Abstract
HIV latency is a major barrier to viral eradication from infected individuals. Lusic et al. show that HIV latency is established by spatially positioning the proviral chromatin in close proximity to promyelocytic leukemia (PML) nuclear bodies, a reversible process that recruits the methyltransferase enzyme G9a to the latent viral promoter.
The nucleus of eukaryotic cells is highly organized. Emerging evidence indicates that the location of distinct genes within the nucleus is tightly regulated, can change during different transcriptional states, and affects their transcriptional activity (Bridger, 2011). When viruses, such as HIV, integrate into the host chromatin, their transcription is strongly influenced by the local chromatin environment at the site of integration (Jordan et al., 2001). HIV preferentially infects activated CD4+ T cells, where it generally establishes a highly productive lifecycle that ultimately leads to the destruction of the infected host cell within days after infection. In a small subset of infected cells, mainly central memory T cells, the viral lifecycle is silenced at the transcriptional level, and the virus establishes a stable latent reservoir that remains unaffected by current antiretroviral regimens (Han et al., 2007). This latent reservoir represents one of the major barriers to HIV eradication in infected patients on antiretroviral therapy, but the molecular mechanisms responsible for proviral latency are only incompletely understood.
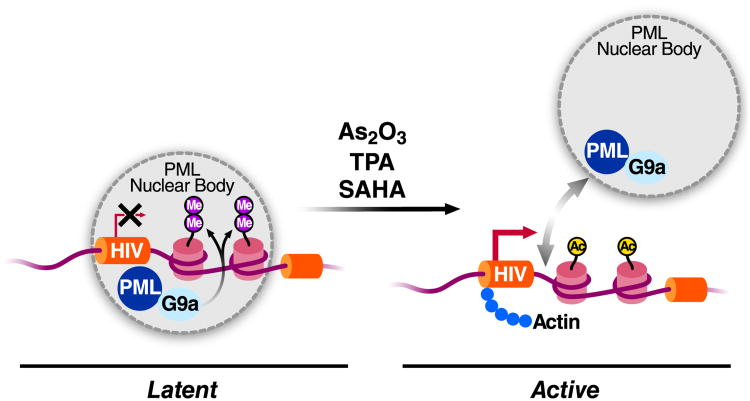
In this issue, Marina Lusic, Mauro Giacca and colleagues present evidence that proximity of the integrated HIV genome to the nuclear body defined by the PML protein plays an important role in latency (CITATION). Using 3D-fluorescent in situ hybridization combined with immunostaining, they examined the location of the integrated HIV genome in latently infected cells with respect to the PML body. Using three different J-LAT clonal cells lines, a model for HIV latency obtained after infection of the Jurkat lymphoid cell line (Jordan et al., 2003), they observed colocalization of latent HIV with the PML nuclear body (Figure 1). This colocalization was lost after reactivation of latent HIV with either the phorbol ester tetradecanoyl phorbol acetate (TPA) or the histone deacetylase inhibitor SAHA. Similar results were obtained using a model for HIV latency in human primary CD4+ T cells developed by Alberto Bosque and Vicente Planelles (Bosque and Planelles, 2009). As further support for a role of the PML nuclear bodies in latency, they observed that PML removal by shRNA-mediated knockdown of PML or by treatment of latent cells with arsenic trioxide (which induces PML degradation) is associated with transcriptional activation of latent HIV. This was also observed in J-LAT cell lines and in the primary CD4+ T cell latency model.
Figure 1. Regulation of HIV latency by proximity to the PML nuclear body.
Under latency conditions, the HIV provirus is positioned in close proximity to PML nuclear bodies. PML recruits the methyltransferase G9a to the HIV promoter leading to dimethylation of lysine 9 in histone H3 and transcriptional silencing. Upon treatment with the phorbol ester TPA, the deacetylase inhibitor SAHA or arsenic trioxide, a compound inducing PML degradation, the HIV provirus is relocated away from PML nuclear bodies, and HIV transcription is activated.
PML nuclear bodies are highly dynamic nuclear structures with extensive contact to chromatin and direct links to transcriptional activation and repression of unique genes. Besides the PML protein, its main structural component, they contain transcriptional corepressors, such as Daxx, that PML helps to recruit to nuclear bodies and gene promoters. Importantly, the authors show that transcriptional repression of latent HIV does not depend on Daxx but on the PML-dependent recruitment of the methyltransferase G9a (Figure 1). G9a mediates dimethylation of histone H3K9 (H3K9me2), a known mark of facultative heterochromatin within euchromatic regions. Using chromatin immunoprecipitation, the authors found G9a and the H3K9me2 modification at the latent, but not the reactivated, HIV provirus, underscoring the link between PML nuclear bodies, epigenetic chromatin modifications and HIV latency.
An interesting aspect of the paper is the connection to actin-mediated chromatin movement. The rapid and potentially non-random relocation of genes within the nucleus implies the presence of molecular motors, similar to those found in the cytoplasm. Actin, long regarded a cytoplasmic contaminant of nuclear preparations, is now an established nuclear regulator of gene expression and is found in many nuclear complexes, including those associating with the RNA polymerase II C-terminal domain and in chromatin-remodeling complexes. In a number of reported cases, repositioning of specific genes to different regions of the genome is dependent on active nuclear actin polymerization (Vartiainen et al., 2012). Importantly, Lusic and colleagues show that treatment of latently infected cells with cytochalasin D, an inhibitor of actin polymerization, prevented the relocalization of the HIV genome away from PML nuclear bodies and suppressed reactivation of latent HIV in response to phorbol esters. In addition, they report that actin itself binds to the transcriptionally activated, and not to the latent, HIV promoter, opposite to what is observed for PML (Figure 1). This observation indicates a direct function for actin in the transcriptional activation of the HIV provirus. Thus, the HIV provirus joins a growing group of genes that are regulated at the transcriptional level by nuclear topology. Despite a significant number of examples connecting gene movement with changes in transcriptional activity, it is still unclear whether intranuclear movement of chromatin is the cause or result of changes in transcriptional activity.
Interestingly, nuclear bodies defined by the PML protein are also implicated in the lifecycle of other nuclear-replicating DNA viruses and some retroviruses. The genomes of herpes simplex virus 1 and cytomegalovirus, two human herpesviruses, also associate with the PML nuclear bodies under restrictive conditions, such as latency (Everett, 2001). The fact that two of the nuclear bodies proteins, PML and Sp100, are induced by interferon has raised the possibility that the PML nuclear body might represent part of the innate immune response to DNA viruses.
How might this insight benefit HIV-infected individuals? One strategy to eradicate HIV from infected individuals is to reverse latency under the cover of intense antiretroviral therapy and to induce the “flushing” of long-lived latently infected T cells. This “shock and kill” approach to purging the latent HIV reservoir is being directly tested in patients with HDAC inhibitors, such as SAHA (vorinostat®), that induce reactivation of latent HIV in cell culture and in infected individuals (Archin et al., 2012; Van Lint et al., 1996). Interestingly, this paper shows that SAHA treatment is associated with the repositioning of the HIV genome away from the closest PML nuclear body, suggesting that PML and histone deacetylases could act in concert to repress HIV transcription under latency conditions. In addition, the authors identify a agent, arsenic trioxide, as a potent reactivator of latent HIV. Arsenic trioxide, a poison used therapeutically in ancient Chinese medicine and today as an anticancer treatment, induces PML degradation. In addition, it induces degradation of the oncogenic PML fusion proteins, such as PML-RARa, and is an approved drug (Trisenox®) for refractory promyelocytic leukemia (Zhang et al., 2010). The finding that it potently activates HIV from latency in cell lines and primary T cells adds an approved drug to the list of latency-reversing compounds with clinical potential.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Archin NM, Liberty AL, Kashuba AD, Choudhary SK, Kuruc JD, Crooks AM, Parker DC, Anderson EM, Kearney MF, Strain MC, et al. Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature. 2012;487:482–485. doi: 10.1038/nature11286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosque A, Planelles V. Induction of HIV-1 latency and reactivation in primary memory CD4+ T cells. Blood. 2009;113:58–65. doi: 10.1182/blood-2008-07-168393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridger JM. Chromobility: the rapid movement of chromosomes in interphase nuclei. Biochem Soc Trans. 2011;39:1747–1751. doi: 10.1042/BST20110696. [DOI] [PubMed] [Google Scholar]
- Everett RD. DNA viruses and viral proteins that interact with PML nuclear bodies. Oncogene. 2001;20:7266–7273. doi: 10.1038/sj.onc.1204759. [DOI] [PubMed] [Google Scholar]
- Han Y, Wind-Rotolo M, Yang HC, Siliciano JD, Siliciano RF. Experimental approaches to the study of HIV-1 latency. Nat Rev Microbiol. 2007;5:95–106. doi: 10.1038/nrmicro1580. [DOI] [PubMed] [Google Scholar]
- Jordan A, Bisgrove D, Verdin E. HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. EMBO J. 2003;22:1868–1877. doi: 10.1093/emboj/cdg188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan A, Defechereux P, Verdin E. The site of HIV-1 integration in the human genome determines basal transcriptional activity and response to Tat transactivation. EMBO J. 2001;20:1726–1738. doi: 10.1093/emboj/20.7.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusic M, Marini B, Ali H, Lucic B, Luzzati R, Giacca M. Cell Host & Microbe. 2013 doi: 10.1016/j.chom.2013.05.006. this issue, *bxs. [DOI] [PubMed] [Google Scholar]
- Van Lint C, Emiliani S, Ott M, Verdin E. Transcriptional activation and chromatin remodeling of the HIV-1 promoter in response to histone acetylation. EMBO J. 1996;15:1112–1120. [PMC free article] [PubMed] [Google Scholar]
- Vartiainen MK, Huet G, Skarp KP. Nuclear actin levels as an important transcriptional switch. Transcription. 2012;3 doi: 10.4161/trns.21062. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XW, Yan XJ, Zhou ZR, Yang FF, Wu ZY, Sun HB, Liang WX, Song AX, Lallemand-Breitenbach V, Jeanne M, et al. Arsenic trioxide controls the fate of the PML-RARalpha oncoprotein by directly binding PML. Science. 2010;328:240–243. doi: 10.1126/science.1183424. [DOI] [PubMed] [Google Scholar]