodomains with therapeutically relevant potencies and are best classified as dual kinase/bromodomain inhibitors. Nanomolar activity on BRD4 by BI-2536 and TG-101348, clinical PLK1 and JAK2/FLT3 kinase inhibitors, respectively, is particularly noteworthy as these combinations of activities on independent oncogenic pathways exemplify a novel strategy for rational single agent polypharmacological targeting. Furthermore, structure-activity relationships and co-crystal structures identify design features that enable a general platform for the rational design of dual kinase/bromodomain inhibitors.
INTRODUCTION
Dysregulation of multiple signaling pathways is a hallmark of cancer development and progression1, and aberrant regulation of kinases in cancer has made these proteins attractive drug targets2. However, the existence of compensating signaling pathways3 has restricted the efficacy of target-selective inhibitors to few cancer subtypes for which a single dominant oncogene has been identified, and the often rapid onset of resistance in responsive patients remains problematic2. These findings suggest that more efficacious inhibitors should target multiple key pathways. The current development approach for multitargeted kinase inhibitors, however, is often based on exploiting incidental “secondary kinase” activities. Most approved kinase drugs potently inhibit multiple targets4, and, as exemplified by imatinib, such accidental off-target activities can extend the use of a drug to other tumor types driven by a distinct kinase. A more robust approach to cancer drug development is to design combinations of inhibitor activities for which a strong rationale has been established, and (pre)clinical studies using combinations of targeted agents in multiple tumor types established proof of principle for this strategy5,6.
While the kinase inhibitor field has overcome challenges related to selectively targeting cancer associated kinases4, a successful strategy for the development of inhibitors conferring high selectivity to multiple rationally selected diverse kinases has not been achieved. We believe that the large size of the kinase family and the similarity of kinase domains make such a strategy a formidable drug discovery challenge. Though this “intra-family” multi-targeting approach remains problematic, we demonstrate in this report the feasibility of an “inter-family” approach for the design of specific inhibitors co-targeting kinases and bromodomain epigenetic reader proteins, which both play key roles in tumorigenesis and inflammatory disease.
Bromodomains (BRDs) are protein interaction modules selectively recruited to ε-N-acetylated lysine containing sequences. Bromodomains are present in diverse nuclear proteins functioning as recruitment platforms for transcriptional regulators, chromatin modulators and chromatin modifying enzymes7. Dysfunction of bromodomain containing proteins has been strongly associated with the development of cancer8. In particular the bromo and extra terminal (BET) proteins (BRD2, BRD3, BRD4, BRDT) have recently received much attention after the development of potent and cell active pan-BET inhibitors7,9,10. BETs are transcriptional regulators that control expression of genes essential for tumor growth (e.g. c-Myc, Aurora B) and survival (e.g. Bcl-2), and BET-specific inhibitors showed efficacy in a number of diverse cancer models7,11–15.
Though bromodomains have only recently been identified as druggable targets, compelling rationale already exists for the development of dual kinase/bromodomain inhibitors as therapeutics for both oncology and inflammatory disease. For example, FLT3 receptor tyrosine kinase and BRD4 are both drivers in acute myelogenous leukemia (AML)16,17, JAK kinase and BRD4 inhibitors show complementary tumor and host microenvironment activities in multiple myeloma models12,18, and both bromodomain and kinase inhibitors have shown compelling efficacy in inflammatory disease9,19. Here we show that several inhibitors developed to target specific kinases also potently inhibit diverse bromodomains. Co-crystal structures of these clinical kinase inhibitors elucidated their binding modes and defined design rules for dual kinase/bromodomain inhibitors. The diversity of the drug binding sites between these target families suggests that specific kinase/bromodomain dual inhibitors can be rationally designed for disease applications where involvement of both target families has been demonstrated. We believe that this will be particularly interesting for designing dual inhibitors that act as single agent therapies on key cancer drivers from these distinct protein families. This “dual-targeted single agent” approach could in principle confer the same benefits as combination therapies (e.g. larger therapeutic windows, more durable responses), while also minimizing several liabilities of combination approaches, including complex and lengthy clinical investigations, the potential for additive/synergistic non-mechanism-based toxicities and drug-drug interactions, and high treatment costs.
RESULTS
Potent and selective dual kinase/bromodomain inhibitors
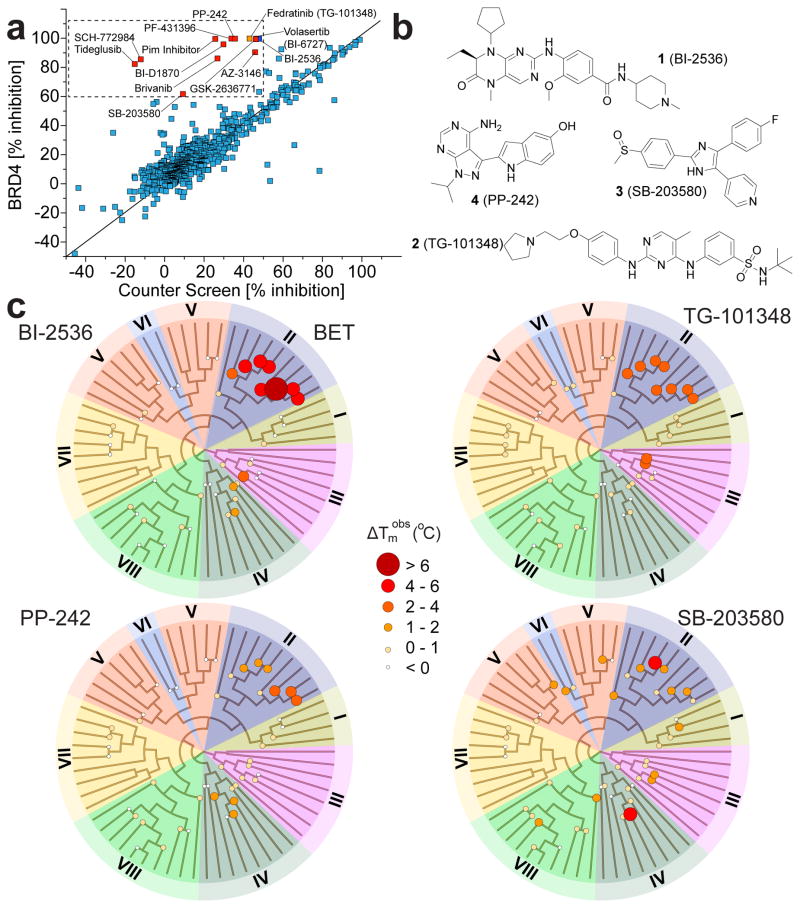
To assess the feasibility of designing dual kinase/bromodomain inhibitors and to find novel lead compounds for bromodomain inhibition in general, we sought to identify cross-reactivity between known kinase inhibitors and bromodomains. In this study we used an AlphaScreen assay format to screen a collection of 628 kinase inhibitors against the first bromodomain of BRD4. The inhibitor collection contained widely used kinase tool compounds as well as more than 200 clinical kinase inhibitors (Fig. 1a). Surprisingly, within this kinase-focused screening set, nine inhibitors were found to strongly inhibit binding of the poly-acetylated histone H4 peptide (>90% at 50 μM) while showing less than 50% inhibition in a counter screen (His6 only) (Supplementary Results, Supplementary Data Set 1). Among the tested kinase inhibitors with potent bromodomain activity were the PLK inhibitor BI-2536 (1) and the related compounds BI-6727 (volasertib) as well as the RSK inhibitor BI-D1870, the JAK inhibitor TG-101348 (2) (fedratinib), the p38 inhibitor SB-203580 (3), the MPS1 inhibitor AZ-3146, an isoxazolo-quinoline developed for targeting PIM kinases, the focal adhesion kinase (FAK) inhibitor PF-431396, and the PI3K/mTOR inhibitors GSK2636771 and PP-242 (4)20–28. The high hit rate that we observed on a target that is structurally and functionally very diverse when compared to protein kinases was surprising and suggests that bromodomains and kinases nevertheless share a high degree of pharmacophore similarity. Importantly, many of the discovered kinase/bromodomain dual inhibitors are widely used as tool molecules or have entered clinical testing. Thus, the consequences of bromodomain activity for these inhibitors will need to be carefully evaluated. Next, we confirmed the BRD4 activity of compounds identified in AlphaScreen assays using a temperature shift assay (ΔTm), a well-established sensitive ligand binding assay for both kinases and bromodomains29,30. We observed large positive temperature shifts for most of the identified kinase inhibitors confirming strong interactions with BRD4(1) (Supplementary Fig. 1). In agreement with this observation, AlphaScreen dose response experiments carried out on selected screening hits against BRD4(1) gave rise to nanomolar IC50 values for BI-2536, BI-D1870 and TG-101348 as well as low μM activity for the closely related p38 inhibitors SB-202190 and SB-203580 and the PI3K inhibitor PP-242 (Supplementary Fig. 1).
Figure 1. Discovery of kinase inhibitors that potently cross-react with bromodomains.
a, AlphaScreen data measured at 50 μM compound concentration against BRD4(1). Shown is a correlation of observed inhibition (% inhibition) with counter screen inhibition (using a His6 peptide lacking the bromodomain). Hits (less than 50% inhibition in counter screen and more than 60% inhibition) are highlighted in the boxed area and are labeled. b, Inhibitor structures. c, Dendrograms of the human bromodomain family temperature shift data collected on BI-2536 (BI), TG-101348 (TG), PP-242, and SB-203580 using a panel of 46 bromodomains. Bromodomains have been grouped in 8 sub-families as described7. A fully labeled dendrogram is shown in Supplementary Figure 3. Temperature shifts are represented by spot size and color as indicated.
Isothermal titration calorimetry (ITC) was used to determine BRD4(1) binding constants (Kds) for the identified inhibitors in solution. These experiments confirmed strong interaction with BRD4(1) for the PLK inhibitors BI-2536 (Kd: 37 ±3 nM) and BI-6727 (Kd: 79 ±3 nM) and the related RSK inhibitor BI-D1870 (Kd: 3.5 ±0.17 μM). The closely related JAK inhibitors TG-101348 and TG-101209 bound BRD4(1) with nanomolar affinities (Kds: 164 ±10 nM and 123 ±18 nM, respectively). Nanomolar affinities were also determined for PF-431396 (Kd: 445 ±42 nM) and for the PIM inhibitor (Kd: 565 ±63 nM). The p38 inhibitor SB-202190 and the PI3K inhibitor PP-242 showed slightly weaker activity (Kds: 3.4 ±0.13 μM and 1.7 ±0.076 μM, respectively). ITC data were in good agreement with temperature shift data and IC50 values measured by AlphaScreen assays. Representative ITC experiments and thermodynamic binding data are compiled in Supplementary Figure 2.
To assess the activity of the identified inhibitors against bromodomains outside the well -studied BET family member BRD4(1) we screened six selected inhibitors against a panel of 46 bromodomains that comprised representative members of all subfamilies (Fig. 1b, c and Supplementary Fig. 3). This comprehensive cross-screening panel showed that bromodomains outside the BET family were also inhibited by the identified kinase/BRD4 dual inhibitors. PP-242 for instance showed a 1.7°C ΔTm for BRD1 and other BRPF family members, TG-101348 also interacted with the bromodomains of the histone acetyl transferases CREBBP and EP300, and SB-202190 showed 1.3°C ΔTm shifts for the CECR2 bromodomain and for BRD7. Thus, it is likely that the activities on non-BET bromodomains will serve as starting points for the development of more potent and selective bromodomain inhibitors outside the BET family. Temperature shift data are summarized in Supplementary Table 1.
Cellular activity of dual inhibitors on BET bromodomains
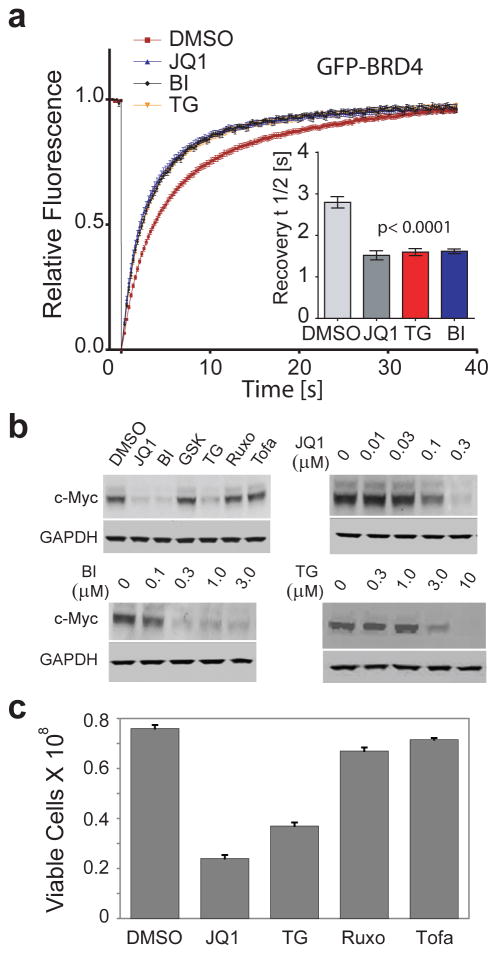
The ability of BI-2536 and TG-101348 to displace BRD4 from chromatin, thus interfering with its function as a recruitment module, was demonstrated by accelerated FRAP (fluorescent recovery after photo-bleaching) recovery times in the same range as measured for the pan-BET inhibitor JQ129 (Fig. 2a, and Supplementary Fig. 4a). BI-2536 and TG-101348 also potently suppressed c-Myc expression in MM.1S multiple myeloma cells, which is significant because c-Myc is a well-established cellular marker of BET inhibition as demonstrated in diverse tumor types with multiple selective BET inhibitors10–13,15. In contrast, c-Myc was not repressed by other potent and selective inhibitors of PLK or JAK lacking BET activity, including ruxolitinib, tofacitinib, and GSK-461364A (Fig. 2b, Supplementary Data Set 1, and Supplementary Fig. 5). Dose-response data show that BI-2536 has similar potency (IC50 ~ 100 nM) to a best-in-class BET inhibitor (JQ1) and that TG-101348 (IC50 ~ 1–3 μM) could potentially inhibit BETs at well tolerated clinical doses26 (Fig. 2b). TG-101348, but not the other JAK inhibitors, also inhibited MM.1S proliferation (IC50 1–3 μM) (Fig. 2c). Interestingly, JAK inhibition has been reported to inhibit proliferation of multiple myeloma cell lines (e.g. MM.1S) based on studies with the TG-101348 analog TG-10120931, which we now show to have potent BET activity by ITC (Supplementary Fig. 2). Our data therefore suggest that BET and not JAK family proteins are the key TG compound targets in MM.1S and likely in the other multiple myeloma cell lines as well.
Figure 2. BI-2536 and TG-101348 displace BRD4 from chromatin and suppress c-Myc expression.
a, Fluorescent Recovery After Photo-bleaching (FRAP) experiments using full-length GFP-BRD4. Time dependence of fluorescence recovery after targeted nuclear photobleaching of U2OS cells transfected with GFP-BRD4 that were untreated or exposed to the pan-BET inhibitor JQ1, BI-2536 (BI) or TG-101348 (TG) at 500 nM. Averaged recovery half times are shown in the insert and images of fluorescent nuclei are shown in Supplementary Figure 4a. b, Western blots showing down regulation of the c-Myc by JQ1 and kinase inhibitors. Top left panel: BI-2536 (BI) and TG-101348 (TG) show strong c-Myc down regulation in MM.1S cells. In contrast the PLK and JAK inhibitors GSK-461364A (GSK), ruxolitinib (Ruxo), and tofacitinib (Tofa), which are inactive against BETs, did not affect c-Myc levels. All inhibitor concentrations were 5 μM, except for JQ1 (1 μM). The other three panels show dose responses for c-Myc downregulation in MM.1s cells for JQ1, BI-2536 (BI) and TG-101348 (TG). Inhibitor concentrations are indicated. GAPDH measured on replicate blots served as the normalization control. Full blot images are shown in Supplementary Figure 4b. c, TG-101348, but not JAK inhibitors that lack BET activity, inhibits proliferation of MM.1S multiple myeloma cells. MM.1S cells were cultured for 72 h in the presence of DMSO, JQ1 (positive control at 0.5 μM), TG-101348 (TG), and the potent JAK inhibitors tofacitinib (Tofa) and ruxolitinib (Ruxo) (all at 3 μM). Error bars are the standard deviations for duplicate measurements.
Mechanism of BI-2536 and TG-101348 BRD4 interactions
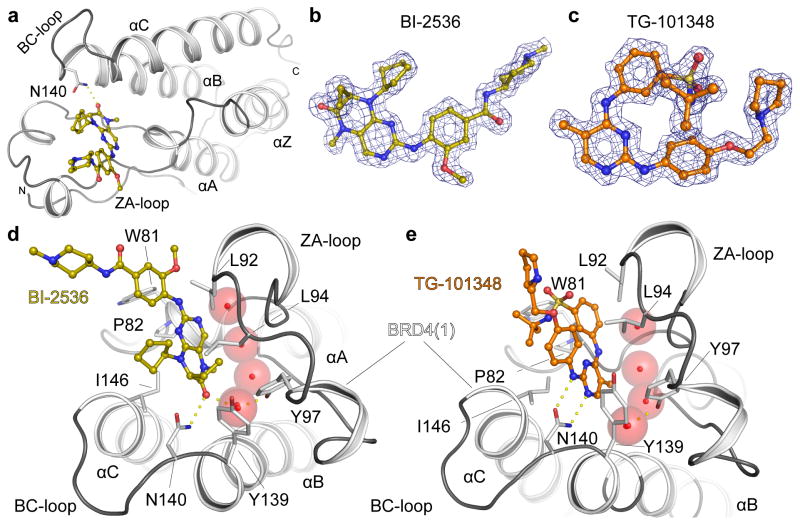
To understand the structural basis for the observed dual kinase/bromodomain activity we determined co-crystal structures of BI-2536 and TG-101348 with BRD4(1) (Fig. 3). The ligands bound to the acetyl-lysine binding site and were well defined by electron density (Fig. 3a–c). The BI-2536 complex revealed that the dihydropteridinone carbonyl together with the methylamino group act as an acetyl-lysine mimetic moiety forming a hydrogen bond with the conserved asparagine N140 (Fig. 3d). Water molecules that are typically present in BET bromodomain ligand complexes are also conserved in the BI-2536 complex. One of these structural waters bridges the dihydropteridinone carbonyl to the hydroxyl group of the conserved tyrosine Y97. The ethyl group that branches off the asymmetric dihydropteridinone carbon atom which has been shown to be important for the high selectivity of this inhibitor for PLK kinases fits perfectly into a hydrophobic cavity formed by L92, L94 and Y13932.
Figure 3. BRD4 Binding modes of BI-2536 and TG-101348.
a, Ribbon diagram showing the structure of the BRD4(1) bromodomain in complex with BI-2536. The main secondary structure elements and the N- and C-termini are labeled. b, Experimental electron density map contoured at 2σ around the inhibitor BI-2536. c, Experimental electron density map defining the binding mode of TG-101348 contoured as described in b. d, Co-crystal structure of BI-2536 with BRD4(1). Main residues interacting with the inhibitor are shown and labeled. Conserved binding site waters are shown as red spheres. e, Co-crystal structure of TG-101348 with BRD4(1). See Supplementary Table 2 for data collection and refinement statistics.
Co-crystallization of TG-101348 revealed an acetyl-lysine mimetic binding mode in which the secondary amine nitrogen as well as the nitrogen and a methyl group present in the 5-methyl-pyrimidine ring system form two hydrogen bonds with BRD4(1) N140 (Fig. 3e). The methyl group is oriented towards the bottom of the acetyl-lysine binding pocket which is also occupied by conserved water molecules in this structure. In addition, the sulfonamide oxygen and the pyrrolidine nitrogen form additional water mediated hydrogen bonds with the ZA loop backbone (L92), while the t-butyl sulfonamide group points towards the BRD4 shelf region. A summary of data collection and refinement statistics has been compiled in Supplementary Table 2 and known/proposed hinge binding and acetyl-lysine binding moieties for additional dual inhibitors identified in this study are shown in Supplementary Figure 6.
Mechanisms of kinase/bromodomain dual activity
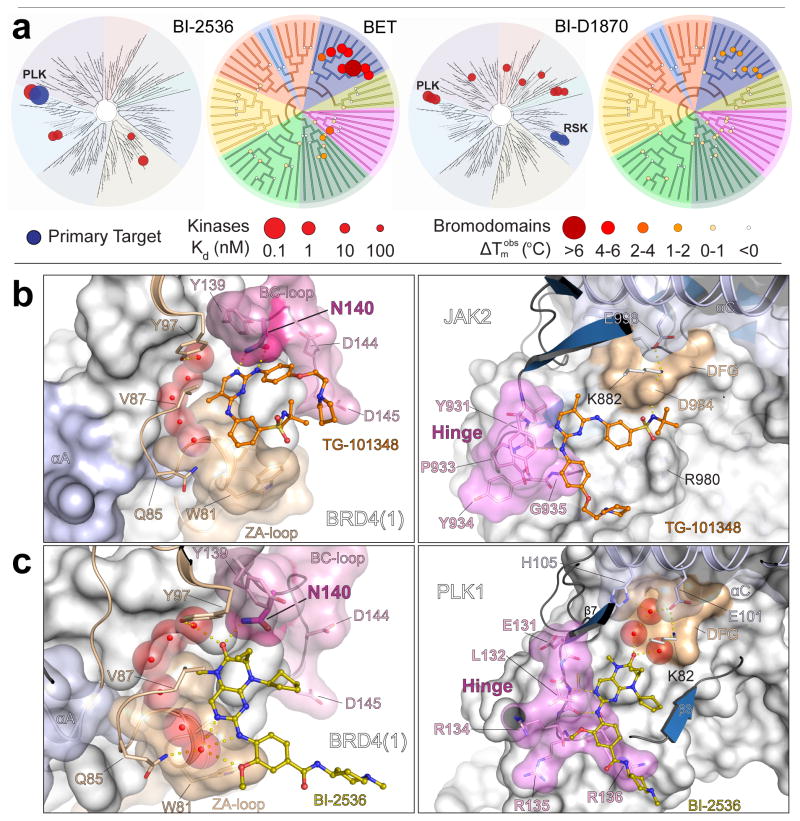
Kinome-wide profiling data4 confirmed the excellent selectivity of BI-2536 for PLK1–3, while its interactions across the bromodomain family were largely restricted to BET bromodomains, with weaker activity observed against BRD7, BRPF1B, and BRWD3(2) (Fig. 4a). The clinically advanced BI-2536 analog BI-6727 (volasertib)33 showed a similar bromodomain selectivity profile, which was expected since the structural differences between these inhibitors are restricted to solvent exposed moieties. Dihydropteridinones have also been developed as RSK kinase inhibitors20. The acetyl-lysine mimetic dihydropteridinone carbonyl methylamino group is conserved in the RSK inhibitor BI-D1870, which showed a bromodomain profile similar to that for BI-2536 (Fig. 4a). The lower affinity of BI-D1870 for BET bromodomains is likely due to the substitution of the dihydropteridinone ethyl group in BI-2536 by a methyl group, which reduces the hydrophobic interaction surface. The activity of BI-D1870 on BRD4(1) was confirmed by ITC (Supplementary Fig. 2).
Figure 4. Structure-activity relationships for dihydropteridinone interactions with kinases and bromodomains and structural comparison of inhibitor binding modes.
a, Dendrograms of the human kinase and bromodomain families showing inhibitor structures interactions with BI-2536 and BI-D1870 (see Supplementary Fig. 6 for BI-D1870 structure and hinge/acetyl-lysine binding moieties). Selectivity against the kinase family is shown in the left panels for each of the two compounds. Temperature shifts against bromodomains are shown in the right panels. The primary, intended kinase targets are indicated by blue spheres. The radii of the spheres correspond to inhibitor potency (Kd) or temperature shift at 10 μM inhibitor concentration as indicated. b, Details of the binding mode of TG-101348 with BRD4(1) (left panel) and JAK2 kinase (right panel). The JAK2/TG-101348 structure is a model consistent with the co-crystal structure of the closely related inhibitor TG-101209 bound to JAK2 (PDB ID 4J19)37. c, Details of the binding mode of BI-2536 with BRD4(1) (left panel) and PLK1 kinase (PDB ID 2RKU)32 (right panel). Critical elements of inhibitor interaction are labeled and highlighted. Conserved water molecules are shown as transparent cpk spheres and key residues are depicted in stick representation.
In contrast to the bromodomain profiling data, kinome-wide data for BI-2536 and BI-D1870 differed dramatically. These data confirmed that BI-D1870 is a potent RSK family kinase inhibitor (Kds: 10–100 nM) with 10–100 fold increases in RSK potencies relative to BI-2536, whereas the PLK family potencies for BI-D1870 (Kds: ~10 nM) were 10–100 fold lower than for BI-2536 (Fig. 4a and Supplementary Table 3). Taken together, these data reveal 100 – 10,000 fold overall selectivity differences between these two closely related inhibitors for the PLK and RSK families. This binding profile comparison demonstrates that BET activity can be maintained as the kinase profile is altered, thus showing the feasibility of optimizing these inhibitors for a diverse kinase target while maintaining a similar bromodomain profile.
The closely related pyridinyl imidazole p38 inhibitors SB-203580 and SB-202190 are highly specific for p38α/β and differ by only substitution of the phenylethanethione in SB-203580 by a hydroxyl group in SB-20219028. This substitution has no effect on the kinase selectivity profile and interactions across the BET family were maintained as well (Supplementary Tables 1 and 3). Though not shown experimentally, comparison to dimethylsulfoxide/bromodomain complexes suggests that the SB-203580 ethanethione acts as an acetyl-lysine mimetic moiety34. For SB-202190 the phenolic OH group is likely the acetyl-lysine mimetic. Indeed, high selectivity of methylphenol head groups for BET bromodomains has recently been demonstrated for the RVX-208 analog RVX-OH, with the OH group forming a hydrogen bond with the conserved asparagine residue N140 in BRD4(1)35. The SB-203580 ethanethione is oriented toward the solvent in co-crystal structures with p38α explaining why this group can be altered without affecting kinase activity36. However, since this position is important for bromodomain binding, it may be possible to optimize these types of inhibitors for diverse bromodomains outside of the BET family while holding the kinase profile constant.
The co-crystal structure of TG-101348 with BRD4(1) and a structural model of its complex with JAK2 (see Fig. 4 legend) show that the hydrogen bonds typically anchoring ATP mimetic inhibitors to the kinase hinge region also interact with the acetyl-lysine coordinating conserved asparagine (N140 in BRD4(1))37 (Fig. 4b). In contrast, the BI-2536/PLK1 co-crystal structure reveals that the acetyl-lysine mimetic dihydropteridinone carbonyl moiety binds to the back pocket of the kinase active site forming water mediated hydrogen bonds with the conserved VIAK motif lysine (K82) and the DFG backbone32. Interactions with the conserved active site lysine are critical for the potency of a number of inhibitors that make either direct or water mediated contacts with this residue. In some cases the back pocket even serves as the lone hydrogen bond anchor point for inhibitors38. The amine linker and a nitrogen atom present in the pyrimidine of the dihydropteridinone ring system form the canonical hinge interactions (Fig. 4c). These hydrogen bonds are critical for the kinase activity of this inhibitor but they face the solvent exposed region in the BRD4(1) co-crystal structure (Fig. 4c). This suggests that methylation of the linker nitrogen would yield bromodomain-selective inhibitors. In addition, the methoxy group in BI-2536 has been shown to be important for PLK selectivity by taking advantage of a PLK-specific binding cavity32, whereas it points toward solvent in the BRD4(1) co-crystal structure. Thus, modifications of this critical position can likely modulate this inhibitor’s ability to recognize different kinases while leaving the bromodomain activity unchanged. Modulation of bromodomain activity can be achieved by modifying the cyclopentyl substitution, which for BI-6727 (volasertib) results in a reduction in BRD4(1) affinity (Supplementary Fig. 2).
FLT3 inhibitor-resistant AML cells retain JQ1 sensitivity
AML provides a compelling example of an oncology indication that could be exploited by the dual targeting of kinases and bromodomains. For instance, AML is often driven by BET bromodomains and mutant FLT316,17 suggesting the utility of inhibitors with dual FLT3/BET activity. Indeed, TG-101348, but not JAK2 inhibitors that lack BET and FLT3 activity, potently inhibits proliferation of MV4-11 AML cells (IC50 = 79 nM) (Fig. 5a), though the relative contributions of the FLT3 and BET activities were difficult to assess in this context. In principle an optimized dual targeted agent or a combination of potent FLT3 and BET inhibitors could delay the rapid onset of drug resistance observed in FLT3 inhibitor treated AML patients17. To explore this concept, we tested JQ1 against Molm14 AML cell lines with acquired resistance to a FLT3 inhibitor39. These resistant lines, Molm14-D835Y and Molm14-F691L, have acquired second site FLT3 mutations (D835Y and F691L, respectively) that confer inhibitor resistance, and these same mutations predominate in AML patients who have become resistant to FLT inhibitor therapy17. JQ1 demonstrated equipotent activity on the parental and resistant Molm14 lines in both proliferation and c-Myc suppression assays (Fig 5b, c) suggesting that cells with acquired resistance to a FLT3 inhibitor retain complete sensitivity to a BET inhibitor. In addition, it was recently reported that an independently derived pool of FLT3 inhibitor-resistant AML cells is even more sensitive to BET inhibition than the parental cell line (55th ASH Annual Meeting (2014), Rodriguez, M. and Bhalla, K.N., abstract 3821). These findings support the use of dual or combined FLT3/BET inhibitors in future AML clinical investigations.
Figure 5. Responses of FLT3 inhibitor-sensitive and -resistant AML cell lines to TG-101348 and BET inhibitors.
a, TG-101348, but not JAK inhibitors that lack FLT3 and BET activity, potently inhibits proliferation of MV4-11 AML cells. MV4-11 cells were tested against the indicated inhibitors for 72 h. Viable cell percent of DMSO control (PoC) is plotted on the y-axis. IC50 values (μM): JQ1 (0.028), TG-101348 (TG, 0.079), I-BET (0.20), ruxolitinib (Ruxo, ~10), and tofacitinib (Tofa, >10). Error bars are the standard deviations for duplicate measurements. b, Molm14 AML cells with acquired resistance to a FLT3 inhibitor retain sensitivity to BET inhibition. Molm14 (parental) and two FLT3 inhibitor-resistant derivatives harboring acquired resistance mutations in the FLT3 kinase domain (D835Y and F691L)39 were treated with JQ1 at multiple concentrations for 72 h and data were analyzed as described in the Online Methods. All Molm14 cell lines were inhibited by JQ1 with similar potencies (IC50’s ~200 nM). c, JQ1 inhibits c-Myc expression with similar potencies in all FLT3 inhibitor-sensitive and -resistant Molm14 cell lines. c-Myc and GAPDH (internal control run on a parallel blot) levels were measured by western blotting as described in Online Methods post 6 h treatment with JQ1 at the indicated concentrations. Full blot images are shown in Supplementary Figure 7.
Dual inhibitor polypharmacology in primary human cells
The results presented above, while confirming dual kinase/bromodomain activity for the identified inhibitors, nevertheless fail to incisively demonstrate polypharmacology from a more global phenotypic perspective. The functional effects of dual kinase/bromodomain inhibition for BI-2536 and TG-101348 were therefore explored across a panel of 12 stimulated primary human cell types and co-cultures designed to recapitulate the complex signaling networks and microenvironment in diseased human tissue (BioMAP)40. This established approach provides a measure of overall phenotypic response of compounds under disease-like conditions and identifies clinically relevant activities across a broad protein biomarker panel40–42. BI-2536 and TG-101348 were tested in parallel with selective benchmark inhibitors of BET, JAK, and PLK. Signature activities for the selective benchmarks included downregulation of TNFα, MCP-1, and IL-8, and anti-proliferative effects on vascular and stromal compartments (BET inhibitors); up-regulation of IL-2, downregulation of Eotaxin-3, anti-proliferative effects on IL-2 stimulated T-cells (JAK inhibitors); downregulation of uPAR and PAI-1 and broad anti-proliferative activity (PLK inhibitors) (Fig. 6a and Supplementary Figs. 8–10). BI-2536 and TG-101348 showed more complex phenotypes across BioMAP than the benchmarks, consistent with their dual kinase/bromodomain activities. To visualize the phenotypic similarities between inhibitors, the BioMAP profiles were analyzed by pairwise correlation analysis and used to generate a distance matrix shown as a function homology map in Fig. 6b, where compounds with similar profiles are connected by lines to form phenotypic clusters. While the selective benchmark inhibitors were confined to distinct target specific clusters, the BI-2536 and TG-101348 profiles clustered with both their BET benchmarks and their respective selective kinase inhibitors. At intermediate doses, BI-2536 and TG-101348 yielded well resolved dual kinase/BET signatures that are represented as nodes linking the target specific clusters (Fig. 6b). Together, these results demonstrate: (i) polypharmacology for BI-2536 and TG-101348 in human disease models, (ii) that, with some exceptions (e.g. IL-2, Supplementary Fig. 8b), the dual inhibitor phenotypes largely reflected the sum of the mono-inhibitor phenotypes – an outcome not necessarily expected due to the complex signaling networks affected, and (iii) that the dual inhibitors affect novel combinations of predictive biomarkers not observed for the mono-inhibitors. For example, TG-101348 inhibited endothelial cell and fibroblast proliferation, immune cell recruitment markers (e.g. MCP-1 and VCAM-1), and production of TNFα, MIG and IL-8 – all hallmarks of BET inhibition (Fig. 6c and Supplementary Figs. 8, 9). In addition, this inhibitor also modulated cytokine and chemokine production (IL-2 and Eotaxin-3) and immune cell proliferation – all hallmarks of the JAK inhibition signature (Fig. 6 and Supplementary Figs. 8, 9). This novel combination of activities would be potentially advantageous for the treatment of hematological malignancies (e.g. multiple myeloma) due to the potential for combined action on both tumor cells and the relevant host vascular and stromal microenvironments18.
Figure 6. Functional activities of dual kinase/bromodomain inhibitors in primary human cell disease models.
a, Test agents and kinase or BET benchmark inhibitors were tested for effects on protein biomarkers (x-axis) in 12 primary human cell-based BioMAP systems (Supplementary Table 4) at 7 doses. The TG-101348 (TG) BioMAP profile in selected systems is overlaid with profiles of inhibitors targeting JAK (tofacitinib (Tofa) and ruxolitinib, (Ruxo) or BET (PFI-1 and I-BET) (all at 3.3 μM). Biomarkers modulated by TG in a manner similar to either JAK or BET inhibitors are labeled in blue and green, respectively. See Supplementary Figure 9 for full BioMAP profile. b, BI-2536 (BI) and TG-101348 (TG) and inhibitors targeting either kinases, including JAK (tofacitinib (Tofa) and ruxolitinib (Ruxo)) or PLK (GSK-461364A (GSK)), or BET bromodomains (JQ1, I-BET, PFI-1, and I-BET 151) were profiled in the BioMAP Diversity PLUS panel at 6 doses to generate dose-specific compound signature activity profiles. Statistical measures of profile similarity were used to cluster compounds with shared phenotypic responses to graphically represent their functional similarity. Dot color and size indicate compound and dose, respectively. Compound profiles similar above the selected threshold (Pearson > 0.8) are represented by connected dots. Target-related clusters reflect a common phenotypic signature in these primary cell models. c, Dose-response plots for inhibitors of JAK (Tofa, Ruxo), BET (JQ1, PFI-1), and the dual inhibitor (TG) against a BET biomarker (TNFα). Biomarker ratio to DMSO control is plotted on the y-axis. Error bars are the SEM (n = 3).
DISCUSSION
The clinical dual kinase/bromodomain inhibitors identified in this study demonstrate proof of principle for the tractability of discovering potent and selective drugs with polypharmacologies anticipated to add significant therapeutic benefit. These inhibitors target diverse key drivers in a number of cancers as well as in inflammatory disease. For instance, the activity of TG-101348 on FLT3 kinase and BET bromodomains, which both drive AML16,17, demonstrates the feasibility of designing next generation targeted AML therapies expected to be less prone to acquired drug resistance (Fig. 5)17. Dual JAK/BET inhibition exemplified by TG-101348 could also be beneficial in the IL7R-driven subset of acute lymphoblastic leukemia where JAK and BET activities promote the same oncogenic pathway43. Dual PLK/BET inhibitors may also be an efficacious combination due to the functions of both targets in mitosis. The potent BET activity of BI-6727 (volasertib) suggests potential applications in highly proliferative BET-driven tumors, and, interestingly, volasertib has been granted a breakthrough therapy designation by the United States Food and Drug Administration for the treatment of AML based on positive clinical trial data (http://www.boehringer-ingelheim.com/news/news_releases/press_releases/2013/17_september_2013oncology.html).
Pyridinyl imidazoles have been identified as anti-inflammatory agents that potently and selectively inhibit p38 kinases44, which also play pivotal roles as regulators of tumor invasion and metastasis45. Similarly, BET bromodomains have shown efficacy suppressing pro-inflammatory cytokine release and confer protection against lipopolysaccharide-induced endotoxic shock and bacteria-induced sepsis9. In addition, these compounds have been used as specific p38 probes in a large number of studies46. Here we identified SB-203580 and SB-202190 as low micromolar inhibitors of BET bromodomains (Supplementary Fig. 2).
The structural models provide a rationale for new polypharmacological drug discovery strategies focused on two complementary target classes. While the identified dual inhibitors have been highly optimized against the targeted kinases, bromodomain potency has not been optimized, and the low nanomolar activity for the discussed JAK and PLK inhibitors on BET bromodomains was an unexpected finding. Comparison of co-crystal structures of the same inhibitors with both target classes identified three distinct structural mechanisms explaining kinase/bromodomain cross-reactivity: 1) The acetyl-lysine mimetic group does not interact with the kinase active site. This is the most straightforward case for removing or modulating kinase and bromodomain inhibitor activities since the critical residues responsible for high affinity bromodomain interaction are dispensable for kinase activity. This is the case for the described p38 inhibitors SB-202380 and SB-202190 where the phenylethanethione or methylphenol groups critical for bromodomain binding are oriented toward the solvent region in p38α. 2) The acetyl-lysine mimetic group forms polar interactions with conserved back pocket residues such as the canonical lysine- αC glutamate salt bridge typically present in active kinases. This is the case for the dihydropteridinone kinase inhibitors targeting PLK and RSK. 3) The acetyl-lysine mimicking group coincides with the hinge binding hydrogen donor/acceptor pair as is the case for TG-101348 and TG-101209. Weak interactions with kinase hinge binding motifs at this site have been reported previously. For instance, a CDK inhibitor (dinaciclib) pyrazolo-pyrimidine hinge binding group forms hydrogen bonds with the acetyl-lysine coordinating asparagine in the BET bromodomain BRDT, which results in a low affinity interaction (Kd: 37 μM)47. We also recently identified low affinity fragment molecules that target both kinases and bromodomains demonstrating that similar anchoring via equidistantly spaced hydrogen bonds can enable inhibitor interactions with both kinases and bromodomains48. Recently, chromen-4-one PI3K inhibitors were also shown to have micromolar activity against BRD4, but in this case the hinge and acetyl-lysine binding moieties are distinct49. Thus, despite the high diversity between the bromodomain acetyl-lysine binding pocket and the kinase ATP binding site, the similar geometry of key inhibitor interactions can explain the observed dual activities.
A rational dual inhibition design strategy can likely yield high affinity compounds with equipotent activity on both the targeted kinase and bromodomain. In addition, the active site diversities allow design of specific bromodomain inhibitors based on optimized drug-like kinase inhibitors. For instance, extension of the sulfonamide decoration with larger substituents would sterically exclude TG-101348 from kinase active sites. Similarly, substitution of the key methoxy group32 in the aromatic ring next to the BI-2536 dihydropteridinone by bulkier moieties or methylation of the nitrogen atoms interacting with the PLK1 hinge region should exclude this compound from kinase active sites. Based on the binding site diversity between bromo and kinase domains, we propose that the rational design of dual kinase/bromodomain inhibitors is more tractable than developing inhibitors specifically targeting two rationally chosen distinct kinases. We are therefore hopeful that the provided design strategies will stimulate the discovery of a new generation of optimized dual kinase/bromodomain inhibitors targeting multiple disease pathways that will achieve improved efficacy and more durable responses than current targeted cancer therapies.
ONLINE METHODS
Compounds
Kinase inhibitors were purchased from A.G. Scientific, Inc. (San Diego, CA), Calbiochem/EMD Chemicals (San Diego, CA), Tocris Bioscience (Ellisville, MO), Archerchem (Mumbai, India), Axon Medchem BV (Groningen, The Netherlands), SYNthesismedchem (Shanghai, China) or Selleckchem (Houston, TX). JQ1 was purchased from BPS Bioscience, Inc. (San Diego, CA) and I-BET 151 and PFI-1 from Tocris. The compound providers confirmed the structures and batch purities (≥ 95%) by using standard analytical methods (HPLC/mass spectroscopy and NMR).
Cell lines
MM.1S, MV4-11, U2OS and Molm14 were purchased from ATCC (Manassas, VA). FLT3 inhibitor-resistant Molm14 lines (Molm14-D835Y and –F691L) were generated as described39.
Recombinant bromodomains
Human bromodomains were expressed in bacteria in fusion with a His6-TEV (Tobacco Etch Virus) protease tag and were purified as described7.
Temperature shift assays
Thermal melting experiments were carried out using an Mx3005p Real Time PCR machine (Stratagene). Proteins were buffered in 10 mM HEPES pH 7.5, 500 mM NaCl and assayed in a 96-well plate at a final concentration of 2 μM in 20 μl volume. Compounds were added at a final concentration of 10 μM. SYPRO Orange (Molecular Probes) was added as a fluorescence probe at a dilution of 1:1000. Excitation and emission filters for the SYPRO-Orange dye were set to 465 nm and 590 nm, respectively. The temperature was raised with a step of 3 °C per minute from 25 °C to 96 °C and fluorescence readings were taken at each interval. Data were analyzed as previously described7.
Biochemical inhibitor screening
AlphaScreen Assays were performed as described previously34 with minor modifications from the manufacturer’s protocol (PerkinElmer, USA). All reagents were diluted in 25 mM HEPES, 100 mM NaCl, 0.1% BSA, pH 7.4 supplemented with 0.05% CHAPS and allowed to equilibrate to room temperature prior to addition to plates. Initial screening was carried out at 50 μM compound concentration. Inhibitors interfering with the assay were identified in counter screens in which the His6 tagged bromodomain was substituted by a His6 peptide. Dose response experiments were carried out using an 11-point 1:2.5 serial dilution of the ligands was prepared over the range of 5000 – 0 μM and 0.1 μl transferred to low-volume 384-well plates filled with 5 μl of the assay buffer (ProxiPlateTM-384 Plus, PerkinElmer, USA), followed by 7 μl of biotinylated peptide [H-YSGRGKacGGKacGLGKacGGAKacRHRK-(Biotin)-OH and His-tagged BRD4(1) to achieve final assay concentrations of 25 nM. Plates were sealed and incubated for a further 30 minutes, before the addition of 8 μl of the mixture of streptavidin-coated donor beads (12.5 μg/ml) and nickel chelate acceptor beads (12.5 μg/ml) under low light conditions. Plates were foil-sealed to protect from light, incubated at room temperature for 60 minutes and read on a PHERAstar FS plate reader (BMG Labtech, Germany) using an AlphaScreen 680 excitation/570 emission filter set. IC50 values were calculated in Prism 5 (GraphPad Software, USA) after normalization against corresponding DMSO controls and are given as the final concentration of compound in the 20 μl reaction volume.
Isothermal titration calorimetry (ITC)
Experiments were carried out on an ITC200 titration microcalorimeter from MicroCal™, LLC (GE Healthcare) equipped with a Washing module, with a cell volume of 0.2003 ml and a 40 μl microsyringe. Experiments were carried out at 15 °C in 50 mM HEPES pH 7.4, 150 mM NaCl. The heat of dilution of the titrated ligand (BRD4(1)) determined by blank titrations corresponded to the heat observed from the last injection, following saturation of ligand binding. The collected data were corrected for protein heats of dilution. Titrations were evaluated using the MicroCal™ Origin software supplied with the instrument to yield enthalpies of binding (ΔH) and binding constants (KB) as described by Wiseman and co-workers50. In all cases a single binding site model was employed. Dissociation constants and thermodynamic parameters are listed in Supplementary Figure 2.
Protein crystallization
Protein expression and purification was carried out as previously described29. Coarse screens were typically setup onto Greiner 3-well plates using three different drop ratios of precipitant to protein per condition (100+50 nl, 75+75 nl and 50+100 nl) and a mosquito® crystallization robot (TTP Labtech, Royston UK). All crystallizations were carried out using the sitting drop vapor diffusion method at 4 °C. Crystals of BRD4(1) with BI-2536 (10 mM final concentration) were grown by mixing 150 nl of the protein (9.9 mg/ml) with 150 nl of reservoir solution containing 0.2 M K3(citrate), 0.1 M BTProp pH 8.5, 20% PEG3350 and 10% ethylene glycol. Crystals of BRD4(1) with TG-1010348 (10 mM final concentration) were grown by mixing 150 nl of the protein (10.2 mg/ml) with 100 nl of reservoir solution containing 0.2 M Na/K tartrate, 0.1 M BTProp pH 7.5, 20% PEG3350 and 10% ethylene glycol.
Data collection and structure solution
In all cases crystals were cryo-protected using the well solution supplemented with additional ethylene glycol and were flash frozen in liquid nitrogen. Data were collected at a Rigaku FRE Superbright using an RAXIS-VI detector. Indexing and integration was carried out using XDS51,52 and scaling was performed with SCALA53. Initial phases were calculated by molecular replacement with PHASER54 using an ensemble of known bromodomain models (PDB IDs 2OSS, 2OUO, 2GRC, 2OO1, 3DAI, 3D7C, 3DWY). Initial models were built by ARP/wARP55 and building was completed manually with COOT56. Refinement was carried out in REFMAC557. Thermal motions were analyzed using TLSMD58 and hydrogen atoms were included in late refinement cycles. Data collection and refinement statistics can be found in Supplementary Table 2. The models and structure factors have been deposited with PDB accession codes: 4OGJ (BRD4(1)/TG-101348) and 4OGI (BRD4(1)/ BI-2536).
Fluorescent recovery after photo-bleaching (FRAP)
FRAP studies were performed using a protocol modified from previous studies29,59. In brief, U2OS cells were transfected (Lipofectamine, Invitrogen, Carlsbad, CA) with mammalian overexpression constructs encoding GFP chimeras with full-length BRD4 (accession: NP_490597.1). SAHA was added after 4 h post transfection and compounds as indicated 16 h post transfection. The FRAP and imaging system consisted of a Zeiss LSM 710 scanhead (Zeiss GmbH, Jena, Germany) coupled to an inverted Zeiss Axio Observer. Z1 microscope equipped with a high-numerical-aperture (N. A. 1.3) 40× oil immersion objective (Zeiss GmbH, Jena, Germany) equipped with a heated chamber set to 37 °C. FRAP and GFP fluorescence imaging were carried out with an argon-ion laser (488 nm) and with a piezomultiplier tube (PMT) detector set to detect fluorescence between 500–550 nm. A 5 μm2 region of a GFP-positive nucleus was selected, and after 5 prescans, the region was bleached. A time-lapse series was then taken to record GFP recovery using 1% of the power used for bleaching with an interval time of ~0.25 s. The image datasets and fluorescence recovery data were exported from ZEN 2010, the microscope control software, into Prism 6. The average intensity at each imaging time point was measured for three regions of interest: the bleached region (It), the total cell nucleus (Tt), and a random region outside of the cell for background subtraction (BG). The relative fluorescence signal in the bleached region was calculated for each time point t, with the following equation:
The baseline was normalized to zero and the prebleach to 1. Half times of recovery were calculated from the individual curves and presented as the mean. P-values were calculated using unpaired t-test.
Western blot analysis of cellular c-Myc levels
MM.1S multiple myeloma cells were cultivated according to ATCC guidelines in RPMI-1640 medium containing 10% fetal bovine serum at 37 °C with 5% CO2. Cells were treated with compounds at the indicated concentrations (0.1% DMSO, final) for 6 h in complete medium at a density of 3 ×105 cells/ml at 37 °C. Treated cells were then harvested by centrifugation, briefly washed with ice-cold PBS, and extracted using RIPA buffer (Thermo Scientific, West Palm Beach, FL) supplemented with 10 mM DTT. Protease inhibitors (Roche, Indianapolis, IN), phosphatase inhibitors (EMD Millipore, Billerica, MA), and benzonase (Sigma-Aldrich, St. Louis, MO) were added at concentrations suggested by the manufacturers. Total protein concentrations were measured using the Bradford assay (Bio-Rad, Hercules, CA). Electrophoresis and western blot analyses were performed according to standard procedures. Duplicate gel blots of identical cell extract samples were processed in parallel using c-Myc or GAPDH antibodies (Cell Signaling, Danvers, MA, catalogue numbers 9402 and 2118, respectively). A similar protocol was followed for measuring c-Myc levels in Molm14 cells.
Cellular proliferation assays
MV4-11 proliferation assays (72 h in the presence of test compound) were performed as described60. MM.1S multiple myeloma cells were cultivated in RPMI-1640 medium containing 10% FBS at 37 °C with 5% CO2. Cells were grown in T75 flasks (3 × 106 cells/flask at a concentration of 2.5 × 105 cells/ml), and the indicated compounds in DMSO (0.1% DMSO, final) were added immediately. Cells were incubated for 72 h at 37 °C with 5% CO2. After the incubation period, cells were counted by trypan blue exclusion using a hemocytometer. Log phase Molm14 cells were plated (20,000 cells/well) in 100 μl of RPMI + 0.5% FCS in each well of a 96-well plate at the indicated concentration of JQ1 in triplicate. Cells were cultured for 72 h and proliferation was assessed using Cell Titer Glo reagent (Promega, Madison, WI) according to manufacturer’s recommendations using a SpectraMax M3 microplate reader and SpectraMax Pro Software (Molecular Devices, Sunnyvale, CA).
BioMAP primary human cell disease model assay systems
All BioMAP assays were performed as previously described40,41, and further information is available at http://bioseekinc.com/.
Deposition of crystallographic data to the PDB database
The models and structure factors have been deposited with PDB accession codes: 4OGJ (BRD4(1)/TG-101348) and 4OGI (BRD4(1)/ BI-2536).
Supplementary Material
Acknowledgments
S.M., S.P., P.F., C.W., O.F., S.M., and S.K. are grateful for support by the SGC, a registered charity (number 1097737) that receives funds from the Canadian Institutes for Health Research, the Canada Foundation for Innovation, Genome Canada, GlaxoSmithKline, Pfizer, Eli Lilly, Takeda, AbbVie, the Novartis Research Foundation, the Ontario Ministry of Research and Innovation and the Wellcome Trust (092809/Z/10/Z). P.F. and S.P. are supported by a Wellcome Trust Career-Development Fellowship (095751/Z/11/Z). This work was supported in part by a grant from the NCI (1R01 CA166616-01) to N.P.S. and NIH National Cancer Institute T32CA108462-07 to E.A.L. N.P.S. is a Leukemia & Lymphoma Scholar in Clinical Research. We thank P. Gallant, T. Wehrman, E.L. Berg, and P. Khanna for critically reading the manuscript and for valuable discussions, the KINOMEscan team for measuring inhibitor Kd values, the BioSeek team for performing BioMAP screens, and A. Rooks, R. Nepomuceno, and B. Belli for performing MV4-11 proliferation assays.
Footnotes
Author contributions P.C., J.P.H., G.P., O.F., S. Martin, and L.M.W. developed in vitro binding assays, executed experiments, and interpreted data. S.P. purified protein and prepared co-crystals. P.F. solved and interpreted co-crystal structures and prepared figures. P.C., C.W., and E.A.L. designed, executed, and interpreted cell-based assay experiments. A.O.M. designed and interpreted all BioMAP studies and prepared figures. D.K.T., S.K., N.P.S. A.O.M., and S. Müller directed the studies and interpreted data. D.K.T. and S.K. wrote the paper with assistance from co-authors.
Competing financial interests D.K.T, P.C., A.O.M., J.P.H., L.M.W, and G.P. are employees of Discoverx Corporation.
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Haber DA, Gray NS, Baselga J. The evolving war on cancer. Cell. 2011;145:19–24. doi: 10.1016/j.cell.2011.03.026. [DOI] [PubMed] [Google Scholar]
- 3.Trusolino L, Bertotti A. Compensatory pathways in oncogenic kinase signaling and resistance to targeted therapies: six degrees of separation. Cancer Discov. 2012;2:876–880. doi: 10.1158/2159-8290.CD-12-0400. [DOI] [PubMed] [Google Scholar]
- 4.Davis MI, et al. Comprehensive analysis of kinase inhibitor selectivity. Nat Biotechnol. 2011;29:1046–1051. doi: 10.1038/nbt.1990. [DOI] [PubMed] [Google Scholar]
- 5.Flaherty KT, et al. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N Engl J Med. 2012;367:1694–1703. doi: 10.1056/NEJMoa1210093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corcoran RB, et al. EGFR-mediated re-activation of MAPK signaling contributes to insensitivity of BRAF mutant colorectal cancers to RAF inhibition with vemurafenib. Cancer Discov. 2012;2:227–235. doi: 10.1158/2159-8290.CD-11-0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Filippakopoulos P, et al. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell. 2012;149:214–231. doi: 10.1016/j.cell.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muller S, Filippakopoulos P, Knapp S. Bromodomains as therapeutic targets. Expert Rev Mol Med. 2011;13:e29. doi: 10.1017/S1462399411001992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nicodeme E, et al. Suppression of inflammation by a synthetic histone mimic. Nature. 2010;468:1119–1123. doi: 10.1038/nature09589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dawson MA, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011;478:529–533. doi: 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Picaud S, et al. PFI-1, a highly selective protein interaction inhibitor, targeting BET Bromodomains. Cancer Res. 2013;73:3336–3346. doi: 10.1158/0008-5472.CAN-12-3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delmore JE, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zuber J, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lockwood WW, Zejnullahu K, Bradner JE, Varmus H. Sensitivity of human lung adenocarcinoma cell lines to targeted inhibition of BET epigenetic signaling proteins. Proc Natl Acad Sci U S A. 2012;109:19408–19413. doi: 10.1073/pnas.1216363109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng Z, et al. Inhibition of BET bromodomain targets genetically diverse glioblastoma. Clin Cancer Res. 2013;19:1748–1759. doi: 10.1158/1078-0432.CCR-12-3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mertz JA, et al. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc Natl Acad Sci U S A. 2011;108:16669–16674. doi: 10.1073/pnas.1108190108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith CC, et al. Validation of ITD mutations in FLT3 as a therapeutic target in human acute myeloid leukaemia. Nature. 2012;485:260–263. doi: 10.1038/nature11016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li J, et al. INCB16562, a JAK1/2 selective inhibitor, is efficacious against multiple myeloma cells and reverses the protective effects of cytokine and stromal cell support. Neoplasia. 2010;12:28–38. doi: 10.1593/neo.91192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kyttaris VC. Kinase inhibitors: a new class of antirheumatic drugs. Drug Des Devel Ther. 2012;6:245–250. doi: 10.2147/DDDT.S25426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sapkota GP, et al. BI-D1870 is a specific inhibitor of the p90 RSK (ribosomal S6 kinase) isoforms in vitro and in vivo. Biochem J. 2007;401:29–38. doi: 10.1042/BJ20061088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hewitt L, et al. Sustained Mps1 activity is required in mitosis to recruit O-Mad2 to the Mad1-C-Mad2 core complex. J Cell Biol. 2010;190:25–34. doi: 10.1083/jcb.201002133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tong Y, et al. Isoxazolo[3,4-b]quinoline-3,4(1H,9H)-diones as unique, potent and selective inhibitors for Pim-1 and Pim-2 kinases: chemistry, biological activities, and molecular modeling. Bioorg Med Chem Lett. 2008;18:5206–5208. doi: 10.1016/j.bmcl.2008.08.079. [DOI] [PubMed] [Google Scholar]
- 23.Han S, et al. Structural characterization of proline-rich tyrosine kinase 2 (PYK2) reveals a unique (DFG-out) conformation and enables inhibitor design. J Biol Chem. 2009;284:13193–13201. doi: 10.1074/jbc.M809038200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weigelt B, Warne PH, Lambros MB, Reis-Filho JS, Downward J. PI3K pathway dependencies in endometrioid endometrial cancer cell lines. Clin Cancer Res. 2013;19:3533–3544. doi: 10.1158/1078-0432.CCR-12-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cai ZW, et al. Discovery of brivanib alaninate ((S)-((R)-1-(4-(4-fluoro-2-methyl-1H-indol-5-yloxy)-5-methylpyrrolo[2,1-f][1,2,4] triazin-6-yloxy)propan-2-yl)2-aminopropanoate), a novel prodrug of dual vascular endothelial growth factor receptor-2 and fibroblast growth factor receptor-1 kinase inhibitor (BMS-540215) J Med Chem. 2008;51:1976–1980. doi: 10.1021/jm7013309. [DOI] [PubMed] [Google Scholar]
- 26.Pardanani A, et al. Safety and efficacy of TG101348, a selective JAK2 inhibitor, in myelofibrosis. J Clin Oncol. 2011;29:789–796. doi: 10.1200/JCO.2010.32.8021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steegmaier M, et al. BI 2536, a potent and selective inhibitor of polo-like kinase 1, inhibits tumor growth in vivo. Curr Biol. 2007;17:316–322. doi: 10.1016/j.cub.2006.12.037. [DOI] [PubMed] [Google Scholar]
- 28.Cuenda A, et al. SB 203580 is a specific inhibitor of a MAP kinase homologue which is stimulated by cellular stresses and interleukin-1. FEBS Lett. 1995;364:229–233. doi: 10.1016/0014-5793(95)00357-f. [DOI] [PubMed] [Google Scholar]
- 29.Filippakopoulos P, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fedorov O, Niesen FH, Knapp S. Kinase inhibitor selectivity profiling using differential scanning fluorimetry. Methods Mol Biol. 2012;795:109–118. doi: 10.1007/978-1-61779-337-0_7. [DOI] [PubMed] [Google Scholar]
- 31.Ramakrishnan V, et al. TG101209, a novel JAK2 inhibitor, has significant in vitro activity in multiple myeloma and displays preferential cytotoxicity for CD45+ myeloma cells. Am J Hematol. 2010;85:675–686. doi: 10.1002/ajh.21785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kothe M, et al. Selectivity-determining residues in Plk1. Chem Biol Drug Des. 2007;70:540–546. doi: 10.1111/j.1747-0285.2007.00594.x. [DOI] [PubMed] [Google Scholar]
- 33.Rudolph D, et al. BI 6727, a Polo-like kinase inhibitor with improved pharmacokinetic profile and broad antitumor activity. Clin Cancer Res. 2009;15:3094–3102. doi: 10.1158/1078-0432.CCR-08-2445. [DOI] [PubMed] [Google Scholar]
- 34.Philpott M, et al. Bromodomain-peptide displacement assays for interactome mapping and inhibitor discovery. Mol Biosyst. 2011;7:2899–2908. doi: 10.1039/c1mb05099k. [DOI] [PubMed] [Google Scholar]
- 35.Picaud S, et al. RVX-208, an inhibitor of BET transcriptional regulators with selectivity for the second bromodomain. Proc Natl Acad Sci U S A. 2013 doi: 10.1073/pnas.1310658110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simard JR, et al. Development of a fluorescent-tagged kinase assay system for the detection and characterization of allosteric kinase inhibitors. J Am Chem Soc. 2009;131:13286–13296. doi: 10.1021/ja902010p. [DOI] [PubMed] [Google Scholar]
- 37.Siu M, et al. 2-Amino-[1,2,4]triazolo[1,5-a]pyridines as JAK2 inhibitors. Bioorg Med Chem Lett. 2013;23:5014–5021. doi: 10.1016/j.bmcl.2013.06.008. [DOI] [PubMed] [Google Scholar]
- 38.Fedorov O, et al. Specific CLK inhibitors from a novel chemotype for regulation of alternative splicing. Chem Biol. 2011;18:67–76. doi: 10.1016/j.chembiol.2010.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith CC, et al. Activity of ponatinib against clinically-relevant AC220-resistant kinase domain mutants of FLT3-ITD. Blood. 2013;121:3165–3171. doi: 10.1182/blood-2012-07-442871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berg EL, et al. Chemical target and pathway toxicity mechanisms defined in primary human cell systems. J Pharmacol Toxicol Methods. 2010;61:3–15. doi: 10.1016/j.vascn.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 41.Bergamini G, et al. A selective inhibitor reveals PI3Kgamma dependence of T(H)17 cell differentiation. Nat Chem Biol. 2012;8:576–582. doi: 10.1038/nchembio.957. [DOI] [PubMed] [Google Scholar]
- 42.Xu D, et al. RN486, a selective Bruton’s tyrosine kinase inhibitor, abrogates immune hypersensitivity responses and arthritis in rodents. J Pharmacol Exp, Ther. 2012;341:90–103. doi: 10.1124/jpet.111.187740. [DOI] [PubMed] [Google Scholar]
- 43.Ott CJ, et al. BET bromodomain inhibition targets both c-Myc and IL7R in high-risk acute lymphoblastic leukemia. Blood. 2012;120:2843–2852. doi: 10.1182/blood-2012-02-413021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee JC, et al. A protein kinase involved in the regulation of inflammatory cytokine biosynthesis. Nature. 1994;372:739–746. doi: 10.1038/372739a0. [DOI] [PubMed] [Google Scholar]
- 45.del Barco Barrantes I, Nebreda AR. Roles of p38 MAPKs in invasion and metastasis. Biochem Soc Trans. 2012;40:79–84. doi: 10.1042/BST20110676. [DOI] [PubMed] [Google Scholar]
- 46.Cuenda A, Alessi DR. Use of kinase inhibitors to dissect signaling pathways. Methods Mol Biol. 2000;99:161–175. doi: 10.1385/1-59259-054-3:161. [DOI] [PubMed] [Google Scholar]
- 47.Martin MP, Olesen SH, Georg GI, Schonbrunn E. Cyclin-dependent kinase inhibitor dinaciclib interacts with the acetyl-lysine recognition site of bromodomains. ACS Chem Biol. 2013;8:2360–2365. doi: 10.1021/cb4003283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vidler LR, et al. Discovery of Novel Small-Molecule Inhibitors of BRD4 Using Structure-Based Virtual Screening. J Med Chem. 2013 doi: 10.1021/jm4011302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dittmann A, et al. The commonly used PI3-Kinase Probe LY294002 is an inhibitor of BET bromodomains. ACS Chem Biol. 2014 doi: 10.1021/cb400789e. [DOI] [PubMed] [Google Scholar]
- 50.Wiseman T, Williston S, Brandts JF, Lin LN. Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal Biochem. 1989;179:131–137. doi: 10.1016/0003-2697(89)90213-3. [DOI] [PubMed] [Google Scholar]
- 51.Kabsch W. Evaluation of single-crystal X-ray diffraction data from a position-sensitive detector. J App Cryst. 1988;21:916–924. [Google Scholar]
- 52.Kabsch W. Automatic indexing of rotation diffraction patterns. J App Cryst. 1988;21:67–71. [Google Scholar]
- 53.SCALA - scale together multiple observations of reflections v. 3.3.0. MRC Laboratory of Molecular Biology; Cambridge: 2007. [Google Scholar]
- 54.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallographica Section D Biological Crystallography. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 55.Perrakis A, Morris R, Lamzin VS. Automated protein model building combined with iterative structure refinement. Nat Struct Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 56.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallographica Section D Biological Crystallography. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 57.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallographica Section D Biological Crystallography. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 58.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallographica Section D Biological Crystallography. 2006;62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 59.Kedersha N, Tisdale S, Hickman T, Anderson P. Real-time and quantitative imaging of mammalian stress granules and processing bodies. Methods Enzymol. 2008;448:521–552. doi: 10.1016/S0076-6879(08)02626-8. [DOI] [PubMed] [Google Scholar]
- 60.Gunawardane RN, et al. Transient exposure to quizartinib mediates sustained inhibition of FLT3 signaling while specifically inducing apoptosis in FLT3-activated leukemia cells. Mol Cancer Ther. 2013;12:438–447. doi: 10.1158/1535-7163.MCT-12-0305. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.