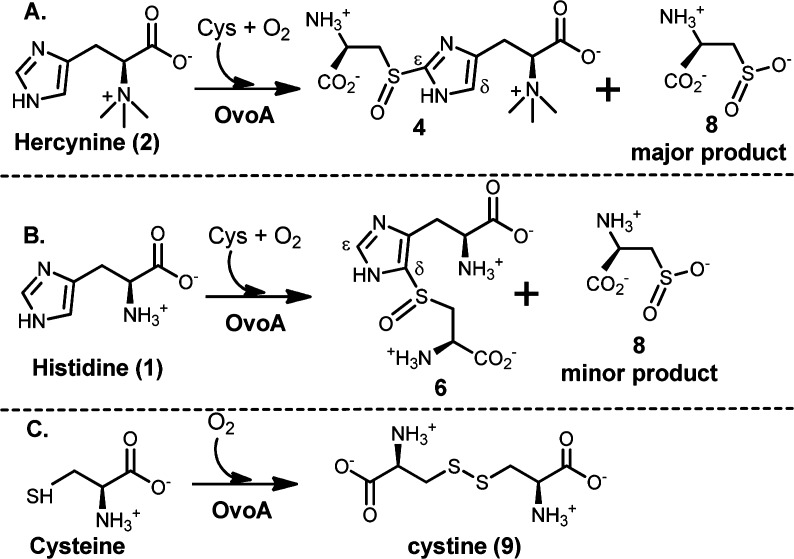
Abstract
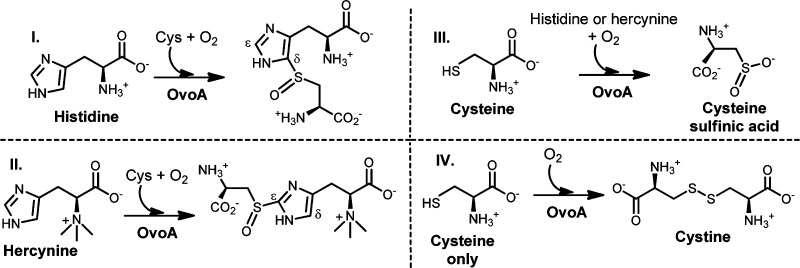
OvoA in ovothiol biosynthesis is a mononuclear non-heme iron enzyme catalyzing the oxidative coupling between histidine and cysteine. It can also catalyze the oxidative coupling between hercynine and cysteine, yet with a different regio-selectivity. Due to the potential application of this reaction for industrial ergothioneine production, in this study, we systematically characterized OvoA by a combination of three different assays. Our studies revealed that OvoA can also catalyze the oxidation of cysteine to either cysteine sulfinic acid or cystine. Remarkably, these OvoA-catalyzed reactions can be systematically modulated by a slight modification of one of its substrates, histidine.
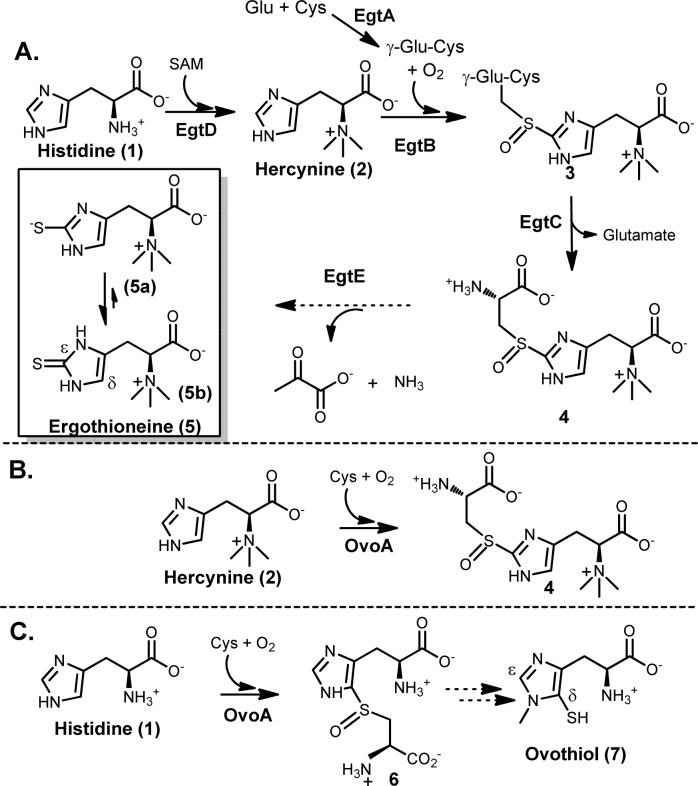
Sulfur-containing molecules are widely distributed in nature.1−9 Among sulfur-containing amino acids, there are two histidine-derived amino acids, ergothioneine (5) and ovothiol (7, Scheme 1). Ergothioneine (5) was isolated from ergot by Tanret in 1909.10 Humans do not synthesize ergothioneine but enrich it from diet to as high as millimolar concentrations in various tissues.11−17 Because ergothioneine is present predominantly in its thione form (5b),3,4,17 it has a much higher reduction potential (E0′ = −0.06 V3) compared to that of glutathione (E0′ = −0.25 V18). As a result, ergothioneine and glutathione play complementary roles in relieving oxidative stress.3,4,10,17−19 Due to ergothioneine’s beneficial roles to human health, there is a strong interest in developing efficient ergothioneine production methods.20,21 In year 2010, Seebeck reported the ergothioneine biosynthetic gene cluster (Scheme 1A) from Mycobacterium smegmatis,22 which has five enzymes. EgtD catalyzes the methylation of histidine to hercynine (2). EgtA condenses glutamate and cysteine to γ-glutamylcysteine (γ-Glu-Cys). EgtB is a mononuclear non-heme iron enzyme, catalyzing oxidative coupling of hercynine (2) and γ-Glu-Cys to form 3, which is then hydrolyzed by EgtC to produce 4; EgtE was proposed to be a PLP-dependent C-S lyase.
Scheme 1. Proposed Ergothioneine Biosynthetic Pathway and Recently Discovered Novel OvoA Reactivities.
To make use of the M. smegmatis pathway (Scheme 1A)22 for ergothioneine production, there is a potential barrier. γ-Glu-Cys is an intermediate in both ergothioneine (Scheme 1A) and glutathione biosyntheses. Due to the high in vivo glutathione concentration (up to 10 mM), such competition is not desirable. To address this issue, we initiated the search for enzymes capable of catalyzing a one-step 2 → 4 transformation (Scheme 1B). Indeed, we discovered that OvoA enzyme has such a novel activity (Scheme 1B).23 The native chemistry of OvoA enzyme is the first step in ovothiol biosynthesis (1 → 6 conversion, Scheme 1C).24 Interestingly, when histidine is replaced by hercynine, the only detectable oxidative coupling product is 4.23 Such a novel OvoA property encouraged us to characterize it in more detail. OvoA catalysis has three different substrates: histidine or hercynine, oxygen, and cysteine.23,24 In this report, we systematically characterized the OvoA enzyme using three different assays: (1) 1H NMR assay to monitor histidine or hercynine consumption, (2) NeoFox oxygen electrode to measure the oxygen consumption, and (3) 13C NMR assay to examine cysteine-related reactions using [β-13C]-cysteine as the substrate. These studies revealed that OvoA also catalyzes two more cysteine oxidation reactions: (1) the oxidation of cysteine to cysteine sulfinic acid and (2) the oxidation of cysteine to cystine. In addition, these OvoA-catalyzed reactions can be systematically modulated by a slight modification of one of its substrates, histidine.
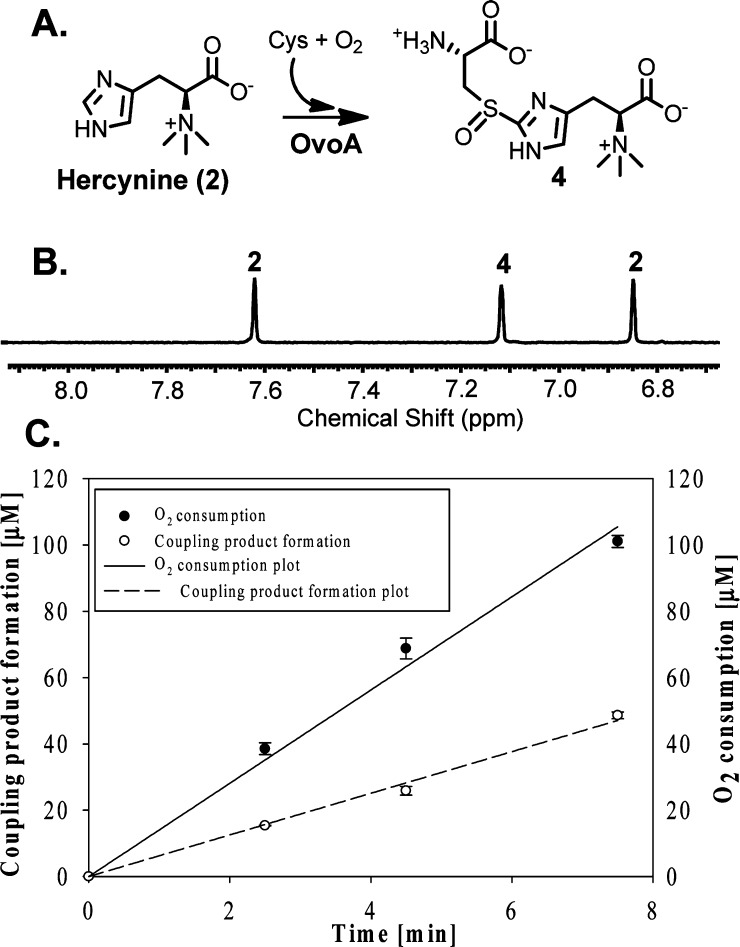
The chemical shifts of the imidazole hydrogen atoms are well-separated from the rest of the enzymatic reaction mixture (Figure 1B). Recently, utilizing this 1H NMR assay, we initiated the search for enzymes capable of catalyzing a one-step 2 → 4 transformation, which led to the discovery of this activity in OvoA enzyme.23 Several lines of evidence suggest that sulfoxide 4 instead of a thioether (13, Supplementary Figure 1S) is the oxidative coupling product: (1) H2O2 was not detected as a side product in this reaction. (2) Thioether 13 was synthesized chemically,20 and under our assay conditions, its oxidation to sulfoxide 4 by either O2 or H2O2 is below our detection limit. (3) When 40× of catalase relative to OvoA was included in the reaction mixture, sulfoxide 4 was still the only detectable oxidative coupling product (Supplementary Figure 2S). OvoA-catalyzed oxidative coupling reaction between hercynine and cysteine was also analyzed quantitatively using an oxygen consumption assay (NeoFox oxygen electrode), and the kinetic parameters obtained are kobs of 270 ± 5 min–1, Km of 395 ± 30 μM for hercynine, and Km of 3.19 ± 0.41 mM for Cys.23 Surprisingly, compound 4 formation rate revealed from 1H NMR assay is only ∼40% of the O2 consumption rate (Figure 1C). In this reaction, compound 4 is the only detectable oxidative coupling product (Figure 1B). Thus, besides compound 4 formation, there must be other uncharacterized reactions.
Figure 1.
1H NMR and oxygen consumption assays of OvoA catalysis. (A) The reaction. (B) 1H NMR of hercynine (2) and compound 4 imidazole ring hydrogens. (C) Quantitative comparison between oxygen consumption and compound 4 formation rates.
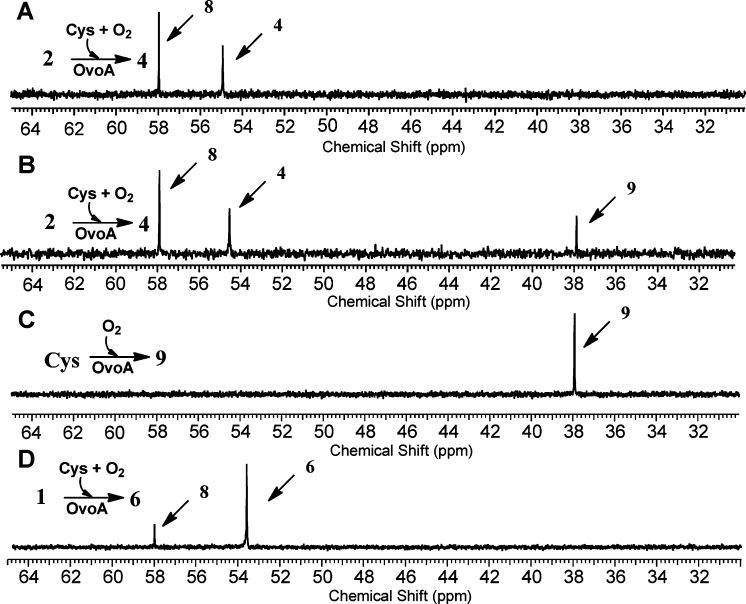
The reaction mixture has several reductants, including tris(2-carboxyethyl) phosphine (TCEP), ascorbate, and one of the OvoA substrates, cysteine. To examine what the additional oxygen consumption reactions were in our assay, we utilized [β-13C]-cysteine as the substrate and monitored the reaction using 13C NMR (Figure 2). When hercynine (2) and [β-13C]-labeled Cys were used as the substrates, besides the formation of compound 4 (54.8 ppm, Figure 2A), there was another compound (8, 57.9 ppm in Figure 2A). We isolated this compound by ion-exchange chromatography, characterized it by 1H NMR, 13C NMR, and high-resolution mass spectrometry. These characterizations allowed us to assign compound 8 as cysteine sulfinic acid (Supplementary Figures 3S–6S), which was further substantiated by comparison to the authentic cysteine sulfinic acid standard purchased from MP Biomedicals.
Figure 2.
13C NMR analyses of OvoA reactions. (A) Formation of compound 4 and cysteine sulfinic acid 8 when hercynine and cysteine were the substrates. (B) Oxidation of cysteine to cystine in reaction A after all hercynine was consumed. (C) Formation of cystine (9) when cysteine was the only substrate. (D) Formation of compound 6 and cysteine sulfinic acid (8) when histidine and cysteine were the substrates.
After cysteine sulfinic acid (8) was identified as another product in OvoA catalysis, a closer examination of 1H NMR spectrum of the reaction, especially the Cys Cβ proton,25 also supports the production of cysteine sulfinic acid OvoA catalysis (Supplementary Figure 7S). By including ethyl viologen as an internal standard and utilizing a 1H NMR signal unique to compound 4 and cysteine sulfinic acid (8), we quantified the ratio between compound 4 and 8 to be ∼1:1.3 (Supplementary Figure 7S). The formation of compound 4 accounts for ∼40% of the oxygen consumption rate (Figure 1C). Oxidation of cysteine to cysteine sulfinic acid (8) also consumes one molecule of oxygen. The results from three assays, namely, 1H NMR assay, oxygen consumption assay, and 13C NMR assay, suggest that when hercynine (2) and cysteine were used as the OvoA substrates, the formation compound 4 and 8 are the two dominant oxidation reactions. A combination of these two reactions accounts for more than 90% of oxygen consumed under our assay conditions (kobs of 137 ± 2.5 min–1 for cysteine sulfinic acid (8) formation and kobs of 106 ± 2.0 min–1 for compound 4 formation, Scheme 2A).
Scheme 2. OvoA-Catalyzed Cysteine Oxidation Reactions.
Formation of cysteine sulfinic acid is OvoA-dependent. Under our assay conditions, the cysteine sulfinic acid formation rate (kobs of 137 ± 2.5 min–1) is several orders of magnitude greater than the uncatalyzed cysteine oxidation process. In addition, the formation of cysteine sulfinic acid (8) depends on the presence of hercynine. When the time course of this reaction was closely monitored, it revealed that cysteine sulfinic acid (8) always forms along with the coupling product (4) and their ratio remains constant throughout the reaction course (Supplementary Figure 8S). Surprisingly, if cysteine was used in excess relative to hercynine, the oxidation of the excess cysteine continues at a much lower rate (kobs of ∼0.1 s–1) without the formation of cysteine sulfinic acid. Instead, cysteine is now oxidized to a compound with a chemical shift at 37.9 ppm in 13C NMR when [β-13C]-labeled Cys was used as a substrate (Figure 2B). This compound was isolated by ion-exchange chromatography and characterized by 1H NMR, 13C NMR, and high-resolution mass spectrometry. Its spectroscopic properties are consistent with those of cystine (9) (Supplementary Figures 9S–12S). The OvoA reaction was also carried out in the absence of hercynine. Consistent with the results in Figure 2B, cystine is the only detectable product (9, Figure 2C and Scheme 2C). In addition, cystine formation also shows OvoA-concentration dependence (Supplementary Figure 13S).
In all of the above OvoA characterizations, hercynine was used as the substrate. Because OvoA’s native substrates are histidine and cysteine (Scheme 1C),24 OvoA catalysis was re-examined using histidine instead of hercynine as the substrate (kobs of 572 ± 20 min–1, Km of 420 ± 31 μM for His, Km of 300 ± 34 μM for Cys).23 Again, the reaction was analyzed using three assays: 1H NMR, oxygen consumption, and 13C NMR assay. Even in native OvoA catalysis, the production of cysteine sulfinic acid (8) was detected (Figure 2D and Scheme 2B). Quantitative analysis using 1H NMR assay indicated that the coupling product 6 and cysteine sulfinic acid (8) were produced at a ratio of ∼8:1 (Supplementary Figure 15S). In native OvoA catalysis (Scheme 2B), cysteine sulfinic acid (8) was a minor product, which is different from the hercynine (2) reaction. Compound 6 formation rate is ∼87% of the oxygen consumption rate (Supplementary Figure 16S). Thus, when histidine and cysteine were used as the substrates, formation of 6 (kobs of 498 ± 2.2 min–1) and cysteine sulfinic acid 8 (kobs of 62 ± 0.3 min–1) accounts for nearly all of the oxygen consumed. When mono- or dimethylated histidine was used as the substrate, cysteine sulfinic acid was also detected as a minor product (Supplementary Figure 17S).
Formation of cysteine sulfinic acid (8) in OvoA catalysis is unexpected because such activity is native to an enzyme called cysteine dioxygenase,26−29 which plays a key role in regulating Cys concentration in vivo.(30) The cysteine dioxygenase activity in OvoA is unique in several aspects. First, the cysteine dioxygenase activity of OvoA depends on the presence of histidine or hercynine. In the absence of histidine/hercynine, OvoA does not produce cysteine sulfinic acid. Instead, cystine is produced as the only product (Scheme 2C). Second, although both OvoA enzyme and cysteine dioxygenase have cysteine dioxygenase activity, they clearly belong to different classes of mononuclear non-heme iron enzymes. When Seebeck and co-workers reported the OvoA activity, it was suggested that OvoA belongs to the 2-His-1-Carboxylate catalytic triad type31−35 of mononuclear non-heme iron enzymes.22,24 Figure 3 shows the sequence alignment of OvoA enzymes from a few species. Mutagenesis studies of H170, H174, and E176 from E. tasmaniensisin are consistent with the initial proposal from Seebeck of having these residues as the iron ligands.22,24 Different from OvoA, cysteine dioxygenase makes use of a 3-His facial triad.36−39 Such a 3-His ligand environment was proposed to be crucial for the cysteine dioxygenase activity.30,40,41 The presence of a significant level of cysteine dioxygenase activity in OvoA enzyme implies that a 3-His catalytic triad is probably not a prerequisite for the cysteine dioxygenase activity as suggested in the literature.30,40,41 To test this hypothesis, we created an OvoA E176H mutant (Supplementary Figure 20S), which would change OvoA from a 2-His-1-Carboxylate to a 3-His ligand environment if the new histidine residue could replace glutamate as the metal ligand. When OvoA E176H mutant was created, characterization of this mutant using the above three assays indicate that it not only lost the cysteine dioxygenase activity but also stopped catalyzing the formation of oxidative coupling product (4 or 6) (Supplementary Figures 21S and 22S). Thus, in this case, mutation of the 2-His-1-Carboxylate catalytic triad disrupts the cysteine dioxygenase activity.
Figure 3.

Sequence alignment of OvoA enzymes from a few species. Mononuclear non-heme iron ligands (2-His-1-carboxylate facial triad) are colored in red.
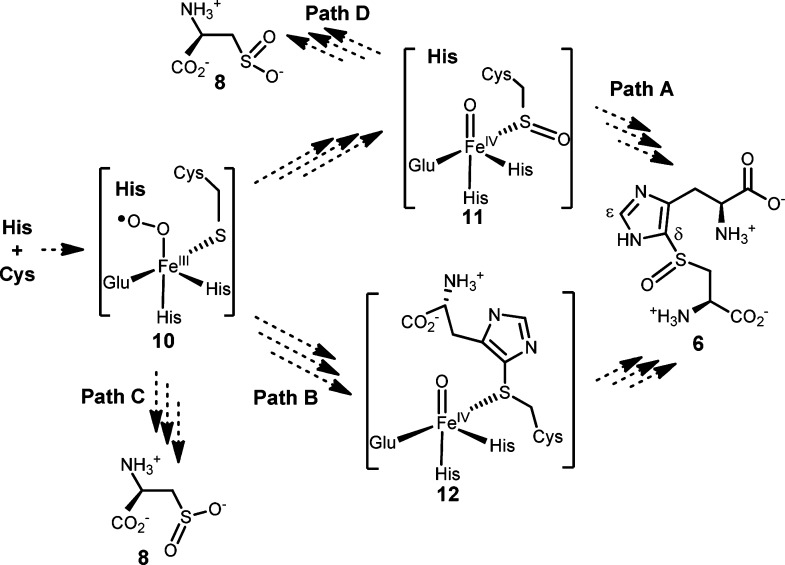
In summary, in addition to catalyzing the formation of oxidative coupling products between histidine/hercynine and cysteine,23,24 OvoA can also catalyze two more cysteine oxidation reactions (Scheme 2). OvoA has cysteine dioxygenase activity. Despite the fact that histidine (or hercynine) is not part of this transformation, cysteine sulfinic acid formation depends absolutely on the presence of histidine (or hercynine). In their absence, OvoA catalyzes the oxidation of cysteine to cystine (Scheme 2). In the literature, several mechanistic models were proposed to account for the oxidative C–S bond formation in OvoA catalysis.23,24,42,43 Our discovery of cysteine dioxygenase activity in OvoA enzyme and the dependence of such an activity on the presence of histidine/hercynine suggest that formation of 6 and cysteine sulfinic acid are probably two OvoA pathways branching out from a common intermediate (Scheme 3). After cysteine and histidine bind to OvoA, oxygen binds and is activated to form a peroxo radical species (11). The two pathways may branch out from either the peroxy radical intermediate (11, Path C) or sulfenic acid intermediate (12, Path D). In the absence of histidine/hercynine, there might be two molecules of cysteine at the OvoA active site. Cystine formation might either involve the production of H2O2 as the side product or have cysteine sulfenic acid as the oxidation product (Supplementary Figure 23S), which then reacts with cysteine to form cystine. Due to the high reactivity of H2O2 and cysteine sulfenic acid under our assay conditions, initial attempts on trapping them were unsuccessful.44−46 Future studies will be focused on trapping and characterization of some of the proposed intermediates.
Scheme 3. Proposed Mechanistic Model.
Formation of compounds 6 and 8 might be two branching pathways in OvoA catalysis. It may involve either a sulfenic acid intermediate (11, Path A) or thiother intermediate (12, Path B). Formations of 6 and 8 may branch out from either the peroxy radical species (10, Path C) or the sulfenic acid intermediate (11, Path D).
Acknowledgments
This work is partially supported by a NSF award (CHE-1309148) and NIH award (GM093903) to PL.
Supporting Information Available
Experimental procedure, enzyme characterization data, copies of the 1H and 13C NMR of compounds 8 and 9, and kinetic data. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Fontecave M.; Ollagnier-de-Choudens S.; Mulliez E. Chem. Rev. 2003, 103, 2149. [DOI] [PubMed] [Google Scholar]
- Kessler D. FEMS Microbiol. Rev. 2006, 30, 825. [DOI] [PubMed] [Google Scholar]
- Hand C. E.; Honek J. F. J. Nat. Prod. 2005, 68, 293. [DOI] [PubMed] [Google Scholar]
- Fahey R. C. Annu. Rev. Microbiol. 2001, 55, 333. [DOI] [PubMed] [Google Scholar]
- Lin C. I.; McCarty R. M.; Liu H. W. Chem. Soc. Rev. 2013, 42, 4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry R. J. In Comprehensive Natural Products Chemistry (I); Barton D., Nakanishi K., Meth-Cohn O., Eds.; Pergamon: Oxford, 1999; Vol. 1, p 825. [Google Scholar]
- Wang L.; Chen S.; Xu T.; Taghizadeh K.; Wishnok J. S.; Zhou X.; You D.; Deng Z.; Dedon P. C. Nat. Chem. Biol. 2007, 3, 709. [DOI] [PubMed] [Google Scholar]
- Mueller E. G. Nat. Chem. Biol. 2006, 2, 185. [DOI] [PubMed] [Google Scholar]
- Jacob C. Nat. Prod. Rep. 2006, 23, 851. [DOI] [PubMed] [Google Scholar]
- Tanret C. C. R. Acad. Sci. 1909, 149, 222. [Google Scholar]
- Grundemann D.; Harlfinger S.; Golz S.; Geerts A.; Lazar A.; Berkels R.; Jung N.; Rubbert A.; Schomig E. Proc. Natl. Acad. Sci. U.S.A. 2005, 102, 5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melville D. B.; Eich S. J. Biol. Chem. 1956, 218, 647. [PubMed] [Google Scholar]
- Fahey R. C.; Newton G. L.; Dorian R.; Kosower E. M. Anal. Biochem. 1981, 111, 357. [DOI] [PubMed] [Google Scholar]
- Genghof D. S.; Inamine E.; Kovalenko V.; Melville D. B. J. Biol. Chem. 1956, 223, 9. [PubMed] [Google Scholar]
- Briggs I. J. Neurochem. 1972, 19, 27. [DOI] [PubMed] [Google Scholar]
- Epand R. M.; Epand R. F.; Wong S. C. J. Clin. Chem. Clin. Biochem. 1988, 26, 623. [DOI] [PubMed] [Google Scholar]
- Hartman P. E. Methods Enzymol. 1990, 186, 310. [DOI] [PubMed] [Google Scholar]
- Scott E. M.; Duncan I. W.; Ekstrand V. J. Biol. Chem. 1963, 238, 3928. [PubMed] [Google Scholar]
- Weaver K. H.; Rabenstein D. L. J. Org. Chem. 1995, 60, 1904. [Google Scholar]
- Erdelmeier I.; Daunay S.; Lebel R.; Farescour L.; Yadan J.-C. Green Chem. 2012, 14, 2256. [DOI] [PubMed] [Google Scholar]
- Xu J. Z.; Yadan J. C. J. Org. Chem. 1995, 60, 6296. [Google Scholar]
- Seebeck F. P. J. Am. Chem. Soc. 2010, 132, 6632. [DOI] [PubMed] [Google Scholar]
- Song H.; Leninger M.; Lee N.; Liu P. Org. Lett. 2013, 15, 4854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunshausen A.; Seebeck F. P. J. Am. Chem. Soc. 2011, 133, 1757. [DOI] [PubMed] [Google Scholar]
- Siakkou E.; Wilbanks S. M.; Jameson G. N. L. Anal. Biochem. 2010, 405, 127. [DOI] [PubMed] [Google Scholar]
- Chai S. C.; Jeerkins A. A.; Banik J. J.; Shalev I.; Pinkham J. L.; Uden P. C.; Maroney M. J. J. Biol. Chem. 2005, 280, 9865. [DOI] [PubMed] [Google Scholar]
- Simmons C. R.; Hirschberger L. L.; Machi M. S.; Stipanuk M. H. Protein Expression Purif. 2006, 47, 74. [DOI] [PubMed] [Google Scholar]
- Crawford J. A.; Li W.; Pierce B. S. Biochemistry 2011, 50, 10241. [DOI] [PubMed] [Google Scholar]
- Chai S. C.; Bruyere J. R.; Maroney M. J. J. Biol. Chem. 2006, 281, 15774. [DOI] [PubMed] [Google Scholar]
- Joseph C. A.; Maroney M. J. Chem. Commun. 2007, 3338. [DOI] [PubMed] [Google Scholar]
- Krebs C.; Fujimori D. G.; Walsh C. T.; Bollinger J. M. Acc. Chem. Res. 2007, 40, 484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu-Omar M. M.; Loaiza A.; Hontzeas N. Chem. Rev. 2005, 105, 2227. [DOI] [PubMed] [Google Scholar]
- Kovaleva E. G.; Lipscomb J. D. Nat. Chem. Biol. 2008, 4, 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon E. I.; Brunold T. C.; Davis M. I.; Kemsley J. N.; Lee S. K.; Lehnert N.; Neese F.; Skulan A. J.; Yang Y. S.; Zhou J. Chem. Rev. 2000, 100, 235. [DOI] [PubMed] [Google Scholar]
- Costas M.; Mehn M. P.; Jensen M. P.; Que L. Chem. Rev. 2004, 104, 939. [DOI] [PubMed] [Google Scholar]
- McCoy J. G.; Bailey L. J.; Bitto E.; Bingman C. A.; Aceti D. J.; Fox B. G.; Phillips G. N. Proc. Natl. Acad. Sci. U.S.A. 2006, 103, 3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons C. R.; Liu Q.; Huang Q. Q.; Hao Q.; Begley T. P.; Karplus P. A.; Stipanuk M. H. J. Biol. Chem. 2006, 281, 18723. [DOI] [PubMed] [Google Scholar]
- Simmons C. R.; Krishnamoorthy K.; Granett S. L.; Schuller D. J.; Dominy J. E. Jr.; Begley T. P.; Stipanuk M. H.; Karplus P. A. Biochemistry 2008, 47, 11390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye S.; Wu X.; Wei L.; Tang D. M.; Sun P.; Bartlam M.; Rao Z. H. J. Biol. Chem. 2007, 282, 3391. [DOI] [PubMed] [Google Scholar]
- Kumar D.; Thiel W.; de Visser S. P. J. Am. Chem. Soc. 2011, 133, 3869. [DOI] [PubMed] [Google Scholar]
- Aluri S.; de Visser S. P. J. Am. Chem. Soc. 2007, 129, 14846. [DOI] [PubMed] [Google Scholar]
- Bushnell E. A. C.; Fortowsky G. B.; Gauld J. W. Inorg. Chem. 2012, 51, 13351. [DOI] [PubMed] [Google Scholar]
- Mashabela G. T. M.; Seebeck F. P. Chem. Commun. 2013, 49, 7714. [DOI] [PubMed] [Google Scholar]
- Thurman R. G.; Ley H. G.; Scholz R. Eur. J. Biochem. 1972, 25, 420. [DOI] [PubMed] [Google Scholar]
- Taylor J. E.; Yan J. F.; Wang J. L. J. Am. Chem. Soc. 1966, 88, 1663. [DOI] [PubMed] [Google Scholar]
- Paulsen C. E.; Carroll K. S. Chem. Rev. 2013, 113, 4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.