Abstract
Genetic diversity, and thus the adaptive potential of invasive populations, is largely based on three factors: patterns of genetic diversity in the species' native range, the number and location of introductions and the number of founding individuals per introduction. Specifically, reductions in genetic diversity (‘founder effects') should be stronger for species with low within-population diversity in their native range and few introductions of few individuals to the invasive range. We test these predictions with Geranium carolinianum, a winter annual herb native to North America and invasive in China. We measure the extent of founder effects using allozymes and microsatellites, and ask whether this is consistent with its colonization history and patterns of diversity in the native range. In the native range, genetic diversity is higher and structure is lower than expected based on life history traits. In China, our results provide evidence for multiple introductions near Nanjing, Jiangsu province, with subsequent range expansion to the west and south. Patterns of genetic diversity across China reveal weak founder effects that are driven largely by low-diversity populations at the expansion front, away from the introduction location. This suggests that reduced diversity in China has resulted from successive founder events during range expansion, and that the loss of genetic diversity in the Nanjing area was mitigated by multiple introductions from diverse source populations. This has implications for the future of G. carolinianum in China, as continued gene flow among populations should eventually increase genetic diversity within the more recently founded populations.
Keywords: Geranium carolinianum, invasive, allozymes, microsatellites, genetic structure, founder effects
Introduction
Research on mechanisms underlying biological invasions has long focused on the genetic diversity of invasive species. Such studies generally measure neutral genetic diversity in invasive populations that is used as a proxy for adaptive potential. Population genetic theory predicts that colonization can result in reduced genetic diversity, known as founder effects; this in turn may lower the evolutionary potential and ultimately the ability of colonizing populations to establish and proliferate. Yet, human-mediated invasions that are, by definition, the result of recent colonization can experience rapid range expansion after only a short time in their new ranges. Reviews and meta-analyses of population genetic studies of invasive species (see, for example, Novak and Mack, 2005; Wares et al., 2005; Dlugosch and Parker, 2008) provide insights into this apparent paradox: that invasions often experience reduced founder effects or none at all.
Colonizing populations are predicted to undergo reductions in genetic diversity because low numbers of founding propagules generally represent only a limited amount of the genetic variation present in source populations (Nei et al., 1975). The severity of founder effects are largely a function of the number and size of introductions, which limit propagule pressure and gene flow with the native range (Simberloff, 2009). The distribution of genetic variation (genetic structure) in the native range, which is influenced by life history characters such as mating system, may also play a large role in determining the proportion of within-species genetic diversity carried by founding propagules (Novak and Mack, 1993; Hamrick and Godt, 1996). For example, if native populations are largely undifferentiated (that is, have low genetic structure), much of the overall diversity is contained within populations and potentially could be transported to the invasive range during a single introduction event, reducing founder effects. Conversely, if native populations are highly differentiated (high genetic structure), a smaller proportion of the native alleles are available to be introduced into the invasive range, enhancing founder effects. Larger founding populations increase initial genetic diversity, and multiple introductions from distinct genetic sources can result in higher diversity in invasive populations relative to native populations (Novak and Mack, 1993).
A clear understanding of the introduction patterns, expansion and gene flow during biological invasions is crucial for making informed management decisions (see, for example, Rollins et al., 2009), especially for species that are transported accidentally via traffic or commerce. Genetic analyses are often the only way to disentangle invasion histories, as historical, observational records on the timing and location of introductions are often lacking and/or misleading (Estoup and Guillemaud, 2010). If known, the colonization history of a particular species can be used to inform a wide range of further work, such as understanding mechanisms of invasion and applications to basic evolutionary theory, as well as control and management programs. For example, such information can be used to test whether phenotypic and/or genetic differences between invasive and native populations are because of adaptive or neutral evolutionary processes (Keller and Taylor, 2008). From a management perspective, an understanding of invasion pathways and genetic connectivity among invasive populations can be useful in designing effective control programs (Rollins et al., 2009) and can help to prevent or limit the introduction and dispersal of similar species by identifying potential routes of introduction and dispersal.
Although invasive species are a global problem, studies based in developing countries in Asia and Africa are underrepresented in the literature (Pyšek et al., 2008). Insufficient research in these geographic regions may prevent many countries from developing effective screening, quarantine and management programs, increasing the potential for economic losses and the invasion of natural areas. Although developing countries tend to harbor fewer invasive species than developed countries because of their relative isolation from the global marketplace (Vilà and Pujadas, 2001), this will likely change as globalization increases overseas trade and, subsequently, the number of accidental introductions.
China, as an emerging global economic power, exemplifies a rapidly developing nation at high risk for new species invasions (Weber and Li, 2008). Trade between China and the United States is rising, and the similar climates of the two countries increases the likelihood that accidental introductions will result in unwanted naturalization and/or invasion of new species (Jenkins and Mooney, 2006). Currently, over half of the invasive plant species in China are native to the Americas (Xu et al., 2006; Weber et al., 2008), and they have already generated serious economic impacts. In 2000, invasive species in China were responsible for US $14 billion in economic losses, or 1.3% of China's gross domestic product (Xu et al., 2006).
Geranium carolinianum L., or Carolina cranesbill, is a weedy winter annual plant native to North America and invasive in East Asia. It has been naturalized in China for almost 90 years, and historical records suggest a single introduction point in or near Nanjing, Jiangsu province. In this study, we address the following questions: Do invasive populations exhibit founder effects? Are the extent of founder effects consistent with the colonization history in China and the genetic structure of native populations? We sampled 40 native and invasive populations to characterize population genetic patterns in its native range, and to reconstruct its introduction history within China. In addition, we assess two classes of biparentally inherited neutral molecular markers with differing mutation rates—allozymes and microsatellites—for their utility in detecting genetic signatures of demographic events associated with colonization and range expansion.
Materials and methods
Species
G. carolinianum (Geraniaceae) is a weedy winter annual herb native to North America. It occurs throughout the continental United States and into Canada; however, most reports are from the mid-Atlantic and southeastern United States (south of 40 °N latitude) west to the Rocky Mountains (Aedo, 2000). In the southeastern United States, where population densities are high, it is a common weed of roadsides, lawns, fields and disturbed areas. In China, G. carolinianum is the only Geranium species listed as invasive, although to date it has had minor environmental impact (Liu et al., 2006). It is reported in 11 provinces in the eastern plains region (Weber et al., 2008) and is largely a roadside and agricultural weed. Early herbarium records in China are from Nanjing (1926), with subsequent reports in the 1930s and 1940s (Chinese Academy of Sciences, 2012). Herbarium records from other cities—Shanghai, Suzhou and Nanchang—do not appear until 1949–1950. Although it has been reported as far north as Saskatchewan, Canada, in North America (49 °N latitude), in China the northern limit of G. carolinianum is 33 °N latitude (Aedo, 2000), despite heavy connectivity by rail and car traffic between colonized areas and the uncolonized, northern regions. G. carolinianum germinates in the fall, overwinters as a rosette and flowers in spring and early summer. Flowers are small (∼5 mm across), insect pollinated and self-compatible (Fiz et al., 2008). Fruits are dry, 5-seeded schizocarps with explosive dispersal.
Collection sites
Collections of ripe fruit and/or leaf tissue were made in May–June 2009 and 2010 from natural populations in the United States and China (Figure 1 and Supplementary Table S1). Population sizes at sampling locations ranged from >50 to several thousand individuals. Ripe fruits and/or leaf tissue were collected from 30 or more plants per population. Seeds were pooled by maternal family and stored in paper envelopes at room temperature until used for genetic analyses; leaf tissue was dried on silica gel and stored at room temperature. In the southeastern United States, collection sites were spaced at least 100 km apart. In China, most collection sites were selected in a hierarchical scheme because of transportation limitations. Major stops (cities) were selected across the range, generally separated by ⩾200 km (Figure 1b). Within cities, one to four populations were sampled at least 2 km apart. In China, 24 populations were sampled from 15 cities and 8 provinces, covering 800 km east/west and 670 km north/south. A total of 21 populations were sampled across 9 southeastern and mid-Atlantic states in the United States, covering ∼1800 km east/west and 1000 km north/south. Locations and sample sizes are given in Supplementary Table S1.
Figure 1.
Sampling locations in native (a) and invasive (b) ranges used for population genetic analysis with allozymes and/or microsatellites. Large open circles mark collection sites. Small filled circles mark major cities in China.
Allozyme extraction and genotyping
All 40 populations with seed collections were used for allozyme genotyping. Because allozymes extracted from mature leaf tissue express poorly in this species (RY Shirk, unpublished data), we were unable to genotype individuals from populations with only leaf tissue samples. For populations with seed collections, 23–55 seeds (mean=38) per population, each from a different maternal family, were scarified with a razor blade and germinated in water for genotyping. At 1 to 2 weeks after germination, when cotyledons were expanded but before true leaves had initiated or expanded, entire seedlings were crushed with the extraction buffer of Wendel and Parks (1982). Allozyme extract was absorbed onto 4 × 6 mm filter paper wicks (Whatman 3 chromatography paper, Piscataway, NJ, USA) and stored at −80 °C until use. Extractions were run on 10% starch gel electrophoresis and genotyped at 28 well-resolved loci. Buffer systems and stains were based on those in Soltis et al. (1983) except where noted: Leucine aminopeptidase (Lap-3), menadione reductase (Mnr-1, Mnr-2; Wendel and Weeden, 1989) and phosphoglucoisomerase (Pgi-2) on buffer system 34; diaphorase (Dia-1; Manchenko, 1994), aspartate aminotransferase (Aat-1, Aat-2) and triosephosphate isomerase (Tpi-1, Tpi-3) on buffer system 8; malate dehydrogenase (Mdh-1, Mdh-2, Mdh-4), fructose-1, 6-diphosphatase (F1,6-1) and adenylate kinase (Ak-3; Wendel and Weeden, 1989) on buffer system 11; and aconitase (Aco-1, Aco-2, Aco-3, Aco-4), aldolase (Ald-2), 6-phosphogluconate dehydrogenase (6P-1, 6P-2), UTP-glucose-1-phosphate (Ugpp-1, Ugpp-2, Ugpp-3, Ugpp-4; Manchenko, 1994), shikimate dehydrogenase (Skdh-1) and isocitrate dehydrogenase (Idh-1, Idh-2) on buffer system 4. In total, 698 individuals from 16 US populations and 856 individuals from 24 Chinese populations were genotyped (Supplementary Table S1).
DNA extraction and microsatellite genotyping
A total of 30 populations were sampled for genotyping at 6 microsatellite loci (Shirk et al., 2013) for comparison with the allozyme data set. We selected 15 invasive populations that spanned the sampled range in China (that is, only one population per city) and 15 native populations that included the 5 populations with leaf tissue collections that we were unable to use for allozyme genotyping (NC-2, TN-1, TX-1, VA-1 and VA-2) and 10 others that spanned the sampled range in the United States (Supplementary Table S1). Total genomic DNA was extracted from 24 individuals per population using either field-collected leaf tissue or seedling tissue germinated from field-collected seeds with a modified CTAB (cetyltrimethylammonium bromide) protocol (Doyle and Doyle, 1990) and amplified using a 3-primer touchdown PCR for G. carolinianum primers GC10, GC38, GC29, GC31, GC39 and GC47. For PCR reaction conditions, see Shirk et al. (2013). PCR products were analyzed on a 3730xl capillary sequencer (Applied Biosystems, Carlsbad, CA, USA) using a ROX-labeled internal size standard (GGF500R, Georgia Genomics Facility, Athens, GA, USA). Allele calls from chromatograms were made using GeneMapper v3.7 (Applied Biosystems) and confirmed by visual inspection. In total, 329 individuals from 15 US populations and 291 individuals from 15 Chinese populations were genotyped at 6 microsatellite loci. Before statistical analysis, the microsatellite data were assessed for scoring errors using Micro-checker (Van Oosterhout et al., 2004). Null allele frequency was estimated for each locus within each population using FreeNA (Chapuis and Estoup, 2007).
Data analyses
For all analyses, microsatellite and allozyme data sets were treated separately. Note that because entire seedlings were needed for DNA and allozyme extractions, we were unable to genotype the same individuals for populations that were sampled in both data sets. For these populations, maternal families sampled for microsatellite genotyping are a random subset of the families represented in the allozyme data set.
Genetic diversity
Standard population genetic statistics were calculated for each population and country using GenAlEx v6.41 (Peakall and Smouse, 2006) and FSTAT (Goudet, 1995): percent polymorphic loci (%P), allelic richness (Rs), private alleles (PA), observed (Ho) and expected (He) heterozygosity and the inbreeding coefficient (F). For the allozyme data set, Pgi-2, Ugpp-4, Mdh-2, Aat-2 and 6P-2 were not used to calculate the inbreeding coefficient (F). Both Pgi-2 and Ugpp-4 appeared to be duplicated loci and, as a result, individuals were scored for four alleles each. Mdh-2 was scored as dominant/recessive because of overlapping bands from a monomorphic, unscored locus that made heterozygotes indistinguishable from homozygotes. Aat-2 and 6P-2 had unusually high heterozygote frequencies in Chinese populations, perhaps also because of duplications, and hence were not used in calculating F-values. These loci were retained for genetic diversity calculations as the allele frequency estimates calculated for these loci are accurate.
To test for founder effects, we compared diversity statistics between the United States and China. Differences in mean population Rs and He between native (United States) and invasive (China) populations were assessed with permutation tests in FSTAT. Percent polymorphic loci were averaged across loci and populations and compared across countries with a two-sample t-test. Differences in total Rs and He (populations pooled by country) were compared across loci using paired t-tests. To test for founder effects during range expansion in China, He and Rs of the Chinese populations were regressed against distance from Nanjing (the putative introduction point) using single linear regressions implemented in R (R Development Core Team, 2012). In addition, we used BOTTLENECK (Cornuet and Luikart, 1996; Piry et al., 1999) to test for evidence of founder effects in each Chinese population separately, based on signatures of excess heterozygosity and mode shifts in allele frequencies. BOTTLENECK was run separately for allozymes and microsatellites, under the infinite allele model with 1000 replicates each.
Genetic structure
To examine the distribution of genetic diversity across the entire study, we performed hierarchical analyses of molecular variance in GenAlEx. Total genetic variation was partitioned between countries, among populations within each country and among all populations (global FRT, FSR and FST, respectively), and within and among populations for each country separately (US FST and China FST). Differences between US and China FST values were tested using a permutation test in FSTAT. Mantel tests comparing geographic distance and FST/(1−FST) (Rousset, 1997) were used to assess isolation by distance in GenAlEx. In addition, to provide a more in-depth analysis of patterns of genetic diversity in China and to assess whether the hierarchical sampling scheme would bias estimates of genetic structure, we performed a second analysis of molecular variance on Chinese populations to partition variance within and among cities.
We further investigated genetic structure using the Bayesian clustering program STRUCTURE (Pritchard et al., 2000). For both data sets, cluster membership coefficients were estimated using the correlated allele frequencies model and 500 000 Markov chain Monte Carlo iterations after 200 000 burn-in. The optimal number of clusters (K) was estimated using the delta K method described by Evanno et al. (2005) and implemented in Structure Harvester (Earl, 2011). Once optimal K was determined, cluster membership coefficients were aligned across 10 replicate runs using CLUMPP (Jakobsson and Rosenberg, 2007) and plotted in Distruct (Rosenberg, 2004). We also attempted to use the program InStruct (Gao et al., 2007) to infer clustering. InStruct is an extension of STRUCTURE that eliminates the assumption of random mating and Hardy–Weinberg equilibrium within clusters and thus may be more suitable for self-compatible species such as G. carolinianum.
Finally, to visualize relationships among all invasive and native populations, we performed a principal coordinates analysis (PCoA) in GenAlEx based on pairwise population Cavalli-Sforza chord measures (Cavalli-Sforza and Edwards, 1967). Analyses were run separately for the allozyme and microsatellite data.
Results
Genetic diversity
Five allozyme loci were monomorphic across both countries: Mdh-1, 6P-1, Ugpp-1, Idh-1 and Idh-2. Four additional loci were monomorphic in China: Aco-1, Aco-2, Ugpp-2 and Tpi-3, resulting in 19 polymorphic allozyme loci in China and 23 polymorphic loci in the United States. Monomorphic loci were included when calculating genetic diversity statistics.
All microsatellite loci were polymorphic in both countries. Roughly half (94 of 180) of the possible locus-by-population combinations had no evidence for null alleles or null alleles estimated to be at low frequency (<0.05); 57 had null alleles estimated to be at intermediate frequency (0.05–0.20); and 29 had an estimated null allele frequency between 0.20 and 0.38. On average, null alleles were at a somewhat lower frequency in Chinese populations (mean=0.06) than in the US populations (mean=0.12). Four populations had null alleles estimated to be absent or at low frequency across all loci: VA-1, CN-8, CN-9 and CN-17.
Null alleles were identified by testing for heterozygote deficiencies in FreeNA. Many populations in both the United States and China had few or several loci with null alleles estimated to be at intermediate or high frequencies, a pattern consistent with true null alleles. Inbreeding will also result in heterozygote deficiency, but should affect all loci equally. Because G. carolinianum is self-compatible, it is likely that the null allele signal in our data was inflated by self-fertilization. Mating system analyses based on allozyme data show that G. carolinianum populations vary in their outcrossing rate, and US populations tend to be more highly selfing than Chinese populations (RY Shirk and JL Hamrick, in review), that is consistent with a higher apparent null allele frequency found in the United States as compared with China.
Genetic diversity statistics consistently reveal the signature of founder effects in China, both at the population and regional levels. With populations pooled by country, China has lower diversity than the United States as measured by He and Rs for both the allozyme and microsatellite data sets, and the differences are significant for all comparisons except microsatellite heterozygosity (allozyme total He: China=0.152; United States=0.222, P=0.006; allozyme total Rs: China=58.37, United States=72.96, P=0.005; microsatellite total He: China=0.290; United States=0.525, P=0.064; microsatellite total Rs: China=18.26, United States=37.54, P=0.003). There are also more private alleles in the United States relative to China (11 vs 0 for allozymes; 21 vs 0 for microsatellites). At the within-population level, Chinese populations have on average fewer polymorphic loci and significantly lower He and Rs values than US populations for both marker types (allozyme mean %P: China=42.4, United States=61.6, P<0.0001; allozyme mean He: China=0.134, United States=0.178, P=0.001; allozyme mean Rs: China=38.84, United States=42.81, P=0.001; microsatellite mean He: China=0.222; United States=0.324, P=0.009; microsatellite mean Rs: China=7.32, United States=7.92, P=0.005). Mean observed heterozygosity is equivalent in both countries for allozymes (China Ho=US Ho=0.162).
The average inbreeding coefficient F calculated from the allozyme data is slightly positive in US populations (0.110) and slightly negative in Chinese populations (−0.039), although F is not significantly different from 0 for most Chinese populations (Supplementary Table S2). One exception is population CN-14, which has F=−0.163. It is possible that this population, which appears to be newly founded in a weedy, roadside area, consists of a high proportion of F1 individuals that would show increased heterozygosity if the founding propagules had different genetic backgrounds.
Because null alleles reduce observed heterozygosity, we do not report inbreeding coefficients for the microsatellite data set for populations with null alleles estimated to be at intermediate or high frequencies (>0.05). The four populations with low estimated null allele frequencies across all loci all had negative inbreeding coefficients: VA-1, −0.142 (s.e. 0.039); CN-8, −0.166 (s.e. 0.080); CN-9, −0.144 (s.e. 0.073); and CN-17, −0.315 (s.e. 0.094). Summary statistics by country are given in Table 1. Population-level values are in Supplementary Tables S2 and S3.
Table 1. Summary statistics by country for allozyme and microsatellite data.
| Region/marker | %P | Rs | PA | Ho | He | F |
|---|---|---|---|---|---|---|
| (a) Allozymes | ||||||
| China mean | 42.39 (1.65) | 38.84 (0.47) | 0.167 | 0.162 (0.010) | 0.134 (0.008) | −0.039 (0.003) |
| China total | 65.52 | 58.37 | 0 | — | 0.152 (0.038) | — |
| US mean | 61.64 (1.13) | 42.81 (0.47) | 0.25 | 0.162 (0.010) | 0.178 (0.010) | 0.110 (0.023) |
| US total | 82.76 | 72.96 | 11 | — | 0.222 (0.040) | — |
| (b) Microsatellites | ||||||
| China mean | 73.3 (3.56) | 7.32 (0.06) | 0 | 0.167 (0.023) | 0.222 (0.020) | — |
| China total | 100 | 18.26 | 0 | — | 0.290 (0.083) | — |
| US mean | 83.3 (4.60) | 7.92 (0.18) | 0.6 | 0.173 (0.020) | 0.324 (0.023) | — |
| US total | 100 | 37.54 | 21 | — | 0.525 (0.060) | — |
Abbreviations: F, fixation index; He, expected heterozygosity; Ho, observed heterozygosity; %P, percent polymorphic loci; PA, private alleles; Rs, allelic richness.
Standard errors are in parentheses.
BOTTLENECK identified founder effects in several Chinese populations based on the allozyme data. Seven populations had a significant excess of heterozygotes based on both the sign test and the Wilcoxon signed-rank test, whereas the eighth population was significant for the Wilcoxon signed-rank test and marginal for the sign test. Six populations had a mode shift in their allele frequency distribution, indicating a recent bottleneck (Table 2).
Table 2. Results of tests for recent bottlenecks in Chinese populations: P-values from sign tests and Wilcoxon signed-rank tests for excess heterozygosity across loci; and mode shifts in allele frequency distributions.
| Population |
Allozymes |
Microsatellites |
||||
|---|---|---|---|---|---|---|
| Sign | Wilcoxon | Mode shift? | Sign | Wilcoxon | Mode shift? | |
| CN-1 | 0.164 | 0.068 | Yes | 0.367 | 0.109 | No |
| CN-10 | 0.307 | 0.160 | No | 0.681 | 0.563 | No |
| CN-11 | 0.031 | 0.032 | No | 0.044 | 0.031 | Yes |
| CN-12 | 0.031 | 0.015 | No | 0.538 | 0.406 | Yes |
| CN-13 | 0.046 | 0.006 | Yes | 0.397 | 0.844 | No |
| CN-14 | 0.050 | 0.008 | No | — | — | — |
| CN-15 | 0.571 | 0.207 | No | 0.318 | 0.906 | No |
| CN-16 | 0.329 | 0.103 | No | 0.210 | 0.969 | No |
| CN-17 | 0.080 | 0.065 | No | 0.733 | 0.250 | No |
| CN-18 | 0.057 | 0.087 | Yes | 0.566 | 0.438 | No |
| CN-19 | 0.234 | 0.151 | No | — | — | — |
| CN-2 | 0.231 | 0.188 | No | — | — | — |
| CN-20 | 0.075 | 0.053 | Yes | 0.623 | 0.688 | Yes |
| CN-21 | 0.054 | 0.040 | No | — | — | — |
| CN-22 | 0.026 | 0.009 | Yes | 0.492 | 0.500 | No |
| CN-23 | 0.152 | 0.120 | No | — | — | — |
| CN-24 | 0.154 | 0.014 | No | — | — | — |
| CN-3 | 0.026 | 0.013 | No | — | — | — |
| CN-4 | 0.105 | 0.086 | No | 0.589 | 0.891 | No |
| CN-5 | 0.215 | 0.072 | No | 0.620 | 0.156 | No |
| CN-6 | 0.209 | 0.080 | No | — | — | — |
| CN-7 | 0.229 | 0.149 | No | — | — | — |
| CN-8 | 0.562 | 0.160 | No | 0.625 | 0.688 | No |
| CN-9 | 0.044 | 0.014 | Yes | 0.573 | 0.688 | No |
P<0.05 are in bold. Simulations were performed under the infinite allele model separately for allozymes and microsatellites.
In contrast, BOTTLENECK only identified founder effects for a single Chinese population using microsatellite data based on excess heterozygosity, and three populations had a mode shift in their allele frequency distribution (Table 2). The discrepancy between the allozyme and microsatellite data sets may be due in part to differences in the number of loci sampled. For example, the Wilcoxon signed-rank test requires a minimum of four polymorphic loci to achieve P<0.05. Populations ranged from 9 to 16 polymorphic allozyme loci, but only had either 4 or 5 polymorphic microsatellite loci.
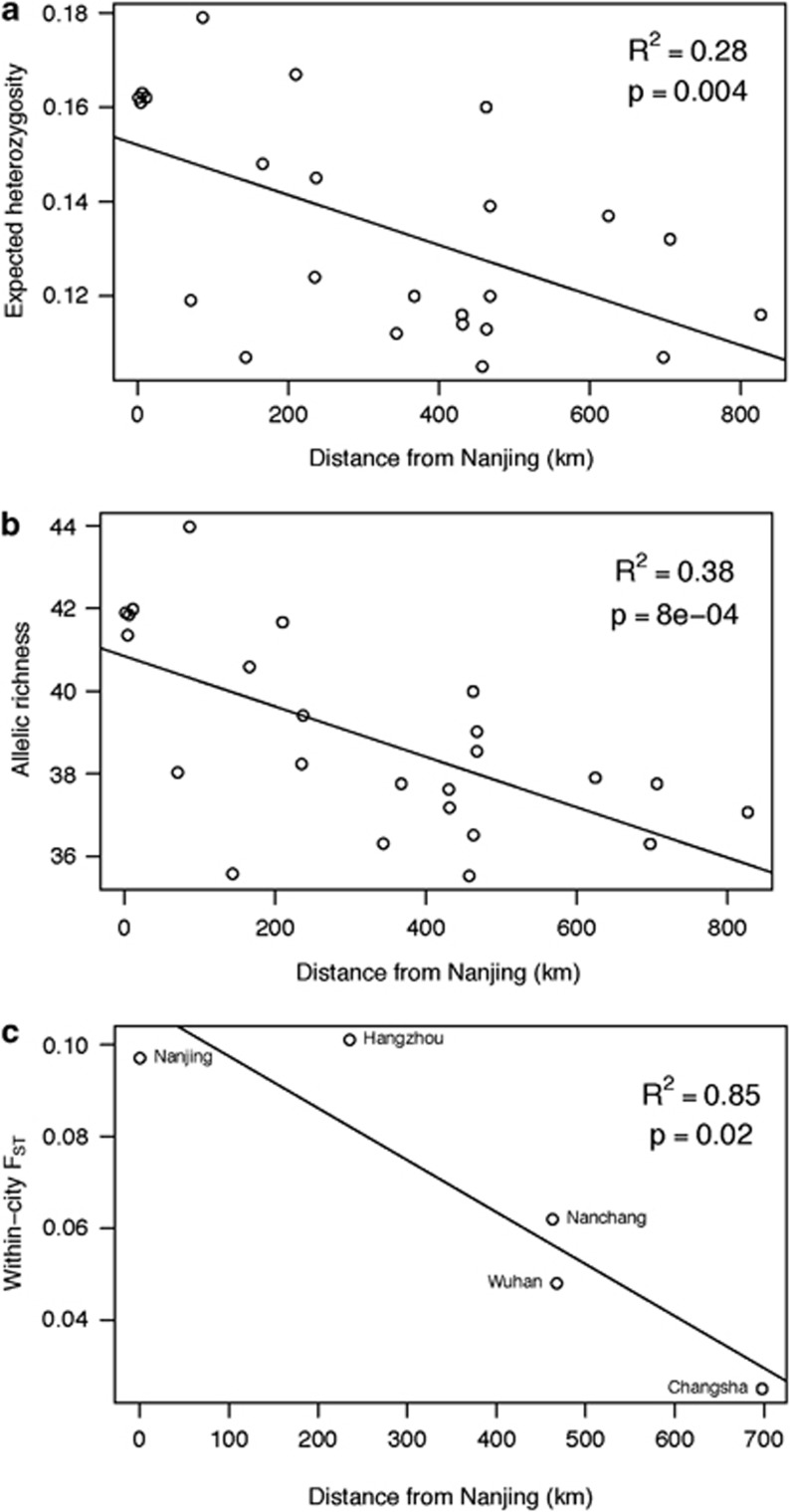
Patterns of genetic diversity within China support Nanjing as an introduction point, consistent with herbarium records. Distance from Nanjing is a significant predictor of both He and Rs for allozymes (He: R2=0.282, P=0.004; Rs: R2=0.377, P=0.0008; Figure 2) but not microsatellites (He: R2=0.018, P=0.629; Rs: R2=0.012, P=0.701). When the allozyme data set is reduced to only the 15 populations shared with the microsatellite data set, only the regression with Rs approaches significance (He: R2=0.122, P=0.20; Rs: R2=0.231, P=0.07). In addition, most populations identified by BOTTLENECK as having a signature of founder effects are located in the western part of the sampled area, away from Nanjing. Thus, the allozyme data support a scenario of serial founder effects as the range of G. carolinianum expanded from Nanjing.
Figure 2.
Correlation between population estimates of allozyme genetic diversity (expected heterozygosity and allelic richness) and within-city population differentiation in invasive populations, and distance from the putative introduction point in Nanjing.
Genetic structure
Both allozymes and microsatellites have significant genetic structure at all hierarchical levels (P<0.001; Table 3). Although structure is greater with microsatellites, a general pattern is consistent across data sets. Divergence between countries is low (allozyme FRT=0.055; microsatellite FRT=0.111). Within countries, structure is significantly higher in the United States than in China (allozyme US FST=0.206 and China FST=0.100; microsatellite US FST=0.338 and China FST=0.209). There is weak but significant isolation by distance in both countries for allozymes (China, R2=0.025, P=0.05; United States, R2=0.120, P=0.03) but none in either country for microsatellites (China, R2=6e−06, P=0.46; United States, R2=0.003, P=0.50).
Table 3. Results of a hierarchical analysis of molecular variance (AMOVA) partitioning variance between countries, among invasive populations in China, and among native populations in the United States.
|
Global |
China |
US |
||||
|---|---|---|---|---|---|---|
| % Variance | P-value | % Variance | P-value | % Variance | P-value | |
| (a) Allozymes Between countries (FRT) | 0.055 | 0.001 | — | — | — | — |
| Among populations within countries (FSR) | 0.156 | 0.001 | — | — | — | — |
| Among populations (FST) | 0.202 | 0.001 | 0.100 | 0.001 | 0.206 | 0.001 |
| (b) Microsatellites | ||||||
| Between countries (FRT) | 0.111 | 0.001 | — | — | — | — |
| Among populations within countries (FSR) | 0.260 | 0.001 | — | — | — | — |
| Among populations (FST) | 0.370 | 0.001 | 0.209 | 0.001 | 0.338 | 0.001 |
Significance of F-statistics was tested with 999 permutations. (a) Allozyme data and (b) microsatellite data.
To further understand colonization and range expansion in China, we assessed genetic structure within and among cities in China. Because of the hierarchical sampling scheme in the allozyme data set, five cities in China—Nanjing, Hangzhou, Wuhan, Nanchang and Changsha—are represented by multiple populations. Within China, genetic structure was significant both among cities (FRT=0.033, P<0.001) and among populations within cities (FSR=0.071, P<0.001). Because variance among populations within cities accounted for ∼70% of the total among-population variation in China, it is unlikely that the hierarchical population sampling scheme was a source of bias in comparisons with the US populations.
The degree of genetic structure within cities was not consistent across China. Within-city FST values range from 0.025 (Changsha) to ∼0.100 (Nanjing and Hangzhou). The degree of within-city genetic structure is also strongly negatively associated with distance from Nanjing (within-city FST regressed against city distance from Nanjing, R2=0.85, P=0.02; Figure 2). Thus, founding events during range expansion reduced genetic diversity both within populations and within cities in China.
Dense sampling within Nanjing (five populations) allowed us to examine patterns within the hypothesized area of introduction. Pairwise differentiation among the five Nanjing populations was highly varied; both the highest and lowest pairwise FST (0.005 and 0.215) values across all Chinese populations were found within Nanjing. The comparatively high differentiation among populations within Nanjing suggests strong founder effects when individual populations were established and/or multiple introductions from divergent sources.
Bayesian clustering analysis using STRUCTURE was performed on the allozyme and microsatellite data sets separately. Because the analysis of molecular variance showed very low genetic divergence between countries, particularly in the allozyme data, we ran STRUCTURE both with and without the LOCPRIOR model. LOCPRIOR uses population information for each individual to assist the clustering algorithm when the genetic signal is weak (Hubisz et al., 2009). Here, the priors consisted of population identity, but not regional identity or geographic location.
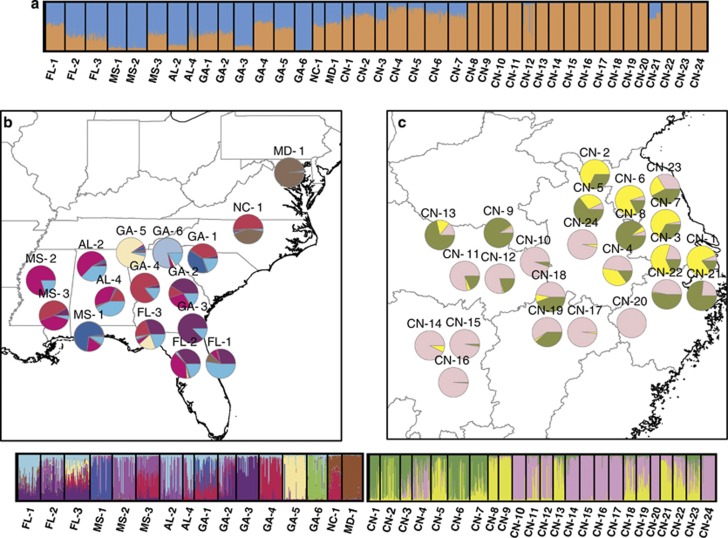
For the global allozyme data set, STRUCTURE identified K=2 as the optimal number of clusters (ΔK=114.99). Although many populations had intermediate membership in each cluster, the US and Chinese populations generally had majority membership in opposite clusters. This division was clearer using the LOCPRIOR model (Figure 3a) than without (data not shown). Because STRUCTURE tends to find the highest level of structure present (Evanno et al., 2005), two subsequent analyses were performed to investigate structure among the US and Chinese populations. Across the 16 US populations, optimal K was 8 (ΔK=12.35; Figure 3b), and across the 24 Chinese populations, optimal K was 3 (ΔK=22.48; Figure 3c).
Figure 3.
STRUCTURE results (allozyme data only) for (a) the global data set (K=2); (b) US subset (K=8); and (c) China subset (K=3).
For the global microsatellite data set, STRUCTURE resolved K=3 (ΔK=4.69) as the optimal number of clusters. Including LOCPRIOR for the microsatellite data set had no qualitative effect on clustering, and hence it was omitted from the model. Because the analysis did not clearly group populations by geographic location, no subsequent analyses were performed. We do not report clustering results from InStruct because we were unable to attain convergence with the global allozyme data set when estimating K, even with long runtimes (1 000 000 iterations after 500 000 burn-in).
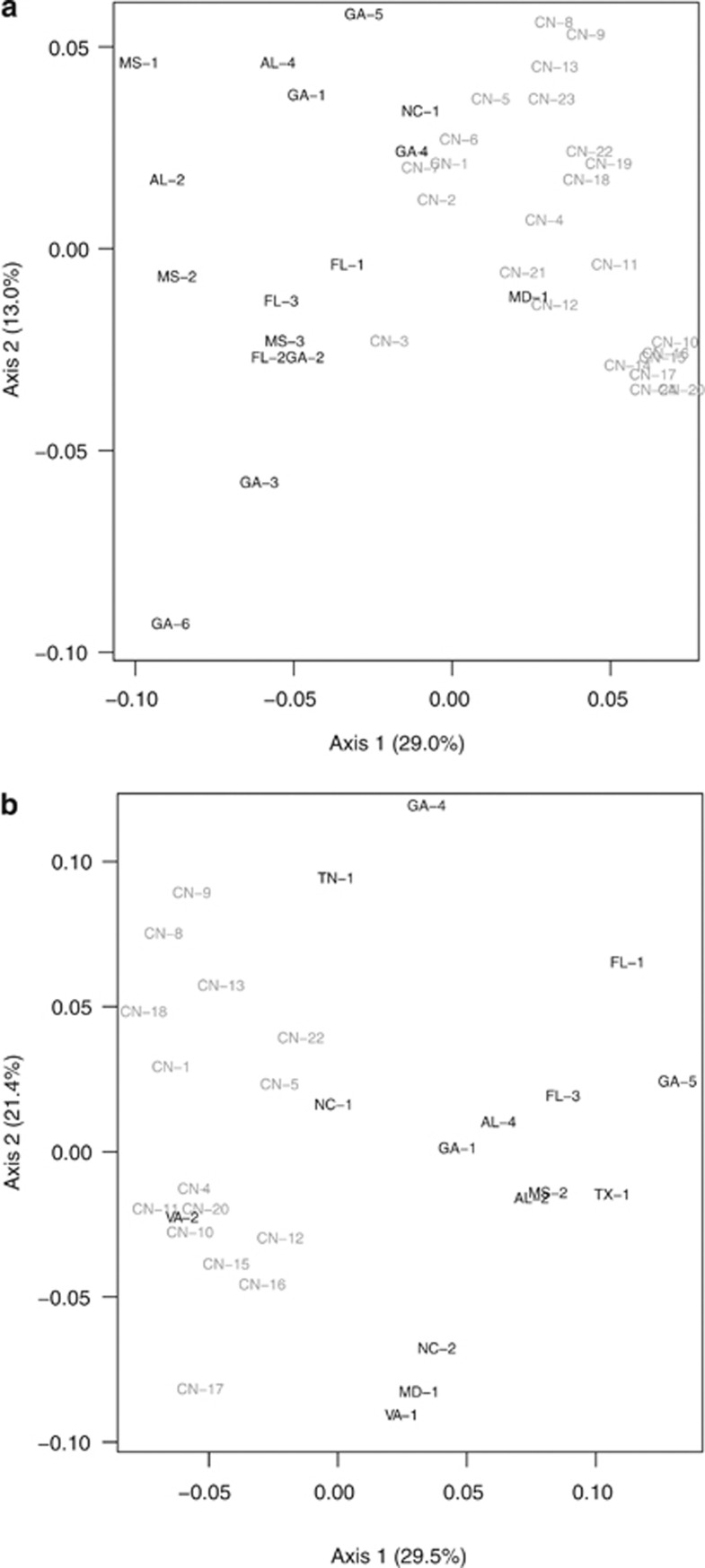
In the PCoA, the US and Chinese populations largely separated along the first axis, which explained 29.0% of the allozyme variance and 29.5% for microsatellites. The second axis explained 13.0% of the allozyme variance and 21.4% of the microsatellite variance (Figure 4). There was more overlap between the United States and China in the allozyme PCoA, and the US populations GA-4, MD-1 and NC-1 were similar to Chinese populations. This is consistent with the global STRUCTURE results (Figure 3a), where these same populations had intermediate membership in both clusters. Chinese populations also grouped in a pattern consistent with the China STRUCTURE results (Figure 3c): populations in the green cluster had more positive values on axis 2, and the pink cluster had more negative values on axis 2. The yellow cluster, which was only found in populations near and around Nanjing (CN-1, CN-2, CN-3, CN-6 and CN-7), were most similar to the US populations (more negative values on axis 1). In the microsatellite PCoA, only VA-2 and NC-1 grouped with the Chinese populations.
Figure 4.
PCoA of pairwise population genetic distances derived from (a) allozyme and (b) microsatellite data. The US populations are in black and the Chinese populations are in gray.
Discussion
Genetic diversity of invasive populations is predicted to be heavily influenced by three factors: genetic structure in the native range, the number of introductions and the number of founding individuals per introduction. We surveyed genetic diversity across 40 native and invasive populations of G. carolinianum, reconstructed the colonization history in China and tested for the signature of founder effects to test the generality of these predictions. Three major points arose. (1) Founder effects, although statistically significant, are not severe. On average, Chinese populations contain 75% of the heterozygosity and 85% of the allozyme allelic diversity present in the US populations. (2) This is consistent with the introduction history of G. carolinianum in China and patterns of diversity in the United States. (3) Successive founding events during range expansion sequentially decreased within-population diversity in populations away from the point of introduction.
The proportion of native genetic diversity introduced by colonizing propagules, especially in cases of single introductions, is dependent on the genetic structure of native populations. If a high proportion of the total diversity in the native range is contained within individual populations (that is, low FST), founder effects will be mitigated even with a low number of colonizing individuals (for example, Epipactis helleborine, Squirrell et al., 2001). In its native range, G. carolinianum has higher allozyme genetic diversity and less genetic structure than expected when compared with similar species, with 80% of the total genetic variation residing within populations (Tables 1a and 3a). Hamrick and Godt (1996) calculated mean species-level estimates of genetic diversity and structure for mixed-mating annual plants as He=0.115 and GST=0.343. If sampling of G. carolinianum in the southeastern United States is representative of the entire native range, this suggests that although populations are genetically distinct, within-population diversity is high enough for founding propagules from a single native population to carry a fair amount of the total genetic diversity in a long-distance dispersal event.
Multiple introductions can ‘rescue' colonizing populations from founder effects (Dlugosch and Parker, 2008); in extreme cases, invasive populations are more diverse than native populations as a result of multiple introductions from genetically distinct source populations (for example, Bromus tectorum, Novak and Mack, 1993). Despite low divergence among Chinese populations, identification of three genetically distinct subgroups in clustering analyses (Figure 3c), as well as relatively high genetic structure within Nanjing, is consistent with a scenario of multiple introductions that mitigated founder events in the Nanjing area. Alternatively, this pattern could be explained by a single introduction from a diverse source population and/or repeated founder events during early stages of range expansion that could have increased divergence of these subgroups as G. carolinianum colonized China.
Our inference of the introduction history of G. carolinianum based on genetic data is in general agreement with the historical record. According to herbarium records, G. carolinianum has been present in China since at least 1926. However, it was not reported outside of the Nanjing area until 1950, implicating Nanjing as the location of its first introduction, and possibly as the source population(s) for subsequent range expansion. Nanjing, which was the capital of the Republic of China before 1949, probably experienced more frequent commercial exchange with the United States. During range expansion, successive founder events would erode genetic diversity as the invasive range expanded outward from the initial point of introduction. This pattern is observed in our allozyme data; eastern (Nanjing and surrounding) populations are the most diverse, and allelic richness and expected heterozygosity decrease significantly with distance from Nanjing (Figure 2). Where multiple populations were sampled per city, we found that pairwise differentiation among Nanjing populations is highly varied, whereas outlying cities are more homogenous. Furthermore, we found a signature of founder effects in several outlying populations. STRUCTURE reveals patterns that are also consistent with a common, eastern entry point for all introduction events. Each invasive cluster contains populations in or near Nanjing. In the western part of the sampled range, there are genetic differences between the north and the south, suggesting two distinct routes of population expansion to the northwest and the southwest. Its pattern of spread northwestward follows the Jinghu and Longhai railways, whereas to the southwest it may have followed a water route along the Changjiang River. Similarly, Chinese populations most similar to the US populations in PCoA clustering were all in or around Nanjing.
The founder effects experienced by Chinese populations at the expansion front are unlikely to persist. Because G. carolinianum is a common agricultural and roadside weed, human-mediated secondary dispersal likely plays a large part in the contemporary patterns of gene flow and genetic structure in this species. This is evident in the native range in the lower-than-expected genetic structure and the large number of geographically widespread distinct genetic groupings. Thus, we can expect that over time, similar patterns of gene flow will act to disperse alleles from the high-diversity Nanjing area to the range edge.
A small number of founders per introduction event also could reduce genetic diversity by limiting the amount of variation introduced, and also because genetic drift is exacerbated when effective population sizes are low (Nei et al., 1975). Although we could not estimate the number of founders, there are reasons to expect that founding populations were small to moderate. First, G. carolinianum is not of horticultural interest in China, and it is likely that introductions were accidental. Secondly, several low-frequency alleles present in the United States were not found in China. This implies that founding populations were not large enough to have contained or maintained the full range of allelic diversity in native populations.
Although not a main focus of this study, our genetic diversity data provide some information on potential sources of the Chinese populations. Depending on the markers used, our data predict that populations from Georgia, Maryland, North Carolina (allozymes) and Virginia (microsatellites) were most similar to the Chinese populations, and thus are putative sources of invasion in China. Correctly identifying source and recipient populations, however, requires comprehensive geographic sampling in the native range. Although G. carolinianum occurs throughout the continental United States, sampling was restricted to the southeastern, Gulf coast and mid-Atlantic United States, and hence it is possible that some or all of the source populations for the Chinese populations came from an unsampled US region with different patterns of genetic diversity and structure. This may have affected our ability to detect founder effects by comparing genetic diversity between countries. However, low divergence between the sampled US and Chinese populations and the lack of private alleles in China suggests that genetic variation observed in the invasive range is represented in the native populations we sampled. In addition, the inability of G. carolinianum to invade latitudes north of 33 °N in China suggests that source populations were located in warmer, southern regions. Further work with more highly variable markers and computational approaches (for example, approximate Bayesian computation; Keller et al., 2012) could be used to test different introduction and expansion scenarios and estimate founder population sizes.
The wide range of molecular markers available for population genetic studies naturally raises the issue of appropriate marker choice (Sunnucks, 2000). Some studies have recently combined data from markers with different patterns of inheritance to test for founder effects and determine introduction history (for example, maternally inherited chloroplast markers and nuclear microsatellites; Gaudeul et al., 2011). However, few studies have compared types of biparentally inherited markers with different mutation rates for their ability to detect genetic signatures of demographic events (but see Kohlmann et al., 2003; Caldera et al., 2008; Hawley et al., 2008). In particular, detection of founder effects has been of great interest in studies of biological invasions (Dlugosch and Parker, 2008). In a meta-analysis, Dlugosch and Parker (2008) compared studies using microsatellite markers and studies using allozymes to test for founder effects. Microsatellite studies showed greater proportional losses in allelic richness because a higher proportion of their alleles are at low frequency, although there were no differences in the magnitude of founder effects when diversity was measured as heterozygosity. This suggests that highly variable markers may be more sensitive for detecting genetic bottlenecks. However, marker comparison is rarely done within the same study, and hence the type II (false negative) error rate for detection of founder effects is difficult to assess. Although recent studies have used DNA-based markers (for example, microsatellites), allozyme-based studies still make up a large part of the literature on this subject because of the many years of allozyme-based genetic analysis before the introduction of microsatellite markers. For example, Dlugosch and Parker (2008) included 44 allozyme studies compared with 25 microsatellite studies.
In this study, we assessed the utility of two biparentally inherited marker types in detecting demographic events in invasive populations. Evidence for founder effects between regions was found for both marker types, but the signature of range expansion in China (tested by regression of within-population diversity against distance from Nanjing and assessing mode shifts and heterozygosity excess in individual populations) was only evident in the allozyme data set. It is possible, but unlikely, that a significant diversity–distance relationship could have been achieved by adding more populations to the microsatellite data set, as no patterns were detectable in the regressions using microsatellite data (P>0.6 and R2<0.02 for both He and Rs). Conversely, when the allozyme data set was reduced to only the 15 populations shared with the microsatellite data set, the negative trend was still present, although not statistically significant. Such low resolution in the microsatellite data set may have been influenced by both the low number of loci (6 compared with 19 polymorphic allozyme loci in China) and the relatively low levels of polymorphism found at some loci.
Highly polymorphic markers such as microsatellites often produce lower FST values as a result of high within-population heterozygosity (Hedrick, 1999). However, microsatellite FST values were higher than allozyme FST values at all hierarchical levels in this study (Table 3). Although within-population microsatellite diversity was higher than allozyme diversity, it was not so high as to artificially deflate FST. Geographic patterns based on the microsatellite data set (for example, isolation by distance; data not shown) did not change when population differentiation was estimated with the unbiased estimator G′ST (Hedrick, 2005), suggesting that any bias arising from using high-diversity markers did not affect the interpretation of our results. However, the presence of null alleles in the microsatellite data may have upwardly biased our FST estimates (Chapuis and Estoup, 2007).
The results of our marker comparison emphasize that it is essential to consider the number of loci available when choosing markers to assess genetic diversity. In our study, 6 microsatellite loci were insufficient to distinguish regional patterns evident with 28 lower-diversity allozyme loci. Although larger panels of loci are becoming the norm in studies of neutral genetic diversity, earlier studies reporting results calculated with small numbers of loci (and reviews and meta-analyses utilizing these data) should be treated with caution.
Conclusions
For good reason, much attention has been paid to prolific invaders with high environmental and/or economic impacts. There is growing evidence that introduced species can be extremely successful despite reductions in neutral genetic diversity, and that many successful invasives have reduced founder effects because of multiple introductions. Although this generally paints the picture that neutral genetic diversity and founder effects are not a strong predictor of establishment success and invasion, there are few studies available on ‘minor' invasives with relatively limited ecological impacts, such as G. carolinianum.
Reduced genetic diversity in Chinese invasive populations of G. carolinianum appears to be because of successive founder events during range expansion after the initial introduction event(s). Loss of diversity during initial colonization, although evident, appears to have been mitigated by moderate population differentiation in the native range and several independent introductions to the Nanjing area. In the future, founder effects observed on the range edges in China may be mitigated by subsequent gene flow among populations that appears to play a relatively large role in the population genetic dynamics of the native range of G. carolinianum. This study of neutral genetic diversity provides a foundation for future work on evolutionary processes in G. carolinianum, as well as investigations of invasion pathways in China.
Data archiving
Data available from the Dryad Digital Repository: http://doi.org/10.5061/dryad.15p8t.
Acknowledgments
This material is based upon work supported by the National Science Foundation's PIRE program under Grant No. OISE 0730218, with additional funding from the National Science Foundation Graduate Research Fellowship Program, the Oberlin College Alumni Fellowship and the Department of Plant Biology at the University of Georgia. We thank S-M Chang and C Deen for lab support, and R Mauricio, K Myhre, B Hu, D Christopher, T Kartzinel, M Poelchau, T Wenner and T Wang for travel and field assistance. Three anonymous reviewers provided helpful comments on the manuscript.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Aedo C. The genus Geranium L. (Geraniaceae) in North America. I. Annual species. Anales Jard Bot Madrid. 2000;51:39–82. [Google Scholar]
- Caldera E, Ross K, DeHeer C, Shoemaker D. Putative native source of the invasive fire ant Solenopsis invicta in the USA. Biol Invasions. 2008;10:1457–1479. [Google Scholar]
- Cavalli-Sforza LL, Edwards AW. Phylogenetic analysis. Models and estimation procedures. Am J Hum Genet. 1967;19:233–257. [PMC free article] [PubMed] [Google Scholar]
- Chapuis M-P, Estoup A. Microsatellite null alleles and estimation of population differentiation. Mol Biol Evol. 2007;24:621–631. doi: 10.1093/molbev/msl191. [DOI] [PubMed] [Google Scholar]
- Chinese Academy of Sciences 2012Chinese Virtual Herbarium ( www.cvh.org.cn ) Beijing, China [Google Scholar]
- Cornuet JM, Luikart G. Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics. 1996;144:2001–2014. doi: 10.1093/genetics/144.4.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dlugosch KM, Parker IM. Founding events in species invasions: genetic variation, adaptive evolution, and the role of multiple introductions. Mol Ecol. 2008;17:431–449. doi: 10.1111/j.1365-294X.2007.03538.x. [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. Isolation of plant DNA from fresh tissue. Focus. 1990;12:13–15. [Google Scholar]
- Earl DA.2011. Structure Harvester v0.6.1 http://taylor0.biology.ucla.edu/structureHarvester/ .
- Estoup A, Guillemaud T. Reconstructing routes of invasion using genetic data: why, how and so what. Mol Ecol. 2010;19:4113–4130. doi: 10.1111/j.1365-294X.2010.04773.x. [DOI] [PubMed] [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Fiz O, Vargas P, Alarcón M, Aedo C, García JL, Aldasoro JJ. Phylogeny and historical biogeography of Geraniaceae in relation to climate changes and pollination ecology. Syst Bot. 2008;33:326–342. [Google Scholar]
- Gao H, Williamson S, Bustamante CD. A Markov Chain Monte Carlo approach for joint inference of population structure and inbreeding rates from multilocus genotype data. Genetics. 2007;176:1635–1651. doi: 10.1534/genetics.107.072371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudeul M, Giraud T, Kiss L, Shykoff JA. Nuclear and chloroplast microsatellites show multiple introductions in the worldwide invasion history of common ragweed, Ambrosia artemisiifolia. PLoS One. 2011;6:e17658. doi: 10.1371/journal.pone.0017658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudet J. FSTAT (Version 1.2): A computer program to calculate F-statistics. J Hered. 1995;86:485–486. [Google Scholar]
- Hamrick JL, Godt MJW. Effects of life history traits on genetic diversity in plant species. Philos Trans R Soc Lond Ser B Biol Sci. 1996;351:1291–1298. [Google Scholar]
- Hawley DM, Briggs J, Dhondt AA, Lovette IJ. Reconciling molecular signatures across markers: mitochondrial DNA confirms founder effect in invasive North American house finches (Carpodacus mexicanus) Conserv Genet. 2008;9:637–643. [Google Scholar]
- Hedrick PW. Perspective: highly variable loci and their interpretation in evolution and conservation. Evolution. 1999;53:313–318. doi: 10.1111/j.1558-5646.1999.tb03767.x. [DOI] [PubMed] [Google Scholar]
- Hedrick PW. A standardized genetic differentiation measure. Evolution. 2005;59:1633–1638. [PubMed] [Google Scholar]
- Hubisz MJ, Falush D, Stephens M, Pritchard JK. Inferring weak population structure with the assistance of sample group information. Mol Ecol Res. 2009;9:1322–1332. doi: 10.1111/j.1755-0998.2009.02591.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson M, Rosenberg NA. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics. 2007;23:1801–1806. doi: 10.1093/bioinformatics/btm233. [DOI] [PubMed] [Google Scholar]
- Jenkins PT, Mooney HA. The United States, China, and invasive species: present status and future prospects. Biol Invasions. 2006;8:1589–1593. [Google Scholar]
- Keller SR, Gilbert KJ, Fields PD, Taylor DR. Bayesian inference of a complex invasion history revealed by nuclear and chloroplast genetic diversity in the colonizing plant, Silene latifolia. Mol Ecol. 2012;21:4721–4734. doi: 10.1111/j.1365-294X.2012.05751.x. [DOI] [PubMed] [Google Scholar]
- Keller SR, Taylor DR. History, chance and adaptation during biological invasion: separating stochastic phenotypic evolution from response to selection. Ecol Lett. 2008;11:852–866. doi: 10.1111/j.1461-0248.2008.01188.x. [DOI] [PubMed] [Google Scholar]
- Kohlmann K, Gross R, Murakaeva A, Kersten K. Genetic variability and structure of common carp (Cyprinus carpio) populations throughout the distribution range inferred from allozyme, microsatellite and mitochondrial DNA markers. Aquat Living Res. 2003;16:421–431. [Google Scholar]
- Liu J, Dong M, Miao SL, Li ZY, Song MH, Wang RQ. Invasive alien plants in China: role of clonality and geographical origin. Biol Invasions. 2006;8:1461–1470. [Google Scholar]
- Manchenko GP. Handbook of Detection of Enzymes on Electrophoretic Gels. CRC Press: Ann Arbor, MI; 1994. [Google Scholar]
- Nei M, Maruyama T, Chakraborty R. Bottleneck effect and genetic variability in populations. Evolution. 1975;29:1–10. doi: 10.1111/j.1558-5646.1975.tb00807.x. [DOI] [PubMed] [Google Scholar]
- Novak SJ, Mack RN. Genetic variation in Bromus tectorum (Poaceae): comparison between native and introduced populations. Heredity. 1993;71:167–176. [Google Scholar]
- Novak SJ, Mack RN.2005Genetic bottlenecks in alien plant species: influence of mating systems and introduction dynamicsIn: Sax DF, Stachowicz JJ, Gaines SD, (eds) Species Invasions: Insights into Ecology, Evolution, and Biogeography Sinauer Associates: Sunderland, MA; 201–228. [Google Scholar]
- Peakall R, Smouse PE. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes. 2006;6:288–295. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piry S, Luikart G, Cornuet JM. BOTTLENECK: A computer program for detecting recent reductions in the effective population size using allele frequency data. J Hered. 1999;90:502–503. [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyšek P, Richardson DM, Pergl J, Jarošík V, Sixtová Z, Weber E. Geographical and taxonomic biases in invasion ecology. Trends Ecol Evol. 2008;23:237–244. doi: 10.1016/j.tree.2008.02.002. [DOI] [PubMed] [Google Scholar]
- R Development Core Team . R Foundation for Statistical Computing. : Vienna, Austria; 2012. [Google Scholar]
- Rollins LA, Woolnough AP, Wilton AN, Sinclair R, Sherwin WB. Invasive species can't cover their tracks: using microsatellites to assist management of starling (Sturnus vulgaris) populations in Western Australia. Mol Ecol. 2009;18:1560–1573. doi: 10.1111/j.1365-294X.2009.04132.x. [DOI] [PubMed] [Google Scholar]
- Rosenberg NA. Distruct: a program for the graphical display of population structure. Mol Ecol Notes. 2004;4:137–138. [Google Scholar]
- Rousset F. Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics. 1997;145:1219–1228. doi: 10.1093/genetics/145.4.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simberloff D. The role of propagule pressure in biological invasions. Annu Rev Ecol Evol Syst. 2009;40:81–102. [Google Scholar]
- Shirk RY, Glenn TC, Chang S-M, Hamrick JL. Development and characterization of microsatellite primers in Geranium carolinianum (Geraniaceae) with 454 sequencing. Appl Plant Sci. 2013;1:1300006. doi: 10.3732/apps.1300006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis DE, Haufler CH, Darrow DC, Gastony GJ. Starch gel electrophoresis of ferns: a compilation of grinding buffer, gel and electrode buffers, and staining schedules. Am Fern J. 1983;73:9–27. [Google Scholar]
- Squirrell J, Hollingsworth PM, Bateman RM, Dickson JH, Light MHS, MacConaill M, et al. Partitioning and diversity of nuclear and organelle markers in native and introduced populations of Epipactis helleborine (Orchidaceae) Am J Bot. 2001;88:1409–1418. [PubMed] [Google Scholar]
- Sunnucks P. Efficient genetic markers for population biology. Trends Ecol Evol. 2000;15:199–203. doi: 10.1016/s0169-5347(00)01825-5. [DOI] [PubMed] [Google Scholar]
- Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P. micro-checker: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes. 2004;4:535–538. [Google Scholar]
- Vilà M, Pujadas J. Land-use and socio-economic correlates of plant invasions in European and North African countries. Biol Conserv. 2001;100:397–401. [Google Scholar]
- Wares JP, Hughes AR, Grosberg RK.2005Mechanisms that drive evolutionary change: insights from species introductions and invasionsIn: Sax DF, Stachowicz JJ, Gaines SD, (eds) Species Invasions: Insights into Ecology, Evolution, and Biogeography Sinauer Associates: Sunderland, MA; 229–257. [Google Scholar]
- Weber E, Li B. Plant invasions in China: what is to be expected in the wake of economic development. Bioscience. 2008;58:437–444. [Google Scholar]
- Weber E, Sun SG, Li B. Invasive alien plants in China: diversity and ecological insights. Biol Invasions. 2008;10:1411–1429. [Google Scholar]
- Wendel JF, Parks CR. Genetic control of isoenzyme variation in Camellia japonica L. J Hered. 1982;73:197–204. [Google Scholar]
- Wendel JF, Weeden NF.1989Visualization and interpretation of plant isozymesIn: Soltis DE, Soltis PS, (eds) Isozymes in Plant Biology Dioscorides Press: Portland, OR; 5–45. [Google Scholar]
- Xu HG, Ding H, Li MY, Qiang S, Guo JY, Han ZM, et al. The distribution and economic losses of alien species invasion to China. Biol Invasions. 2006;8:1495–1500. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.