Abstract
Random peptide libraries are displayed on filamentous bacteriophage as fusions to either the minor coat protein, pIII, or the major coat protein, pVIII. We have devised a means of isolating the peptide displayed on a phage clone by transferring it to the N-terminus of the maltose-binding protein (MBP) of Escherichia coli encoded by malE. Transfer of a peptide sequence to monomeric MBP eliminates phage-encoded amino acids downstream of the insert peptide as well as avidity effects caused by multivalent display on phage. Peptide:MBP fusions are also easily affinity purified on amylose columns. The pMal-p2 vector was engineered to accept phage DNA encoding pIII- and pVIII-displayed peptides fused to their respective leader sequences. Both types of leader sequence were shown to target the peptide:MBP fusions to the periplasm of E. coli. A streamlined procedure for transferring peptides to MBP was applied to clones that had been isolated from a panel of pVIII-displayed peptide libraries by screening with an HIV-1-specific monoclonal antibody (Ab). By enzyme-linked immunosorbent assay, the Ab bound each of the peptide:MBP fusions and required the presence of a disulfide bridge within each peptide. Some of the peptide:MBP fusions were also analyzed using surface plasmon resonance. Thus, our study shows the value of malE fusion vectors in characterizing phage-displayed peptides.
Phage display continues to grow as a technology for selecting peptides that bind to a variety of biomolecules including (Abs),2 enzymes, receptors, DNA, and RNA (for reviews, see Refs. 1 and 2). The two phage proteins used for peptide display are the minor coat protein, pIII, and the major coat protein, pVIII. pIII is present in four or five copies that are closely clustered at one tip of the virion, whereas pVIII is present in thousands of copies that are arranged in a fish-scale-like pattern forming the body of the virion. Thus, multivalency can complicate the determination of the affinity of a selecting molecule for its cognate peptide displayed on either pIII or pVIII. In pIII display, avidity effects are produced by the close clustering of peptides. In pVIII display, the potential avidity effects vary between clones, as there is variation in the level of incorporation of recombinant peptide:pVIII fusions into the hybrid virion coat; this appears to be governed by the rate of processing of the pro-coat (3). Furthermore, in both cases there is the potential for contribution of the phage-coat proteins to peptide affinity, either by sequences flanking a peptide or by conformational stabilization of the peptide induced by the coat milieu. By transferring peptides from phage to maltose-binding protein (MBP), the binding of an antibody (Ab) to a peptide can be measured in the absence of potential “phage effects” (i.e., avidity and/or conformational effects). Although our approach of genetically transferring phage-displayed peptides to MBP can be applied to virtually any phage display system, in the present study it was applied to the pVIII-display libraries of Bonnycastle et al. (4), which were derived from the vector f88.4 (5), as well as the “Ph.D.” pIII-display libraries of New England Biolabs, Inc. (NEB).
MBP provides a useful, monovalent scaffold for peptide display for several reasons. First, it is easily purified by chromatography on amylose columns. Second, like the phage-coat proteins, MBP is secreted, allowing disulfide formation to occur in the periplasmic space. Third, MBP has no cysteines that could form disulfide bonds with cysteines within the fused peptide. Finally, with MBP fusions, there is less concern about peptide solubility, since the peptide is already “conjugated” to soluble MBP. This allows phage-derived peptides to be transferred to MBP and tested for activity in the absence of flanking phage sequence. This is a useful step before designing peptides for chemical synthesis, as we often observe significant variations in affinity on moving from a fusion protein to a synthetic peptide. Moreover, synthetic peptides may not be required for several applications, such as immunization (see Discussion); MBP offers an alternative means of testing peptide affinity.
The commercially available vectors pMal-p2 and pMal-c2 (NEB) are designed for fusions to the C-terminus of MBP (6), and fusions of short peptides to the C-terminus of MBP have been described for peptides derived from lac-repressor-fusion libraries (7). Since the peptides in phage-display libraries are N-terminally displayed, we engineered pMal-p2 to produce N-terminal fusions. A previous study showed that not only could single-chain Fv fragments be fused to the N-terminus of MBP, but in some cases, N-terminal display was superior to C-terminal display because it resulted in fewer incomplete translational products (8). Our initial studies demonstrated that both the pVIII and pIII leader sequences were capable of transporting peptide:MBP fusions to the periplasm of Escherichia coli for subsequent signal peptide cleavage. Encouraged by these results, we designed a streamlined strategy for transferring the peptides from phage clones that had been affinity selected from a panel of pVIII-displayed peptide libraries. The monoclonal antibody (MAb) used to screen the phage libraries, loop2, binds to a conserved sequence within the V3 loop of HIV-1 gpl20 (9). Four loop2-selected peptides were chosen for fusion to MBP. Enzyme-linked immunosorbent assays (ELISAs) showed that the binding of Ab to the phage-derived peptides was largely retained with MBP display. Surface plasmon resonance (SPR) analysis (10) resulted in well-behaved dose–response curves for three of the peptide:MBP fusions probed with MAb loop2.
MATERIALS AND METHODS
Reagents
All reagents, unless otherwise specified, were from NEB. Construction of pVIII-display peptide libraries and procedures for selection, amplification, and purification of phage clones are described in Bonnycastle et al. (4). Oligonucleotide sequences are as follows: No. 1,5′-TATGAAAAA(ATT)3CGCAATTCC-TTTAGTGGTACCTTTCTATTCTCACTCGGCCGA-3′; No. 2, 5′-TATCGGCCGAGTGAGAATAGAAAGGT-ACCACTAAAGGAATTGCG(AAT)3TTTTTCA-3′; No. 3, 5′-TTCCCCGTCAAGCTCTAAATCG-3′; No. 4, 5′-GCGGGCTGGGTATCTGAGTTC-3′; and pMal sequencing primer, 5′-ACCGTTATAGCCTTTATCGC-3′. IgG1κ and Fab forms of loop2 are as described (9), as is the cyclic loop2-specific MN peptide,  (11); the peptide was used as a conjugate to bovine serum albumin (BSA), MN-BSA. The MAb Pf2A.10 was a kind gift of Dr. R. Wirtz (WRAIR, Washington, DC; SmithKline Beecham and New York University). Polyclonal rabbit anti-phage Ab was prepared as described (4). The vector pPR1068 was a gift of P. Riggs. The E. coli strain AR182 [araD139 Δ(araABC-leu)7696 thr ΔlacX74 galU galK hsdR mcrB rpsL-(strA) thi prlA4 zhc::Tn10kan] was kindly provided by P. J. Schatz (Affymax Research Institute, Palo Alto, CA). DH5αF′ [F′ Φ80dlacZΔM15 Δ(lacZYA-argF)-U169 deoR recAl endAl hsdR17 (rK−, mK+) supE44 λ-−
thi-1 gyrA96 relA1] was kindly donated by R. Donahue (GibcoBRL, Life Technologies, Inc., Gaithersburg, MD). NM522 [F′ lacIq Δ(lacZ)M15 proA+B+ /supEthi Δ(lac-proAB) Δ(hsdMS-mcrB)5 (rK−mK− McrBC−)] was from NEB. G. P. Smith (University of Missouri-Columbia) kindly supplied the phage vector f88.4 (5).
(11); the peptide was used as a conjugate to bovine serum albumin (BSA), MN-BSA. The MAb Pf2A.10 was a kind gift of Dr. R. Wirtz (WRAIR, Washington, DC; SmithKline Beecham and New York University). Polyclonal rabbit anti-phage Ab was prepared as described (4). The vector pPR1068 was a gift of P. Riggs. The E. coli strain AR182 [araD139 Δ(araABC-leu)7696 thr ΔlacX74 galU galK hsdR mcrB rpsL-(strA) thi prlA4 zhc::Tn10kan] was kindly provided by P. J. Schatz (Affymax Research Institute, Palo Alto, CA). DH5αF′ [F′ Φ80dlacZΔM15 Δ(lacZYA-argF)-U169 deoR recAl endAl hsdR17 (rK−, mK+) supE44 λ-−
thi-1 gyrA96 relA1] was kindly donated by R. Donahue (GibcoBRL, Life Technologies, Inc., Gaithersburg, MD). NM522 [F′ lacIq Δ(lacZ)M15 proA+B+ /supEthi Δ(lac-proAB) Δ(hsdMS-mcrB)5 (rK−mK− McrBC−)] was from NEB. G. P. Smith (University of Missouri-Columbia) kindly supplied the phage vector f88.4 (5).
Preparation of Neutravidin and Biotinylated Horseradish Peroxidase Complex
Complexes of neutravidin (Pierce Chemical Co., Rockford, IL) and biotinylated horseradish peroxidase were prepared as a detection reagent for biotinylated Abs. Briefly, 500 µg of horseradish peroxidase (Cat. P-6782; Sigma Chemical Co., St. Louis, MO) was dissolved in 250 µl phosphate-buffered saline (PBS; 137 mM NaCl, 2.7 mM KC1, 5.37 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.4; Ref. 12) and divided into five aliquots. To each 50-µl aliquot were added 11 µl 1 M NaHCO3 and 50 µl 0.5 mg/ml NHS-LC Biotin (Pierce) in 2 mM sodium acetate, pH 6.0. The solution was incubated for 2 h at room temperature, and then 500 µl 1 M ethanolamine was added. The mixture was incubated for another 2 h at room temperature, and then 20 µl of Tris-buffered saline (TBS; 50 mM Tris-HCl, pH 7.5, 150 mM NaCl) containing 1 mg/ml BSA (Fraction V, Sigma) was added. The mixture was transferred to a Centricon-30 device (Amicon, Inc., Beverly, MA) and washed twice with TBS and then once with TBS containing 0.02% NaN3. The optimal ratio of neutravidin to biotinylated horseradish peroxidase was determined by ELISA to be 50 ng:12 ng per microwell. This was prepared by adding 1 µl 1.4 mg/ml stock of neutravidin and 1 µl of a 0.33 mg/ml stock of biotinylated horse-radish peroxidase to 1 ml of TBS containing 0.1% Tween 20 (ICN Biomedicals, Inc., Aurora, OH).
Vector Construction
pMal-pIII
This vector was derived from pPR1068 (P. Riggs, NEB), which is identical to the MBP fusion vector pMal-p2, except for the following changes: (i) the region encoding the MBP leader sequence was deleted; (ii) the unique NdeI site was removed; (iii) a new, unique NdeI site (CATATG), part of which encodes the initiator methionine of the MBP leader sequence, was added; and (iv) the unique NcoI site and one of two AvaI (4566) sites were removed. To construct pMal-pIII, oligonucleotides 1 and 2 were annealed to form a duplex encoding the pIII leader sequence of the Ph.D. vector and the unique Acc65I/KpnI and EagI cloning sites. This duplex was then spliced into the NdeI site of pPR1068 (Fig. 1A).
Figure 1.
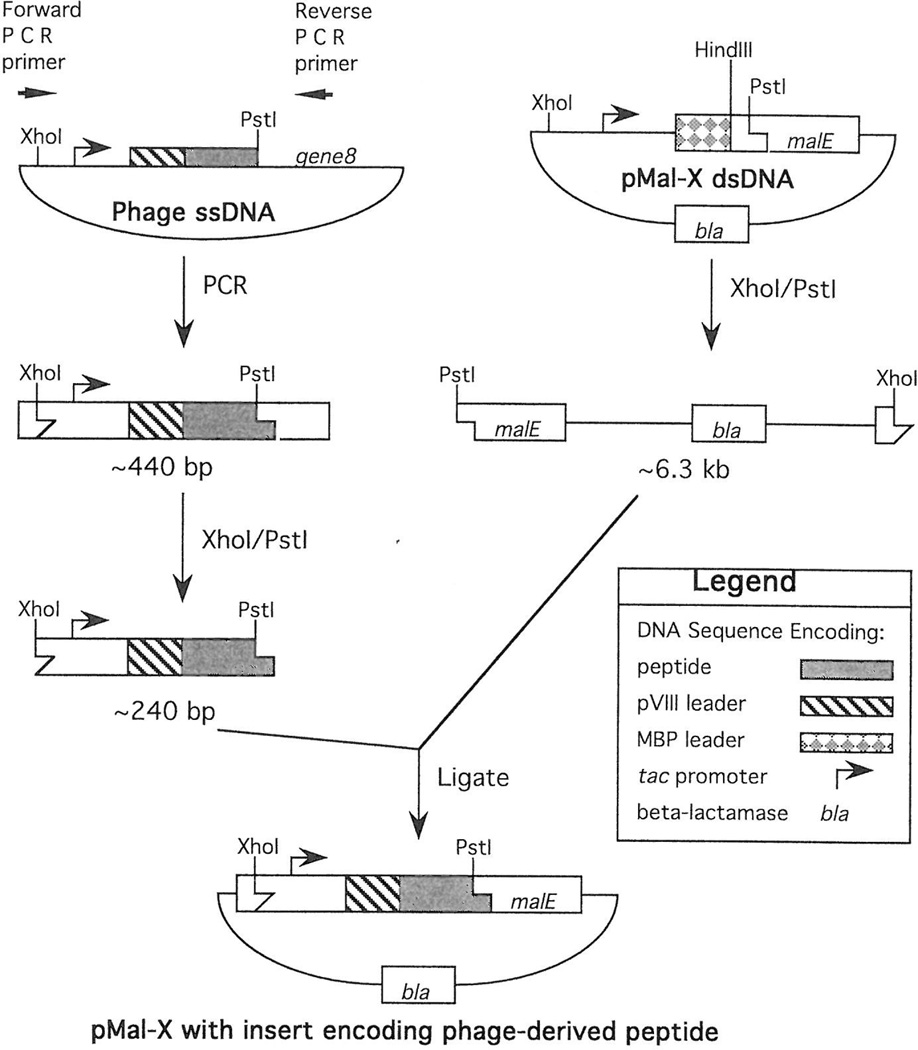
Nucleotide and amino acid sequences for constructs described in this paper. (A) pMal-pIII vector with pIII leader sequence, including KpnI(Acc65I) and EagI sites. (B) Ph.D. phage display library with random peptide sequence [Xn, (NNK)n; K = G or T] followed by a GGGS linker, which is inserted between the pIII leader sequence and the mature protein. (C) pMal-X and pMal-KHP vectors with MBP:pVIII chimeric leader sequence with pVIII-derived residues in bold. (D) Phage-display vector f88.4 with pVIII leader sequence. (E) pVIII-displayed peptide library [Xn, (NNK)n] in f88.4. Leader peptidase cleavage sites (^) are indicated.
pMal-X
This construct was also derived from the vector pMal-p2 with the following changes: (i) the HindIII and PstI sites in the polylinker cloning region (located at the 3′ end of the MBP coding region) were replaced with a unique KpnI site; (ii) a unique HindIII site, followed by a sequence encoding the influenza hemagglutinin peptide tag YDVPDYA (13, 14), were added immediately 3′ to the region encoding the MBP leader (see Fig. 1C); (iii) a unique PstI site was added 3′ to the region encoding the hemagglutinin tag and 5′ to that encoding mature MBP; and (iv) a unique XhoI site was added 5′ to the tac promoter and 3′ to lacIq (1212 G, 1213 A). The pMal-KHP construct is identical to pMal-X, except it lacks the unique XhoI site (1212 A, 1213 T). The nucleotide sequences of the pMal-X and pMal-pIII vectors have been deposited in GENBANK (Accession Nos. AF031813 and AF031088, respectively).
Engineering pMal-X to Express Peptide:MBP Fusions
Purified pMal-X vector DNA was digested with XhoI and PstI according to the manufacturer’s instructions, and the resulting DNA fragments were separated on a 0.8% agarose gel in TAE buffer (12). A 6.3-kb band of DNA encoding the tac promoter, leader sequence, and hemagglutinin tag was extracted and purified with a GeneClean kit (BIO101, Inc., Vista, CA) following the manufacturer’s instructions. Phage clones were purified by precipitation in polyethylene glycol (4) and the polymerase chain reaction (PCR) was used to amplify a segment of viral DNA encoding the peptide insert (Fig. 2). The forward PCR primer (No. 3) anneals immediately upstream of the XhoI site, which is 5′ to the tac promoter of f88.4. The reverse PCR primer (No. 4) anneals 3′ to the PstI site within the synthetic gene 8. The resulting PCR product encodes a unique XhoI site, the phage tac promoter, the pVIII leader sequence, the phage-displayed peptide, and a unique PstI site.
Figure 2.
PCR-subcloning strategy for transferring pVIII-displayed peptides from phage to MBP fusion vector pMal-X (see text for details).
The PCR was performed in a total volume of 100 µl and consisted of ThermoPol reaction buffer containing 200 µM each dNTP, 300 nM each forward and reverse primer, 1 U Vent DNA polymerase, 2 mM MgSO4, and 83 pM phage (5 × 109 particles). Samples were pre-soaked at 95°C for 120 s and then cycled 30 times with the following parameters: 95°C for 60 s, 56°C for 30 s, and 72°C for 25 s. Amplified products were purified using a GeneClean kit (BIO 101) and then digested with XhoI and PstI. DNA fragments from the digest were separated on a 6% polyacrylamide gel in TBE buffer (12), and bands of the correct size (~240 bp) were excised. The gel slices were crushed and soaked in sterile water at 37°C for 2 h and then the DNA was eluted from the gel pieces by centrifugation through a Micropure Separator (0.45 µm, Amicon). The eluates were desalted on Ultrafree-MC filters (30 000 NMWL; Millipore Corp., Bedford, MA) prior to their addition to ligation reactions containing the 6.3-kb fragment from the vector pMal-X.
Approximately 20 ng (0.3 pmol) of purified insert was ligated to 30 ng (15 fmol) of the purified pMal-X fragment in T4 DNA-ligase buffer (GibcoBRL) containing 0.25 units of T4 DNA ligase (GibcoBRL; total volume, 7 µl) for 16 h at 15°C. CaCl2-competent DH5αF′ cells were transformed with the ligation products using heat shock (12), and ampicillin-resistant colonies were picked and amplified in LB medium containing 100 µg/ml ampicillin (15). To verify the presence of the insert, plasmid DNA was sequenced by the University of Calgary Core DNA Services (Alberta) using the pMal sequencing primer.
Engineering pMal-pIII to Express Peptide:MBP Fusions
The Ph.D.-7 heptapeptide library was screened with the Erk1/2 MAP kinase (NEB), following the direct-coating method described in the manufacturer’s instructions. After washing, bound phage were incubated for 30 min with the substrate peptide TGPLSPGPF (0.1 mM) in TBS; this “competitive elution” specifically inhibited phage from rebinding the substrate-binding site. Two ELISA-positive phage clones were selected from this screening, and their displayed peptides were transferred to MBP via the vector pMal-pIII. Recombinant constructs were prepared by annealing the −96 gIII sequencing primer (Cat. No. 1259, NEB) to single-stranded phage DNA, extending through the peptide-encoding region with Klenow fragment and dNTPs, and digesting the resulting duplex DNA with Acc65I and EagI. (The isoschizomer Acc65I is used in place of KpnI to facilitate simultaneous double digestion with EagI.) Fragments encoding the peptide inserts (53 bp) were purified from an 8%, nondenaturing, polyacrylamide gel as above, subcloned into pMal-pIII that had been digested with Acc65I and EagI and then ligated as above. The ligation products were used to transform E. coli NM522 cells, and the presence of the inserts was verified by DNA sequencing using the pUC/M13 reverse primer (Cat. No. 1233, NEB).
Peptide:MBP Fusion Protein Production and Purification
A single, fresh, ampicillin-resistant colony was used to prepare an overnight culture in LB medium containing 0.2% glucose, 100 µg/ml ampicillin, and 25 µg/ml kanamycin. Twenty milliliters of this culture was used to inoculate 2 L fresh LB medium containing the same antibiotics, and the culture was grown at 37°C with shaking at 250 rpm to OD600 ~0.5. Isopropylthio-β-d-galactoside (IPTG, GibcoBRL) was then added to a final concentration of 0.2 mM, and the culture was shaken at 25°C for another 2 h. The cells were harvested by centrifugation at 4000g for 20 min at 4°C. The periplasmic fraction was prepared by cold osmotic shock (16) according to the Protein Fusion and Purification System manual (NEB). Briefly, 600 ml of 30 mM Tris-HCl, 20% sucrose, pH 8.0, were added to the bacterial pellet, and then 0.5 M Na2EDTA, pH 8.0, was added to a final concentration of 1 mM. The pellet was resuspended by repeatedly pipetting the solution over the pellet and then by shaking the mixture at ~200 rpm for 5–10 min at room temperature. The suspension was centrifuged at 8000g for 20 min at 4°C and then the cells were osmotically shocked by resuspending the pellet in 400 ml ice-cold, 5 mM MgSO4, and vigorously rocking for 10 min at 4°C. The mixture was then centrifuged at 8000g for 20 min at 4°C. The supernatant (referred to as the cold osmotic-shock fluid) was stabilized by the addition of 8 ml 1 M Tris-HCl, pH 7.4, preserved with sodium azide (final concentration = 1 mM), and stored at −20°C.
Peptide:MBP fusions were purified by amylose affinity chromatography following the instructions provided in the Protein Fusion and Purification System manual (NEB). Briefly, the column was prepared by pouring amylose resin in a glass-wool plugged, 60-ml syringe (2.5 cm diameter 10–15 ml bed volume). The column was washed with 8 column volumes of column buffer (PBS, 1 mM EDTA, pH 7.4). The osmotic shock fluid was defrosted in a cold-tap-water bath, passed through a bottle-top filter (0.45 µm, 500 ml; Corning Glass Works, Corning, NY), and loaded onto the column at a flow rate of about 1 ml/min. The column was washed with 12 column volumes of column buffer. The fusion protein was eluted within the first 10 fractions with filter-sterilized column buffer containing 10 mM maltose (fraction size, 3 ml or one-fifth column volume); the fractions were monitored by UV absorbance at 280 nm. The protein-containing fractions were pooled, washed, and concentrated using a Centriprep-30 concentrator (30,000 MWCO, Amicon). The packed resin was regenerated with the following sequence of washes, which were carried out at room temperature: 3 column volumes deionized water, 3 volumes 0.1% SDS, 1 volume water, and 3 volumes column buffer. Columns were reused three to five times.
ELISA
One microgram of MBP fusion, anti-phage Ab, or MN-BSA (11) was diluted in 35 µl of 0.1 M NaHCO3, added to microwells (96 Well Easy Wash; Corning), and gently rocked overnight at 4°C. For all the washing steps, 200 µl TBS containing 0.1% Tween 20 (ICN) was dispensed by an EL-403 plate washer (Bio-Tek Instruments, Inc., Winooski, VT); the plate was shaken for 5 s on low setting and set to stand 5 s. The wells were washed three times and blocked for 1 h at 37°C with 200 µl TBSB [TBS containing 2% BSA (Fraction V, Sigma)]. Phage clones (5 × 109 particles/well in TBSB) were added to wells that had been coated with anti-phage Ab, whereas other wells remained blocked for 90 min at 4°C. Wells were washed four times, and “+dithiothreitol (DTT)” wells were treated with 35 µl TBS containing 15 mM DTT, whereas “–DTT” wells were treated with TBS alone for 30 min at 4°C. The wells were aspirated and 35 µl of the following Abs was immediately added: 30 nM biotinylated MAb loop2, 50 nM rabbit anti-phage Ab (4), or a 1:2500 dilution of rabbit anti-MBP Ab. The Abs were diluted in TBSB alone, or in TBSB containing 5 mM DTT, and incubated in the appropriate wells for 3 h at 4°C. The wells were washed five times, and 35 µl goat anti-rabbit IgG:horseradish-peroxidase conjugate (Pierce), diluted 1:800 in TBSB, was added to wells containing nonbiotinylated Ab; 35 µl of a complex of Neutravidin and biotinylated horseradish peroxidase (see above) diluted in TBS containing 0.1% Tween 20 (ICN) was added to wells containing biotinylated Ab. ABTS solution was prepared by mixing 3.07 ml 0.1 M citric acid and 1.93 ml 0.2 M Na2HPO4, adding 2 mg of 2,2′-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid) (Sigma), mixing again, and adding 5 µl 30% (w/w) H2O2 (Sigma). After incubating the plate 30 min at room temperature, the wells were washed seven times, and 35 µl of freshly-prepared ABTS solution was added. After 45 min, the absorbance at 405 and 450 nm was measured on an EL 312e Bio-Kinetics Reader (Bio-Tek). The results are reported as (A405–A490) × 1000.
Western Blotting
The Phototope-Star Western Blot Detection Kit (NEB) was used according to the manufacturer's instructions. Immobilon-P polyvinylidene fluoride membrane (Millipore) was used for blotting; rabbit anti-MBP Ab (1:10,000 dilution) and MAb loop2 (1:5000) were used as primary Abs, and alkaline phosphatase-conjugated anti-rabbit or anti-human IgG (1:1000) were used as secondary Abs. For multiple probings, membranes were stripped by incubation in 62.5 mM Tris-HCl, pH 6.6, 100 mM β-mercaptoethanol, 2% SDS at 50°C for 30 minutes and washed twice in PBS prior to blocking for the second probing.
Surface Plasmon Resonance Analysis
A BIAcore 1000 instrument (BIAcore Inc., Uppsala, Sweden) was used to determine the surface-binding affinities of the peptide:MBP fusions. The sensor chip (research grade CMS, BIAcore) was activated by injecting 40 µl of activation solution [50% 0.2 M N-ethyl-N′-(3-diethylaminopropyl)carbodiimide (BIAcore) and 50% N-hydroxysuccinimide (BIAcore)] at a flow rate of 5 µd/min. Peptide:MBP fusion protein was coupled to the surface by injection of 35 µl of 10 µg/ml sample in 10 mM sodium acetate, pH 4.0. The number of resonance units immobilized for each peptide:MBP fusion were: 1574 (C6), 1485 (D5), 2538 (G5), and 1945 (J5). The remaining activated sites on the chip were blocked by injecting 40 µl of 1 M ethanolamine, pH 8.5. Fab loop2 or MAb loop2 was injected in PBS, pH 7.4, at concentrations ranging from 200 nM to 1.2 µM, and at a flow rate of 10 µl/min during the association phase and 50 µl/min during the dissociation phase. The surface was regenerated by injection of 50 µl of 20 mM HC1 at a flow rate of 5 µl/min. The association and dissociation rates (kon and koff) were calculated assuming a one-to-one pseudo-first-order kinetic model using BIA evaluation Software Version 2.1 (BIAcore). Both kon and koff were determined by analyzing the curves obtained at four or five different concentrations of loop2 MAb or Fab.
RESULTS
Engineering pMal Vectors to Produce MBP Fusions to Peptides Derived from Phage-Display Libraries
The peptide:MBP vectors were designed for the transfer to the N-terminus of MBP of pIII- and pVIII-displayed peptides that had been selected from phage display libraries. In the Ph.D. and f88.4 systems, random peptides are expressed at the N-terminus of the mature pIII and pVIII coat proteins, respectively. Thus, these phage vectors were designed for a library-construction strategy that uses an upstream cloning site within the leader-peptide coding sequence and a downstream cloning site that is at or beyond the sequence encoding the signal peptidase cleavage site (Figs. 1B, 1D, and 1E). As a result, when DNA fragments encoding selected peptide sequences are transferred from a phage to a malE fusion vector via these cloning sites, residues from the C-terminal end of the leader sequence are transferred as well. This creates a problem, in that the malE vectors must have similar cloning sites within the region encoding the leader sequence. To circumvent this problem, we constructed vectors in which the MBP leader sequence was replaced with the pIII or pVIII leader sequence, or fragments thereof, and were thus engineered to contain the same cloning sites as the Ph.D. and f88.4 phage vectors, respectively (Figs. 1A and 1C).
MBP:Peptide Fusions Are Correctly Processed and Secreted via Foreign Leader Sequences
The pIII leader sequence
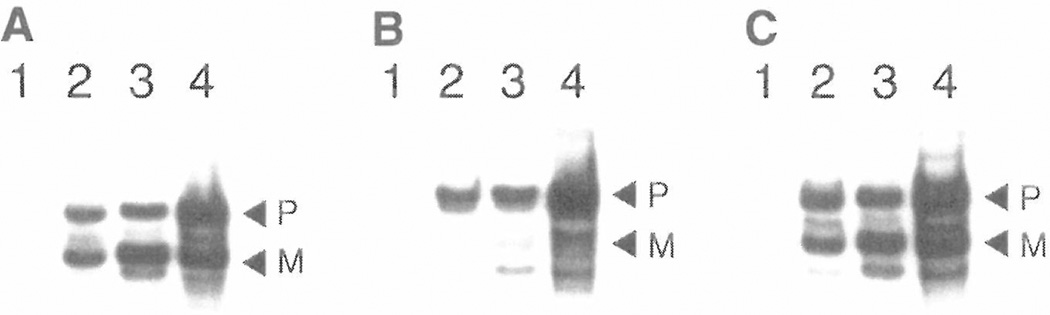
The pMal-pIII vector was designed for transferring to MBP peptide sequences from Ph.D. phage-display libraries. To test whether the pIII leader sequence can substitute for the native MBP leader sequence in directing the transport and processing of peptide:MBP fusions, peptides selected from a representative panning experiment were transferred to pMal-pIII, and expression and processing of the resulting peptide-MBP fusions were monitored on im-munoblots using anti-MBP Ab (Fig. 3). In this panning experiment, the target was the Erk1/2 MAP kinase, and peptides specific for its active site were selected. Two sequences that produced positive ELISA signals (data not shown) were transferred to pMal-pIII: N11 (FHKPLKR) and N15 (NPAHSPW). As shown in Fig. 3, the N15:MBP fusion (Fig. 3C) was exported and processed at levels that are comparable to the pMal-p2 control (Fig. 3A), in which MBP processing is directed by its own leader sequence. In contrast, the N11 peptide, which carries multiple positive charges, greatly inhibits proper processing of its MBP fusion (Fig. 3B), as would be expected for secY-dependent processing (Ref. 17; see Discussion).
Figure 3.
Western blots of expression of MBP (A) produced by pMal-p2, and Erk1/2 MAP kinase-binding peptides N11 (B) and N15 (C) fused to MBP produced by pMal-pIII. Blots were probed with rabbit anti-MBP polyclonal Ab. (Lane 1) Whole-cell extract harvested immediately prior to IPTG induction or (lane 2) four hours after IPTG induction. (Lane 3) Osmotic shock supernatant. (Lane 4) resuspended osmotic shock pellet. P, precursor protein containing a leader sequence; M, mature processed protein without leader sequence.
The pVIII and MBP. pVIII chimeric leader sequences
A peptide:MBP fusion was tested for correct processing and secretion via a chimeric leader sequence that is identical to the MBP leader sequence, except for two amino acid changes that are present near the C-terminus of the pVIII leader sequence. A well-characterized malarial epitope was chosen as a control peptide for fusion to the N-terminus of MBP. To engineer the fusion, purified replicative-form (RF) DNA from a phage clone bearing the peptide sequence NANPN-VDP(NANP)3GGPA (termed the NANP sequence) was cleaved with HindIII and PstI and ligated into similarly cleaved pMAL-KHP. Thus, the NANP sequence was inserted between the chimeric leader sequence and the N-terminus of mature, wild-type MBP (see Fig. 1C). The fusion, NANP:MBP, was produced and isolated from osmotic-shock fluid (see Materials and Methods). SDS-PAGE analysis of the osmotic-shock fluid revealed a band of correct size. This was confirmed to be NANP:MBP on Western blot analysis using the MAb Pf2A.10 (18). The expected sequence of the correctly processed NANP:MBP protein was also verified by N-terminal amino-acid sequencing. Thus, the NANP:MBP fusion was secreted and correctly processed via the chimeric leader peptide.
Previously, we had determined that the MBP leader peptide could substitute for the pVIII leader peptide in exporting the NANP:pVIII fusion protein to the periplasm and, thus, allowed its assembly into the phage coat (data not shown). To determine if the pVIII leader could conversely substitute for the MBP leader in exporting NANP:MBP, the analogous fusion having the full pVIII leader peptide would have to be produced. Thus, a plasmid was engineered to encode this fusion protein using the cloning strategy outlined in Fig. 2. SDS–PAGE and Western blot analysis of this fusion revealed that both leader sequences produce similar levels of NANP:MBP (data not shown).
Transferring pVIII-Displayed Peptides from Phage to MBP by PCR Subcloning
We sought to improve the cloning procedure due to inefficiencies in the method described above. As a first step, we chose to increase the size of the insert to include DNA encoding not only the insert peptide, but also the pVIII leader peptide and the phage-derived tac promoter; this DNA fragment of ~240 bp is easily gel purified before ligation into pMal-X. We chose to PCR-amplify the fragment from phage, rather than using RF DNA, for four reasons: (i) high yields of fragment could be obtained by PCR; (ii) RF DNA preparations have low yields due to relatively low copy numbers in E. coli (this is especially true for fd-tet-derived vectors); (iii) RF DNA is more difficult to prepare than viral DNA; and (iv) phage selected from libraries are usually stored as virions, which can be used directly in the PCR. Thus, the PCR-based cloning strategy, depicted in Fig. 2, circumvents several of the problems involved in the transfer of peptides to MBP.
MBP Display and Characterization of Phage-Derived Peptides Selected by the Human Anti-HIV-1 MAb loop2
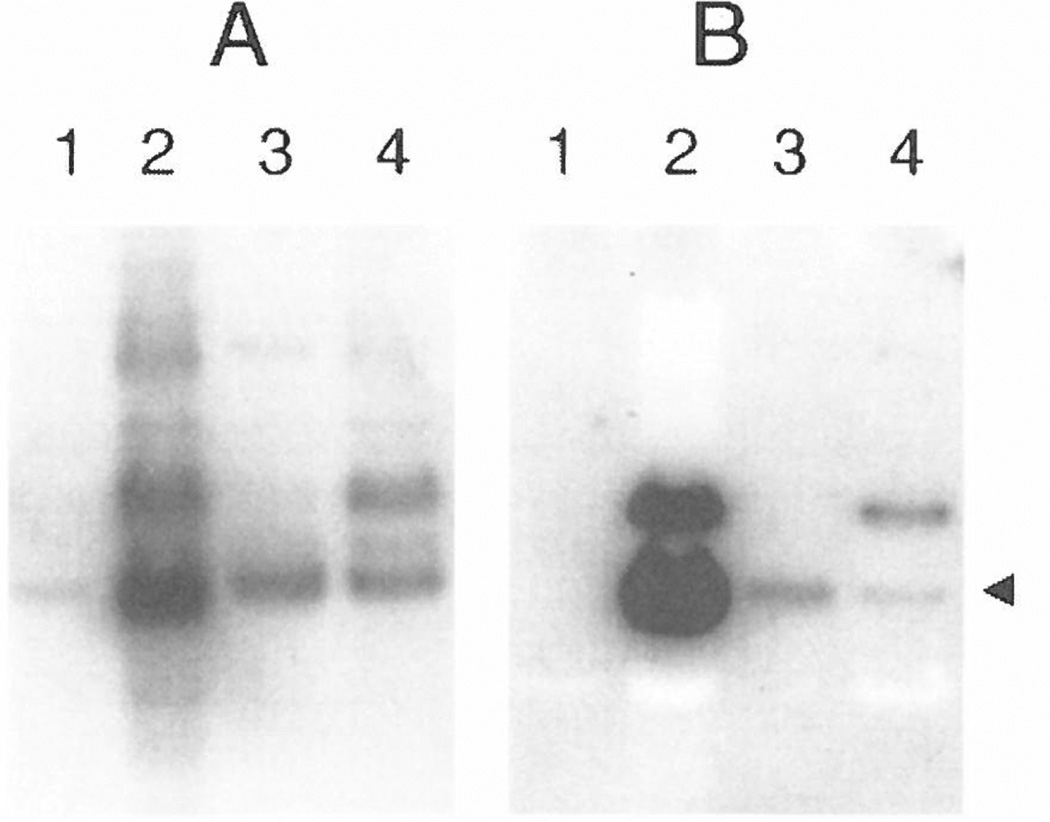
The MAb loop2 is a recombinant human IgG1κ that was originally isolated by screening a phage-displayed, human Fab library with a hydrazone-bridged cyclic peptide that corresponds to a conserved sequence within the V3 loop of the MN strain of HIV-1 gp120 (the MN peptide; see Ref. 9). Peptides corresponding to portions of the V3 loop of gpl20 have been shown to elicit neutralizing Abs against several strains of HIV-1 (19–21). MAb loop2 was used to screen a panel of 12 phage-displayed peptide libraries (4). Table 1 shows the sequences of peptides displayed by the selected phage clones. Alignment of these sequences reveals the consensus motif GPXR, which is also present in the constrained MN peptide. Using the pMal-X vector, the peptides C6, D5, G5, and J5 were transferred from the phage to MBP, and the resulting MBP fusions were purified. Figure 4 shows a Western blot of the G5:MBP fusion, which was probed first with MAb loop2 and then the membrane was stripped and probed again with rabbit anti-MBP Ab. The Western blot indicates that only the processed form of the G5:MBP fusion is detected in the osmotic-shock (periplasmic) fraction.
TABLE 1.
Representative Sequences of Peptides Selected from the Fourth Round of Affinity Selection with MAb loop2
| RCMMPAGPGRCLa | CVEVGPGRCLRLTSAPA9 (J5) |
| ACMMQLGPGRCI | CVPLGPGRCWLRTDHSM2 |
| RCNMPLGPARCF2b (G5) | MAKACTIGPHRCLGTP6 |
| KCAMPLGPVRCF (D5) | RKAVCMGTRGPCWPNG |
| GCLRKLGPSRCL2 | SLKRCLGHTKTCVRIG2 |
| RCTLPMGPTRCL2 | RKEVCMGTRGPCWPNG |
| DCTVRMGPNRCL | GDRRCDSRWMLCTPST3 |
| RCQISIGRGRCL (C6) | |
| KCAHTLGHGRCI | |
| ACGPWGRYWICT |
Underlining indicates consensus sequence.
Superscripted numbers indicate the number of times that clones having that sequence were selected. Peptide sequences C6, D5, G5, and J5 were transferred to MBP.
Figure 4.
Western blot of the G5:MBP fusion expressed by the vector pMal-X. The blot was first probed with rabbit anti-MBP polyclonal Ab (A) and then stripped and reprobed with MAb loop2 (B). (Lane 1) Whole-cell extract harvested immediately prior to IPTG induction or (lane 2) four hours after IPTG induction. (Lane 3) Osmotic-shock supernatant containing processed, G5:MBP protein (indicated by arrow). (Lane 4) Resuspended osmotic-shock pellet containing mostly unprocessed protein.
The ELISA data in Table 2 show that signals produced by the loop2 MAb for the peptides fused to MBP correlate well with those produced with fusions to the pVIII protein. Furthermore, side-by-side testing under reducing and nonreducing conditions revealed that the requirement for disulfide bridging was retained for the phage- and the MBP-displayed peptides. The ELISA controls indicated that signals produced by anti-MBP and anti-phage Abs were not affected by DTT. Although there was some decrease in the ELISA signal of MAb loop2 for the positive control (the MN peptide conjugated to BSA; MN-BSA), the signals were reduced to background levels for the MBP- and phage-displayed peptides. Taken together, the ELISA data indicate that the loop2-binding peptides behave similarly when displayed on phage and on MBP.
TABLE 2.
ELISA Data and SPR-Derived Kinetic Constants and Kd Values of IgG loop2 for Selected Peptides Displayed on Phage and on MBP in the Presence of and Absence of DTT
| ELISA | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| loop2 | Anti-MBP | Anti-phage | SPR | ||||||
| Antigen | −DTT | +DTT | −DTT | +DTT | −DTT | +DTT | kon (M−1 s−1) | koff(S−1) | Kda (nM) |
| G5:MBP | 975b | 31 | 1189 | 1130 | —c | — | 7.8 × 103 | 3.7 × 10−4 | 48 |
| G5-phage | 1113 | 95 | — | — | 1021 | 1014 | — | — | — |
| C6:MBP | 712 | 21 | 1083 | 1197 | — | — | 8.3 × 103 | 2.0 × 10−3 | 230 |
| C6-phage | 1097 | 104 | — | — | 1064 | 1055 | — | — | — |
| J5:MBP | 266 | 19 | 1162 | 1172 | — | — | nmd | nm | nm |
| J5-phage | 595 | 175 | — | — | 1041 | 1021 | — | — | — |
| D5:MBP | 133 | 15 | 1130 | 1065 | — | — | 7.7 × 103 | 1.4 × 10−3 | 180 |
| D5-phage | 462 | 81 | — | — | 1065 | 1049 | — | — | — |
| MN-BSA | 983 | 471 | — | — | — | — | — | — | — |
| MBP | 13 | 11 | 1170 | 1165 | — | — | — | — | — |
| BSA | 13 | — | 58 | — | 12 | — | — | — | — |
| f88.4 | 59 | 73 | — | — | 1011 | 1022 | — | — | — |
Apparent kinetic and affinity constants determined as on solid phase.
Results are reported as optical density (A405 – A490) × 1000.
—, not determined.
nm, not measurable.
SPR analysis was performed to determine whether the peptide:MBP fusions would produce well-behaved binding responses with Fab and MAb loop2. The sensorgram curves obtained were well-behaved, and a dose response was observed indicating specific binding (data not shown). Soluble Fab loop2 was used at several concentrations and bound only G5:MBP (Kd = 570 nM). This is consistent with G5:MBP producing the highest signal with MAb loop2 by ELISA. The fusions were also probed with MAb loop2, and apparent Kd values were determined for three of the four fusions (Table 2). MAb loop2 binding to J5:MBP was not measurable, most likely due to an artifact resulting from the immobilization procedure. Such artifacts due to covalent coupling of proteins to the BIAcore sensor chip are not rare (pers. comm., K. Dickerson, BIAcore). The surface affinity measured for MAb loop2 and G5:MBP (Kd = 48 nM) was ~10-fold lower than that measured for the Fab, indicating that the kinetics of MAb binding are strongly affected by avidity. There was a poor correlation between the surface affinities determined by SPR analysis with MAb loop2 and the ELISA data. Given the ~10-fold avidity boost in the SPR assay involving MAb loop2, the poor correlation is most likely due to differences in the spacing of the MBP:peptide fusions on the chip compared to the ELISA plate. Chemical coupling results in a more disperse spacing, whereas simple adsorption creates “islands” of closely grouped proteins.
Use of AR182 (prlA−) Cells for the Production of Peptide:MBP Fusions Containing Multiple Positive Charges
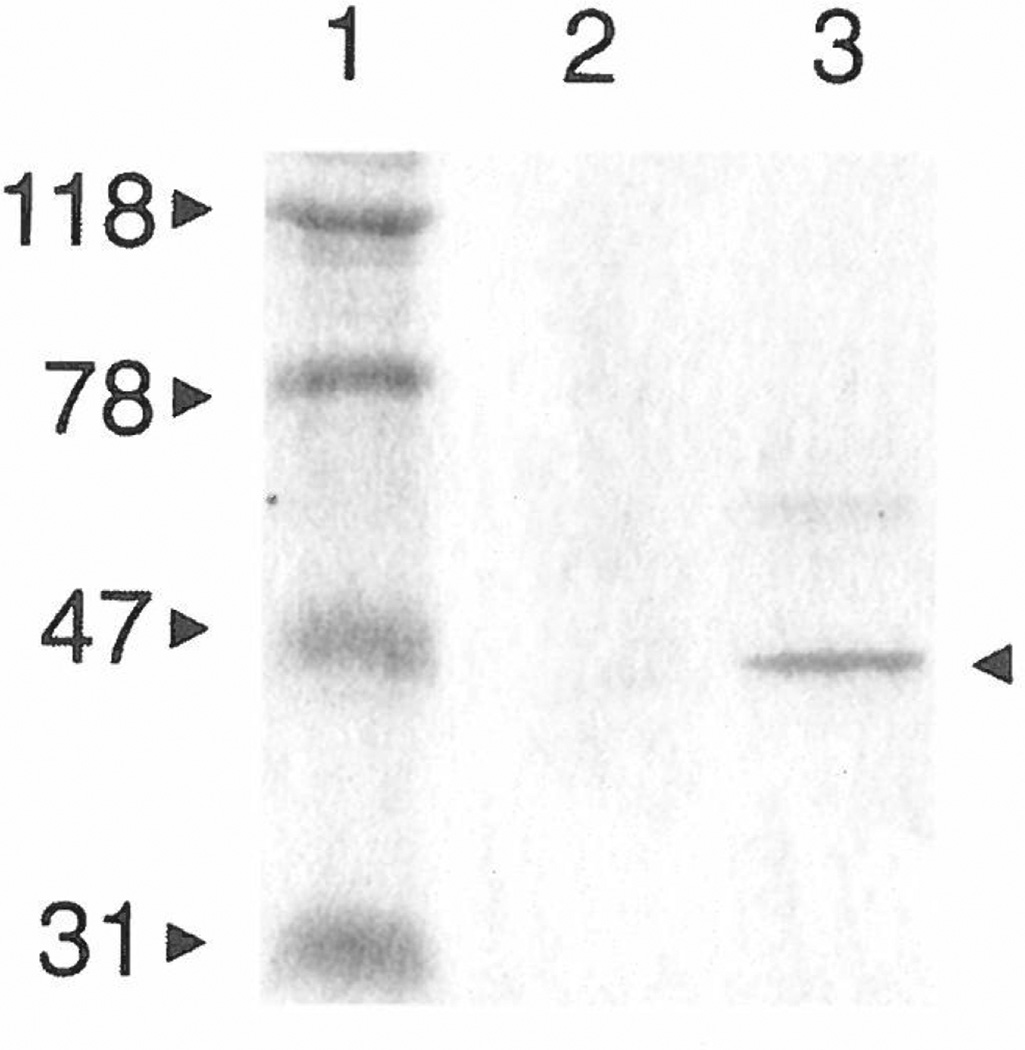
As with most protein-production systems, we found that the yield varied from one peptide:MBP fusion to the next. In particular, the yield of the C6:MBP fusion, when produced by the E. coli strain DH5αF′, was approximately 100-fold lower than that of the other fusion proteins. The C6 peptide sequence contains two closely spaced arginine residues and has a net charge of +3, whereas the D5, G5, and J5 sequences have net charges of +2, +2, and +1, respectively. Similarly, in the pMal-pIII system, the N11 Erk1/2 binding sequence, with a net charge of +3.5, was barely processed in E. coli NM522 (Fig. 3B). In contrast, the N15 peptide, whose net charge is +0.5, was processed at levels comparable to native MBP (cf. Figs. 3A and 3C). Peters et al. (17) reported that, for pIII-displayed peptides, a high density of positively charged residues near the signal-peptide cleavage site inhibits proper insertion of pIII into the E. coli inner membrane. In addition, they found that suppressor mutations in the SecY component of the protein-export apparatus (e.g., prlA4) dramatically reduce the inhibition of export caused by these positively charged, N-terminal residues. Our SDS–PAGE analysis of osmotic-shock fluids showed that production of the C6:MBP fusion was increased in the E. coli strain AR182 (prlA4) compared to its production at the same time and under the same conditions in DH5αF′ cells (Fig. 5). Based on this observation, we now produce all peptide:MBP fusions in AR182 cells; this is a conservative measure, as the prlA+ E. coli strains perform equally well for MBP fusions whose peptides do not have multiple, positively charged residues.
Figure 5.
SDS–PAGE showing the effect of the prlA4 mutation on periplasmic levels of the C6:MBP fusion in E. coli. Osmotic-shock supernatants containing C6:MBP fusion protein were simultaneously produced in prlA+ DH5αF′ (lane 2) and AR182 (prlA4) cells (lane 3). The sizes (in kDa) of the molecular weight standards (lane 1) are indicated (left), as is the mature C6:MBP fusion protein (arrowhead, right).
DISCUSSION
Using genetic engineering, we have transferred phage-displayed peptides to the N-terminus of MBP for subsequent use as a biologically produced, monovalent form of peptide for ELISA and SPR analysis. MBP was chosen over other protein-display scaffolds because it is easily affinity-purified, free of cysteines, secreted, and monomeric. Alkaline phosphatase has also been used for N-terminal display of pIII-derived peptides. In one case, Grihalde et al. (22) chose alkaline phosphatase as a means of characterizing peptides in the absence of phage and in another, Yamabhai et al. (23) used the alkaline phosphatase fusions as a one-step detection probe of peptide ligand:SH3 domain interactions. MBP has several advantages over alkaline phosphatase: It can be affinity purified rather simply; it is usually monomeric (whereas alkaline phosphatase is dimeric and thus has potential for avidity effects); and unlike alkaline phosphatase, MBP does not contain internal cysteines that could form disulfide linkages with cysteines within a peptide fusion. Recently, Tudyka et al. (24) engineered glutathione S-transferase from Shisto-soma japonicum to display a recombinant protease inhibitor at its N-terminus and to be functionally secreted by E. coli cells. This system might also be useful for the display of phage-derived peptides. The enzyme can be purified on glutathione columns, and it contains no exposed cysteines; however, like alkaline phosphatase, it forms a dimer.
In many instances, a synthetic peptide is desired as the final ligand. MBP display may act as a useful prescreen for identifying peptides whose reactivity does not depend on the phage carrier and thus would potentially be active as free peptides. We have found that the “peptide-on-pins” method of producing and analyzing peptides (Chiron Mimotopes PTY Ltd., Clayton Victoria, Australia) often results in inconsistencies with the data derived from phage libraries. Similarly, others have reported that the phage itself can be important for the affinity of a selected peptide, since the corresponding synthetic peptides have reduced affinity (25–28).
To date, we have transferred to MBP six different pIII-displayed peptides and more than 20 different pVIII-displayed peptides. The peptides ranged from 6 to 22 amino acids in length, and all were recognized by their cognate proteins; in many cases the peptides contained disulfide constraints. Moreover, for all cases studied, the pattern of ELISA reactivity was similar between MBP fusions and their phage-displayed counterparts. Significantly, all of the fusions retained amy-lose-binding activity. Thus, MBP is a reliable, monovalent vehicle for the N-terminal display of short peptides selected from phage libraries.
A study of the leader peptides from a few proteins secreted by E. coli have demonstrated that subsegments of the leader peptides, including those of MBP and pVIII, are not always interchangeable (29). Our results indicate that both the filamentous phage pIII and pVIII leader sequences allow the export of many peptide:MBP fusions to the periplasm, even in the prlA+ strains DH5αF′ and NM522. In general, our results support a model of MBP export that permits changes in leader sequence, as well as diverse sequence variation in the N-terminal region of the mature protein. We observed significant variation in the yield of peptide:MBP fusions in prlA+ cells for fusions bearing the highly positively charged N11 and C6 peptides, which were preceded by the pIII and pVIII leader sequences, respectively. The periplasmic level of the C6:MBP fusion was returned to near-normal when produced in cells bearing the prlA4 mutation. Derman et at. (30) reported that in E. coli strains having the prlA4 mutation, MBP and alkaline phosphatase are exported to the periplasm, even if each lacks its entire signal sequence. The improvement in C6:MBP production we observed indicates that the pVIII leader, which is normally associated with SecY-independent membrane translocation of pVIII (31), can be directed into a SecY-dependent pathway by particular sequences that follow it, and that SecY-induced restrictions on secretion are relaxed by the prlA4 mutation.
One group has reported that dimerization of MBP can occur under certain purification conditions, and that this can be reversed in the presence of 1 mM maltose (32, 33). We did not observe this, as assessed by nondenaturing PAGE, although we found varying amounts of intermolecular disulfide bridging for some cysteine-containing peptides using nonreducing SDS–PAGE. This may have the effect of raising apparent affinities due to an avidity boost or lowering the apparent affinity if intermolecularly bridged peptides become nonreactive. In either case, such effects are expected to be rare compared to the avidity effects caused by multivalent display on phage or strictly bivalent display, such as occurs with alkaline phosphatase display.
The loop2 MAb identified the consensus motif GPXR with a few exceptions (i.e., the C6 peptide, which has an arginine in the place of proline). Phage display technology has been used by several groups to identify peptides that bind to MAbs other than loop2, but which are also specific for the V3 loop of gpl20 on HIV (22, 34–36). The sequences selected by MAb loop2 in the present study, and those selected by each of the an-ti-V3 loop MAbs used in other reports, have different consensus elements, most probably reflecting differences in Ab specificity.
The order of reactivity of MBP fusions to the peptides selected by MAb loop2 differed between the ELISA and SPR analysis. This is not surprising given our results, which showed a large effect of Ab valency on the SPR results. For simplicity, we assumed a 1:1 pseudo-first-order kinetic model for the Kd calculations with the MAb loop2. Thus, the Kds we report are a measure of surface binding affinity and depend intimately on the immobilization density of the peptide: MBP fusions on the sensor chip surface; very likely these densities are different from their counterparts in the ELISAs. A more rigorous affinity analysis, in which SPR is used to measure Ab in equilibrium with different peptide concentrations, has been developed by Nieba et al. (37). The KinExA instrument (Sapidyne Instruments, Inc.) was developed for this same purpose (38, 39). Such in-solution measurements were not made in this current study, but may be used to circumvent the confounding effects of valency, rebinding, and mass transport that are associated with solid-phase SPR biosensor measurements (40).
In the future, MBP display could be exploited for other purposes. In one approach, phage plaque lifts were screened with polyclonal serum as a means of identifying disease-specific epitopes from phage-displayed peptide libraries (41). In a similar fashion, colony immunoblotting (12) or maltodextrin capture ELISAs on cell lysates (42) may be employed after one or two rounds of phage library selections to identify peptides that bind Ab independently of phage. Another potential application of peptide:MBP fusions is as immunogens, since MBP has been used as a fusion carrier to elicit immune responses (43, 44). An MBP hybrid that incorporated a B-cell epitope from the pre-S2 region of Hepatitis B virus into an external α-helix of MBP was used to elicit the production of peptide-specific Abs (45). The X-ray crystal structure of the MBP-preS2 hybrid has been solved (46), as has the 3D structure of wild-type MBP (47). These studies suggest future roles for phage-displayed peptides in the design of structurally defined molecular fusions that can elicit peptide-specific Abs. Moreover, peptides or proteins having other functions can be fused to the C-terminus of peptide:MBP fusions to make “trifunctional” proteins.
In conclusion, we have demonstrated an efficient means of transferring peptides from the filamentous phage coat to the N-terminus of MBP for the characterization of affinity-selected peptides. A variety of peptide sequences and lengths can be displayed, and the peptide:MBP fusions are amenable to analysis by ELISA, Western blot, and SPR. In this way, phage-derived peptides can be characterized in monomeric form and independent of phage-specific effects.
ACKNOWLEDGMENTS
We thank the following people: K. Brown, C. Marchetti, and M. Rashed for excellent technical assistance; J. Benner for N-terminal protein sequencing; P. Riggs for assistance with MBP fusion purifications; L. Saltman for critical reading of the manuscript; and D. Comb for support and encouragement. This work was supported by grants from the Natural Sciences and Engineering Research Council of Canada (NSERC) and the National Health Research and Development Program (J.K.S.). J.K.S. was supported in part by a scholarship from the B.C. Health Research Foundation. M.B.Z. was supported by a scholarship from NSERC. C.F.B. and S.V. were supported by NIH Grant AI 37470. S.V. was supported in part by a grant from The Foundation BLANCEFLOR Boncompagni-Ludovisi, nee′Bildt.
Footnotes
Abbreviations used: MBP, maltose-binding protein; Ab, antibody; MAb, monoclonal antibody; ELISA, enzyme-linked immunosorbent assay; SPR, surface plasmon resonance; DTT, dithiothreitol; BSA, bovine serum albumin; NANP, NANPNVDP(NANP)3GGPA; PBS, phophate-buffered saline; TBS, Tris-buffered saline; IPTG, isopropyl-thio-β-d-galactoside; RF, replicative form.
REFERENCES
- 1.Scott JK, Craig L. Curr. Opin. Biotechnol. 1994;5:40–48. doi: 10.1016/s0958-1669(05)80068-0. [DOI] [PubMed] [Google Scholar]
- 2.Smith GP, Petrenko VA. Chem. Rev. 1997;97:391–410. doi: 10.1021/cr960065d. [DOI] [PubMed] [Google Scholar]
- 3.Malik P, Terry TD, Gowda LR, Langara A, Petukhov SA, Symmons MF, Welsh LC, Marvin DA, Perham RN. J. Mol. Biol. 1996;260:9–21. doi: 10.1006/jmbi.1996.0378. [DOI] [PubMed] [Google Scholar]
- 4.Bonnycastle LL, Mehroke JS, Rashed M, Gong X, Scott JK. J. Mol. Biol. 1996;258:747–762. doi: 10.1006/jmbi.1996.0284. [DOI] [PubMed] [Google Scholar]
- 5.Zhong G, Smith GP, Berry J, Brunham RC. J. Biol. Chem. 1994;269:24183–24188. [PubMed] [Google Scholar]
- 6.Maina CV, Riggs PD, Grandea AGI, Slatko BE, Moran LS, Tagliamonte JA, McReynolds LA, di Guan C. Gene. 1988;74:365–373. doi: 10.1016/0378-1119(88)90170-9. [DOI] [PubMed] [Google Scholar]
- 7.Schatz PJ, Cull MG, Martin EL, Gates CM. Methods Enzymol. 1996;267:171–191. doi: 10.1016/s0076-6879(96)67012-8. [DOI] [PubMed] [Google Scholar]
- 8.Bregegere F, Schwartz J, Bedouelle H. Protein Eng. 1994;7:271–280. [PubMed] [Google Scholar]
- 9.Barbas CFI, Collet TA, Amberg W, Roben P, Binley JM, Hoekstra D, Cababa D, Jones TM, Williamson RA, Pilkington GR, Haigwood NL, Cabezas E, Satterthwait AC, Sanz I, Burton DR. J. Mol. Biol. 1993;230:812–823. doi: 10.1006/jmbi.1993.1203. [DOI] [PubMed] [Google Scholar]
- 10.Karlsson R, Miachaelsson A, Mattsson L. J. Im munol. Methods. 1991;145:229–240. doi: 10.1016/0022-1759(91)90331-9. [DOI] [PubMed] [Google Scholar]
- 11.Satterthwait AC, Chiang L-C, Arrhenius T, Cabezas E, Zavala F, Dyson HJ, Wright PE, Lerner RA. Bull. W.H.O. 1990;68:17–25. [PMC free article] [PubMed] [Google Scholar]
- 12.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 13.Wilson IA, Niman HL, Houghten RA, Cherenson AR, Connolly ML, Lerner RA. Cell. 1984;37:767–778. doi: 10.1016/0092-8674(84)90412-4. [DOI] [PubMed] [Google Scholar]
- 14.Rini JM, Schulze-Gahmen U, Wilson IA. Science. 1992;255:959–965. doi: 10.1126/science.1546293. [DOI] [PubMed] [Google Scholar]
- 15.Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 16.Neu HC, Heppel LA. J. Biol. Chem. 1965;240:3685–3692. [PubMed] [Google Scholar]
- 17.Peters EA, Schatz PJ, Johnson SS, Dower WJ. J. Bacteriol. 1994;176:4296–4305. doi: 10.1128/jb.176.14.4296-4305.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wirtz RA, Zavala F, Charoenvit Y, Campbell GH, Burkot TR, Schneider I, Esser KM, Beaudoin RL, Andre RG. Bull. W.H.O. 1987;65:39–45. [PMC free article] [PubMed] [Google Scholar]
- 19.Gorny MK, Conley AJ, Karwowska S, Buchbinder A, Xu J-Y, Emini EA, Koenig S, Zolla-Pazner S. J. Virol. 1992;66:7538–7542. doi: 10.1128/jvi.66.12.7538-7542.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Javaherian K, Langlois AJ, LaRosa GJ, Profy AT, Bolognesi DP, Herlihy WC, Putney SD, Matthews TJ. Science. 1990;250:1590–1593. doi: 10.1126/science.1703322. [DOI] [PubMed] [Google Scholar]
- 21.White-Scharf ME, Potts BJ, Smith LM, Sokolowski KA, Rusche JR, Silver S. Virology. 1993;192:197–206. doi: 10.1006/viro.1993.1022. [DOI] [PubMed] [Google Scholar]
- 22.Grihalde ND, Chen Y-CJ, Golden A, Gubbins E, Mandecki W. Gene. 1995;166:187–195. doi: 10.1016/0378-1119(95)00658-3. [DOI] [PubMed] [Google Scholar]
- 23.Yamabhai M, Kay BK. Anal. Biochem. 1997;247:143–151. doi: 10.1006/abio.1997.2040. [DOI] [PubMed] [Google Scholar]
- 24.Tudyka T, Skerra A. Protein Sci. 1997;6:2180–2187. doi: 10.1002/pro.5560061012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Balass M, Morag E, Bayer EA, Fuchs S, Wilchek M, Katchalski-Katzir E. Anal. Biochem. 1996;243:264–269. doi: 10.1006/abio.1996.0515. [DOI] [PubMed] [Google Scholar]
- 26.Balass M, Heldman Y, Cabilly S, Givol D, Katchalski-Katzir E, Fuchs S. Proc. Natl. Acad. Sci. USA. 1993;90:10638–10642. doi: 10.1073/pnas.90.22.10638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yayon A, Aviezer D, Safran M, Gross JL, Heldman Y, Cabilly S, Givol D, Katchalski-Katzir E. Proc. Natl. Acad. Sci. USA. 1993;90:10643–10647. doi: 10.1073/pnas.90.22.10643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Felici F, Luzzago A, Folgori A, Cortese R. Gene. 1993;128:21–27. doi: 10.1016/0378-1119(93)90148-v. [DOI] [PubMed] [Google Scholar]
- 29.Laforet GA, Kaiser ET, Kendall DA. J. Biol. Chem. 1989;264:14478–14485. [PubMed] [Google Scholar]
- 30.Derman AI, Puziss JW, Bassford PJJ, Beckwith J. EMBO J. 1993;12:879–888. doi: 10.1002/j.1460-2075.1993.tb05728.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wolfe PB, Rice M, Wickner W. J. Biol. Chem. 1985;260:1836–1841. [PubMed] [Google Scholar]
- 32.Richarme G. Biochem. Biophys. Res. Commun. 1982;105:476–481. doi: 10.1016/0006-291x(82)91459-0. [DOI] [PubMed] [Google Scholar]
- 33.Richarme G. Biochem. Biophys. Ada. 1983;748:99–108. doi: 10.1016/0167-4838(83)90032-8. [DOI] [PubMed] [Google Scholar]
- 34.Jellis CL, Cradick TJ, Rennert P, Salinas P, Boyd J, Amirault T, Gray GS. Gene. 1993;137:63–68. doi: 10.1016/0378-1119(93)90252-x. [DOI] [PubMed] [Google Scholar]
- 35.Keller PM, Arnold BA, Shaw AR, Tolman RL, Van Middlesworth F, Bondy S, Rusiecki VK, Koenig S, Zolla-Pazner S, Conrad P, Emini EA, Conley AJ. Virology. 1993;193:709–716. doi: 10.1006/viro.1993.1179. [DOI] [PubMed] [Google Scholar]
- 36.Laisney IL, Benjamin H, Gefter M, Strosberg AD. Eur. J. Immunol. 1996;26:1634–1640. doi: 10.1002/eji.1830260734. [DOI] [PubMed] [Google Scholar]
- 37.Nieba L, Krebber A, Pluckthun A. Anal. Biochem. 1996;234:155–165. doi: 10.1006/abio.1996.0067. [DOI] [PubMed] [Google Scholar]
- 38.Blake DA, Khosraviani M, Pavlov AR, Blake RC. Immunol. Technol. Environ. Appl. 1997;657:49–60. [Google Scholar]
- 39.Craig L, Sanschagrin PC, Rozek A, Lackie S, Kuhn LA, Scott JK. J. Mol. Biol. 1998 doi: 10.1006/jmbi.1998.1907. in press. [DOI] [PubMed] [Google Scholar]
- 40.Schuck P. Curr. Op. Biotechnol. 1997;8:498–502. doi: 10.1016/s0958-1669(97)80074-2. [DOI] [PubMed] [Google Scholar]
- 41.Folgori A, Tafi R, Meola A, Felici F, Galfre G, Cortese R, Monaci P, Nicosia A. EMBO J. 1994;13:2236–2243. doi: 10.1002/j.1460-2075.1994.tb06501.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Felleisen R, Zimmermann V, Gottstein B, Muller N. BioTechniques. 1996;20:616–620. doi: 10.2144/19962004616. [DOI] [PubMed] [Google Scholar]
- 43.Zhang T, Stanley SLJ. Infect. Immun. 1996;64:1526–1531. doi: 10.1128/iai.64.5.1526-1531.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seong SY, Huh MS, Jang WJ, Park SG, Kim JG, Woo SG, Choi MS, Kim IS, Chang WH. Infect. Immun. 1997;65:1541–1545. doi: 10.1128/iai.65.4.1541-1545.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martineau P, Guillet J-G, Leclerc C, Hofnung M. Gene. 1992;113:35–46. doi: 10.1016/0378-1119(92)90667-e. [DOI] [PubMed] [Google Scholar]
- 46.Saul FA, Vulliez-le Normand B, Lema F, Bentley GA. Proteins. 1997;27:1–8. doi: 10.1002/(sici)1097-0134(199701)27:1<1::aid-prot2>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 47.Spurlino J, Lu G-Y, Quiocho F. J. Biol. Chem. 1991;266:5202–5219. doi: 10.2210/pdb1mbp/pdb. [DOI] [PubMed] [Google Scholar]