Abstract
Despite extensive studies on cultivated rice, the genetic structure and subdivision of this crop remain unclear at both global and local scales. Using 84 nuclear simple sequence repeat markers, we genotyped a panel of 153 global rice cultivars covering all previously recognized groups and 826 cultivars representing the diversity of Chinese rice germplasm. On the basis of model-based grouping, neighbour-joining tree and principal coordinate analysis, we confirmed the widely accepted five major groups of rice cultivars (indica, aus, aromatic, temperate japonica and tropical japonica), and demonstrated that rayada rice was unique in genealogy and should be treated as a new (the sixth) major group of rice germplasm. With reference to the global classification of rice cultivars, we identified three major groups (indica, temperate japonica and tropical japonica) in Chinese rice germplasm and showed that Chinese temperate japonica contained higher diversity than that of global samples, whereas Chinese indica and tropical japonica maintained slightly lower diversity than that present in the global samples. Particularly, we observed that all seasonal, drought-tolerant and endosperm types occurred within each of three major groups of Chinese cultivars, which does not support previous claims that seasonal differentiation exists in Indica and drought-tolerant differentiation is present in Japonica. It is most likely that differentiation of cultivar types arose multiple times stemming from artificial selection for adaptation to local environments.
Keywords: population structure, diversity, classification, Oryza sativa , rice germplasm
Introduction
Asian cultivated rice (Oryza sativa L.) is one of the most important crops in the world. Rice not only serves as a primary food source for more than half of the world's population (Khush, 1997), but also provides an excellent model system for studying a wide range of biological questions (Shimamoto and Kyozuka, 2002; Wing et al., 2005; Zhang et al., 2008). As a result of both natural and artificial selection, rice has evolved into a crop containing high levels of morphological, physiological and genetic diversity, with >1 20 000 distinct rice varieties recognized worldwide (Oka, 1988; Khush, 1997; Sang and Ge, 2007; Vaughan et al., 2008). An assessment and classification of this diversity is important for effective utilization of the germplasm and will provide a powerful evolutionary framework for many future biological studies using rice as a model.
To date, substantial investigations have been undertaken on the classification of rice cultivars (see reviews in Oka, 1988; Sang and Ge, 2007; Sweeney and McCouch, 2007). Although two major subspecies (Indica and Japonica) have been widely accepted without dispute, the genetic subdivision or substructure of the crop has been inconsistent (for example, Matsuo, 1952; Glaszmann, 1987; Ni et al., 2002; Garris et al., 2005; Ebana et al., 2010). In a landmark study using 15 polymorphic enzyme loci on 1688 landraces, Glaszmann (1987) identified six varietal groups (I to VI), with two of the largest groups (groups I and IV) corresponding to typical Indica and Japonica varieties (subspecies) (Khush, 1997). By screening a sample of 234 rice accessions using 169 nuclear simple sequence repeats (SSRs) and two chloroplast loci, Garris et al. (2005) detected five distinct groups and referred to them as indica, aus, aromatic, temperate japonica and tropical japonica, corresponding to Glaszmann's (1987) groups I, II, V and VI (temperate japonica and tropical japonica), respectively. This classification was subsequently supported by analyses of genome-wide single-nucleotide polymorphism (SNP) data (Caicedo et al., 2007; Zhao et al., 2010; Huang et al., 2012). Nevertheless, a survey of whole-genome single-nucleotide polymorphisms in 140 diverse rice accessions from a world plus a Japanese core collection yielded slightly different result (Ebana et al., 2010). This study identified seven rice groups, three of which were defined as tropical japonica, one as temperate japonica, two as indica and one as aus in terms of Garris et al.'s (2005) classification. Numerous studies on rice cultivars at a local scale have also been conducted and found different subdivisions of rice germplasm (de Oliveira Borba et al., 2009; Zhang et al., 2009, 2011b; Jin et al., 2010; Li et al., 2010). Thus, the genetic subdivision and classification of rice germplasm remain unclear, with three to seven major groups identified by different studies.
One problem arising from previous studies is that some groups of major varieties (for example, groups III and IV recognized by Glaszmann, 1987) were not included in all analyses of global rice germplasm (Garris et al., 2005; Ebana et al., 2010; Zhao et al., 2010; Huang et al., 2012). This may have lead to unreliable estimate of the substructure and lineage affinities among rice cultivars. Another issue is that almost all previous studies on global rice germplasm did not include sufficient samples from China, which might have biased the estimates of both genetic diversity and population structure given that 71 970 rice accessions are recognized as present in China (Wei et al., 2000). For example, of 1688 accessions surveyed by Glaszmann (1987), only 263 (15.6%) were Chinese and classified exclusively into groups I and VI. In the study by Garris et al. (2005), only 15 accessions from mainland China were included and these were grouped into just two groups (indica and temperate japonica). Recent studies have provided convincing evidence that more than two distinct groups were present in Chinese rice germplasm (Zhang et al., 2009, 2011b; Huang et al., 2010, 2012; Jin et al., 2010), implying that greater sampling of Chinese rice cultivars is required for a better resolution of rice population genetics. Finally, as the largest rice genepool, the Chinese rice germplasm has been a focus of investigations for a long time. However, different studies have obtained inconsistent opinions and thus yielded conflicting conclusions regarding cultivar grouping and subdivisions (for example, Ting, 1957; Cheng, 1985; Zhang et al., 2009, 2011b; Huang et al., 2010; Jin et al., 2010). Hence, there is a need to clarify the classification of rice cultivars in China and to place this within a global classification of rice germplasm.
Here, we used 84 SSR markers to genotype 153 rice accessions representing all major groups resolved in previous studies plus an additional 826 accessions representing Chinese rice cultivars. Our specific objectives were (1) to investigate the population genetic structure of global rice cultivars by sampling all known cultivar groups and sufficient Chinese accessions, so as to provide a better understanding of classification of the global rice germplasm; (2) to elucidate the population structure of Chinese rice germplasm and to re-assess its subdivision and classification with reference to the grouping of global rice cultivars; and (3) to evaluate the genetic diversity and differentiation of major groups of Chinese cultivars. In this way, we aimed to obtain an improved understanding of rice germplasm at both global and local scales, which will enhance its exploitation and utilization in rice genetics and breeding.
Materials and methods
Plant materials
A total of 826 accessions of Chinese rice cultivars were sampled, including 194 accessions from the rice core collection of the National Mid-term Genebank for Rice (Hangzhou, China; Supplementary Table S1). Of them, 480 were labelled as Indica and 346 as Japonica. The cultivars included 747 landraces and 79 modern varieties, and represent different seasonal (early, medium and late), drought-tolerant (lowland and upland) and endosperm (waxy and non-waxy) types (Table 1, Supplementary Table S1). In addition, we constructed a set of 153 ‘global' accessions that included 43 representative Chinese accessions (recognized in Zhang et al., 2009; 2011a) and 110 accessions representing rice germplasm from countries other than China. The latter accessions consisted of the major rice groups identified by Glaszmann (1987) and Garris et al. (2005) and were obtained from the Germplasm Resource Center of the International Rice Research Institute (Supplementary Table S2). Of these, 31 accessions were selected based on Glaszmann's (1987) classification, including 7 from group II (aus), 8 from group III (ashina), 11 from group IV (rayada) and 5 from group V (aromatic) accessions. The remaining 79 accessions were chosen based on Garris et al.'s (2005) classification, including 25 indica, 14 aus, 10 aromatic, 16 temperate japonica and 14 tropical japonica accessions.
Table 1. Number (percentage) of accessions belonging to different cultivar types in three groups of Chinese germplasm used in this study.
| Group | Sample size a |
Seasonal type
|
Drought-tolerant type
|
Endosperm type
|
||||
|---|---|---|---|---|---|---|---|---|
| Early | Medium | Late | Lowland | Upland | Waxy | Non-waxy | ||
| indica | 495 | 43 (8.7%) | 245 (49.5%) | 207 (41.8%) | 401 (81.0%) | 94 (19.0%) | 72 (14.5%) | 423 (85.5%) |
| temperate japonica | 201 | 23 (11.4%) | 77 (38.3%) | 101 (50.3%) | 178 (88.6%) | 23 (11.4%) | 83 (41.3%) | 118 (58.7%) |
| tropical japonica | 82 | 3 (3.7%) | 27 (32.9%) | 52 (63.4%) | 10 (12.2%) | 72 (87.8%) | 45 (54.9%) | 37 (45.1%) |
| Total | 778 | 69 (8.9%) | 349 (44.8%) | 360 (46.3%) | 589 (75.7%) | 189 (24.3%) | 200 (25.7%) | 578 (74.3%) |
Admixed accessions among three groups were excluded.
DNA extraction and SSR genotyping
Total genomic DNA was extracted from tender plant shoots following a modified sodium dodecyl sulphate ‘mini-extraction' protocol developed by Zheng et al. (1995). DNA extracts were quantified using a spectrophotometer, checked by 1.5% agarose gel electrophoresis and finally dissolved in 0.1 × Tris-EDTA buffer solution and stored at −20 °C. On the basis of available information on genome-wide SSR markers in rice (McCouch et al., 2002), we initially screened a total of 125 SSR primer pairs and finally chose 84 primer pairs that were consistently amplified in our analysis. The SSR loci are randomly distributed throughout the 12 rice chromosomes and their detailed information is provided in Supplementary Table S3. Primers were synthesized by ABI (Applied Biosystems, Foster City, CA, USA), with the forward primers labelled with blue (FAM), yellow (NED), red (PET) and green (VIC) fluorophores, respectively.
DNA amplification was carried out using a 2720 thermal cycler (Applied Biosystems) in a 10 μl reaction mixture. Each reaction contained 10 × buffer 1.0 μl, 2 mmol l–1 dNTPs 1.0 μl, 25 mmol l–1 MgCl2 1.0 μl, 0.6 μl each of the forward and reverse primer (10 μmol l–1), 5 U μl–1 Taq polymerase 0.1 μl and 20 ng template DNA. The PCR cycle profile was: 94 °C, 2 min; 35 cycles of 94 °C, 45 s, 55 °C (50 °C for RM6091, 61 °C for RM161 and RM185, 67 °C for RM119, RM132, RM169 and RM176, respectively), 45 s, and 72 °C, 1 min; and 72 °C, 8 min.
PCR products (2 μl) were diluted in 8 μl of ultra pure water, purified in the solution mixed with ethanol and sodium acetate for 30 min, and scoured in 100% alcohol for 15 min. The diluted DNA was dried and mixed with highly deionized-formamide (Applied Biosystems), and then submitted for fragment analysis by capillary electrophoresis using an Applied Biosystems 3130xl DNA analyser (Applied Biosystems). DNA fragment size analysis and allele calling were performed using GeneScan and GeneMapper software (Applied Biosystems), followed by the manual allele binning. The matrix standard kit was used to generate the ‘multicomponent matrix' when analyzing the FAM-, NED-, PET-, VIC- and Liz-labelled DNA fragments with the ABI3130 series systems. The multicomponent matrix was used by Data Collection Software (Applied Biosystems) for the instrument to automatically analyse the five different coloured fluorescent dye-labelled samples in a single capillary. In addition, reference lanes were further verified by polyacrylamide gel electrophoresis. In the case of non-amplification, PCR was repeated to exclude technical failure and a null allele was recorded if both PCRs failed.
Statistical analysis
The model-based program STRUCTURE (Pritchard et al., 2000; Falush et al., 2003) was used to calculate the genetic component for each accession using a burn-in length of 10 000, run length of 100 000, and a model allowing for admixture and correlated allele frequencies. To infer population structure, we used both the LnP(D) value and Evanno's ΔK (Evanno et al., 2005) based on five independent simulations. Genetic distance was calculated using the LogSharedAllele distance, DLS (Chakraborty and Jin, 1993), and phylogenetic reconstruction was based on the neighbour-joining (NJ) method implemented in PowerMarker (Liu and Muse, 2005). We also used NTSYTS-pc version 2.10e (Rohlf, 1997) to perform the principal coordinate analysis (PCA), which summarized the major patterns of variation in multi-locus data set. To investigate the group relationship, FST value was calculated and tested using FSTAT (Goudet, 2001). PowerMarker (Liu and Muse, 2005) was used to calculate the number of alleles per locus (Na) and gene diversity per locus (He) as well as polymorphism information content (PIC).
Results
Genetic structure and subdivision of global rice cultivars
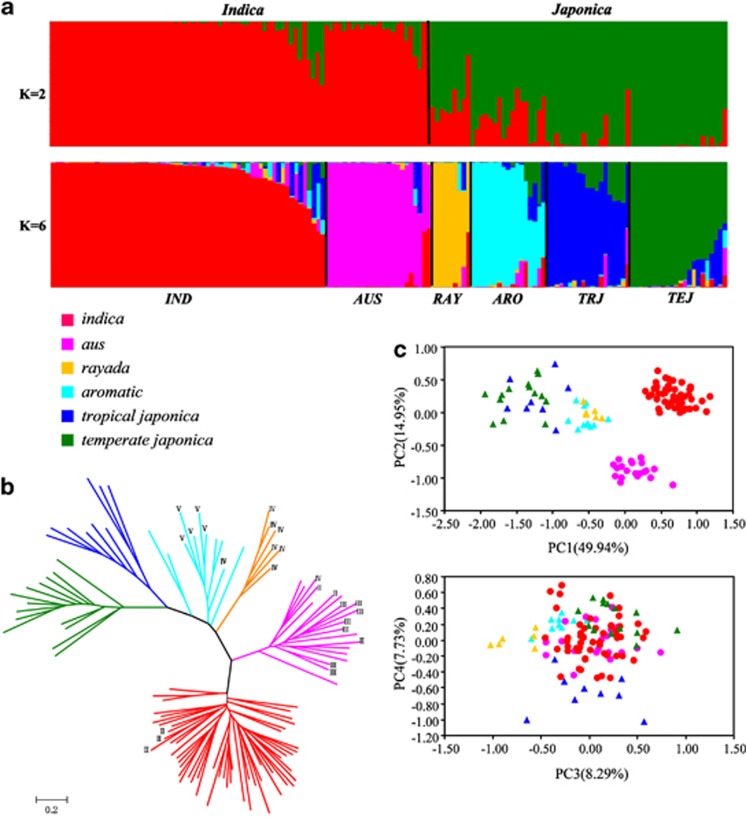
We sampled 153 diverse global rice cultivars representing all major groups identified in previous studies (Glaszmann, 1987; Garris et al., 2005; Zhang et al., 2009, 2011a) to analyse the population genetic structure and the genetic affinities among major sub-populations of rice. We first used STRUCTURE, a model-based approach to calculate the genetic component of each accession and subdivide all cultivars into clusters. We found that the LnP(D) value increased with K from 1 to 10, but showed peaks of Evanno's ΔK at k=2 and k=6, with very similar grouping results at K>6 (Supplementary Figure S1). Thus, we chose K=2 and K=6 for the final analysis of the global accessions. At K=2, two major groups corresponding to two rice subspecies (Indica and Japonica) are apparent; whereas six well-clustered groups are evident at K=6, with two groups under Indica and four under Japonica (Figure 1a). It is noted that all five groups determined by Garris et al. (2005), that is, indica, aus, aromatic, temperate japonica and tropical japonica, were confirmed, corresponding to groups I, II, V and VI of Glaszmann (1987). As 39 (25%) accessions were admixed individuals (with the membership <80% in any one group), we ran STRUCTURE again excluding these admixed accessions and the results remained unchanged (data not shown).
Figure 1.
Population structure of 153 world-wide rice samples. (a) Model-based population assignment using STRUCTURE analysis. (b) NJ tree based on DLS genetic distance. (c) Principal component analysis. Capital roman numbers represent the groups identified by Glaszmann (1987).
Our NJ tree and PCA further confirmed the STRUCTURE results. When admixed accessions were removed, six groups were clearly identified in NJ tree (Figure 1b). Note that all rayada rice accessions form a distinct cluster close to aromatic rice. In the PCA, the first two eigenvectors clearly separate the two major subspecies, Indica and Japonica. The former consists of two well-separated groups (indica and aus); whereas subdivision under Japonica is not apparent. In contrast, the third and fourth eigenvectors divide the Japonica into four groups, that is, rayada, aromatic, temperate japonica and tropical japonica (Figure 1c).
Taken together, all three approaches detected deep population structure of rice germplasm and demonstrated the presence of six distinct groups within Asian cultivated rice, that is, indica (I), aus (II), rayada (IV), aromatic (V), temperate japonica (VI) and tropical japonica (VI) (roman numerals in parentheses are the groups recognized by Glaszmann (1987); Figure 1c). Importantly, for the two groups (III and IV) of Glaszmann (1987) that were not sampled in previous studies, group III (ashina) seems to belong to the same group as aus; whereas group IV (rayada) represents a unique lineage (Figure 1; Supplementary Figure S1).
Genetic structure and subdivision of Chinese rice cultivars
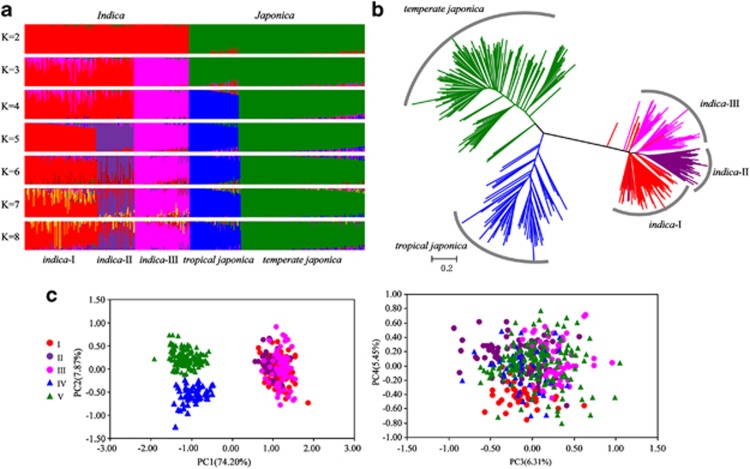
We first ran STRUCTURE on a panel of 826 Chinese rice cultivars to optimize the number of groups (K). By comparing the LnP(D) and Evanno's ΔK values by increasing K from 1 to 10, we found that LnP(D) values increased with K, with the highest log likelihood score at K=2, whereas a peak of Evanno's ΔK occurred at K=5 (Supplementary Figure S2). This indicated that the genetic structure of 826 Chinese accessions had two solutions at K=2 and K=5, supporting the occurrence of two subspecies (Indica and Japonica) and five groups under two subspecies. This substructure was also supported by the NJ tree analysis (Supplementary Figure S3). It is noteworthy from the STRUCTURE analysis that a high proportion of admixed accessions was present in Chinese cultivars, particularly at K>3. Thus, we found 56 (6.7%) admixed individuals at K=2, but 447 (54.1%) admixed accessions at K=5, meaning that for these accessions, <80% of their inferred ancestry arose from one of the five model-based populations. These results suggest that the majority of Chinese rice cultivars are derived from admixture among groups within either Indica or Japonica rather than between the two subspecies. After excluding all admixed accessions detected when K=5, we performed the STRUCTURE, NJ and PCA analyses on the remaining 379 accessions. As shown in Figure 2, two genetically distinct groups (Indica and Japonica) and five groups across the subspecies are again evident. However, the subdivisions within subspecies are less clear, particularly within Indica.
Figure 2.
Population structure of 379 Chinese rice samples. (a) Model-based population assignment using STRUCTURE analysis. (b) NJ tree based on DLS genetic distance. (c) Principal component analysis.
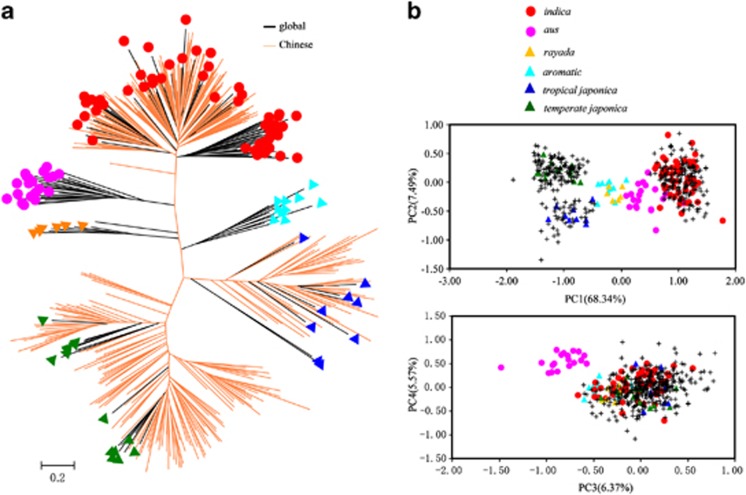
To determine the identity and genetic affinity of Chinese rice cultivars with reference to the global rice groups, we combined the 114 global and 379 Chinese cultivars with membership probabilities over 80% (that is, accessions deemed not to be admixed) and constructed a NJ tree (Figure 3a). We found that three out of six groups identified for the global accessions (indica, temperate japonica and tropical japonica) are present in Chinese rice germplasm, whereas three minor groups identified in the global germplasm (aus, aromatic and rayada) are absent from Chinese germplasm (Figure 3a). When all 826 accessions (that is, admixed and non-admixed) were analysed, we found that 13 Chinese accessions clustered with either aus (8 accessions) or aromatic (5 accessions) (Supplementary Figure S4). However, these accessions were all admixed (Supplementary Table S1) and were probably introduced into China during the process of modern breeding practice. PCA gave similar results (Figure 3b), suggesting that Chinese rice germplasm consists mainly of three major groups as defined for the global cultivars, that is, indica, temperate japonica and tropical japonica.
Figure 3.
Population structure of the combined samples of global and Chinese rice cultivars by excluding the admixed accessions. (a) NJ tree based on DLS genetic distance. (b) Principal component analysis.
To better understand differentiation within each group in terms of seasonal, drought-tolerant and endosperm types, we categorized 778 Chinese cultivars according to their known records by excluding 48 admixed accessions among three groups (Table 1, Supplementary Table S1). It is evident that most Chinese cultivars are medium (44.9%) and late (46.3%) sown types, grow in lowland (75.7%) and are non-waxy (74.3%). However, the three major groups exhibit different characteristics for drought-tolerant type, with more lowland cultivars found in indica (81%) and temperate japonica (88.6%) while more upland cultivars are found in tropical japonica (87.8%). Importantly, the differentiation of cultivar types within subspecies claimed by previous studies (Ting, 1957; Zhang et al. 2009) is not evident. Under Japonica, all cultivar types are found in both temperate japonica and tropical japonica (Table 1). Similarly, all types are present in each of three indica groups, with no correlation found between cultivar types and genetic subdivision within subspecies (Supplementary Table S1).
Level and organization of genetic diversity of rice cultivars
To evaluate the genetic diversity and differentiation of major groups of Chinese cultivars, we calculated several population genetic parameters for both global and Chinese samples. We analysed 778 non-admixed cultivars (Table 2) and found that, for the global cultivars, genetic diversity varied slightly among six major groups, with the lowest found for rayada (He=0.506; PIC=0.465) and the highest for tropical japonica (He=0.639; PIC=0.608). It is noted that two minor groups (aromatic and rayada) maintained comparable diversity to that of other groups despite smaller sample sizes (17 and 9, respectively). These estimates of genetic diversity for global accessions are slightly higher than that of Garris et al. (2005) except for rayada that was not evaluated previously. For Chinese cultivars, we obtained almost the same level of diversity to global samples for indica; whereas the Chinese temperate japonica possessed higher diversity than that of global samples, contrasting to tropical japonica in which slightly higher diversity was found for the global cultivars (Table 2). It is evident that at the subspecies level, Japonica shows higher diversity than Indica for both global (He=0.731 vs 0.694; PIC=0.702 vs 0.665) and Chinese (He=0.643 vs 0.623; PIC=0.619 vs 0.595) samples.
Table 2. Genetic diversity of major groups of global and Chinese rice cultivars inferred by 84 SSR loci (s.d. are presented in parentheses).
| Group/subspecies | N (%) |
Na
|
He
|
PIC
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Global | Chinese total | Chinese CC* | Global | Chinese total | Chinese CC | Global | Chinese total | Chinese CC | Global | Chinese | Chinese CC | |
| indica | 62 (40.5) | 495 (63.7) | 124 (67.4) | 8.8 (4.9) | 13.0 (7.5) | 10.2 (5.8) | 0.624 | 0.623 | 0.615 | 0.593 | 0.595 | 0.587 |
| aus | 24 (15.7) | — | — | 6.2 (3.5) | — | — | 0.604 | — | — | 0.568 | — | — |
| Indica | 83 | 495 | 124 | 10.5 (6.0) | 13.0 (7.5) | 10.2 (5.8) | 0.694 | 0.623 | 0.615 | 0.665 | 0.595 | 0.587 |
| aromatic | 17 (11.1) | — | — | 5.5 (2.7) | — | — | 0.607 | — | — | 0.572 | — | — |
| rayada | 9 (5.9) | — | — | 3.6 (1.9) | — | — | 0.506 | — | — | 0.465 | — | — |
| temperate japonica | 22 (14.4) | 201 (25.8) | 43 (23.4) | 5.7 (3.0) | 11.7 (6.5) | 7.9 (4.3) | 0.573 | 0.608 | 0.609 | 0.542 | 0.585 | 0.583 |
| tropical japonica | 19 (12.4) | 82 (10.5) | 17 ((9.2) | 6.1 (2.7) | 9.5 (5.3) | 5.5 (2.7) | 0.639 | 0.621 | 0.587 | 0.608 | 0.596 | 0.556 |
| Japonica | 70 | 283 | 60 | 10.6 (5.7) | 12.6 (7.0) | 9.1 (5.0) | 0.731 | 0.643 | 0.638 | 0.702 | 0.619 | 0.611 |
| Total | 153 | 778 | 184 | 13.0 (7.4) | 14.0 (8.0) | 12.3 (7.0) | 0.769 | 0.747 | 0.737 | 0.738 | 0.712 | 0.704 |
Abbreviations: CC, core collection; N, sample size; Na, the number of alleles of per locus; He, gene diversity per locus; PIC, polymorphism information content; SSR, simple sequence repeat. Two subspecies and their estimates of the parameters were bold faced.
Table 3 shows the genetic differentiation among the recognized groups for both global and Chinese cultivars. Pairwise FST values showed highly significant differentiation among the six global groups, ranging from 0.129 (temperate japonica vs tropical japonica) to 0.298 (indica vs temperate japonica). For the Chinese cultivars, differentiation between three groups varied from 0.106 (temperate japonica vs tropical japonica) to 0.305 (indica vs temperate japonica), paralleling the results for the comparisons of global cultivars. Note that significantly larger genetic differentiation was recorded between the two subspecies (Indica vs Japonica) at both global (FST=0.167) and China (FST=0.285) scales, consistent with the genetic structure and genealogical analyses. We also calculated the FST value between three subgroups within Chinese Indica and found relatively low differentiation between them (FST=0.076–0.132), consistent with the finding that subdivision was less apparent within Chinese Indica.
Table 3. Pairwise FST values among six groups for the global (above the diagonal) and Chinese (below the diagonal) samples estimated based on 84 SSR loci.
| Group | indica | aus | aromatic | rayada | temperate japonica | tropical japonica |
|---|---|---|---|---|---|---|
| indica | 0.191*** | 0.201*** | 0.234*** | 0.298*** | 0.256*** | |
| aus | — | 0.189*** | 0.244*** | 0.291*** | 0.242*** | |
| aromatic | — | — | 0.182*** | 0.180*** | 0.133*** | |
| rayada | — | — | — | 0.217*** | 0.217*** | |
| temperate japonica | 0.305*** | — | — | — | 0.129*** | |
| tropical japonica | 0.290*** | — | — | — | 0.106*** |
Abbreviation: SSR, simple sequence repeat.
***P<0.001
We further estimated the genetic diversity of the 184 Chinese cultivars that were identified to belong to the core collection (Zhang et al. 2011a). We found that the number of alleles in the core collection is lower than that in total samples, particularly for tropical japonica (9.5 vs 5.5). Nevertheless, over 98% of the diversity in the total samples is retained in the core collection, as measured by heterozygosity and PIC values at either species or group levels (Table 2). This result indicates that the Chinese core collection maintained abundant diversity and efficiently represented most of the genetic diversity present in Chinese rice germplasm.
Discussion
Six distinct and major groups within O. sativa
The classification of rice cultivars can be dated back to the beginning of the twentieth century when Kato et al. (1928) indicated the existence of two main varietal types, denoted as Indica and Japonica, corresponding to the Hsien and Keng rice in China (Ting, 1957; Second, 1982; Oka, 1988). On the basis of gross morphology, Matsuo (1952) recognized a third type of cultivar that was later referred to as Javanica by Morinaga and Kuriyama (1958) according to its geographical distribution. Although Oka (1958) considered Japonica and Javanica as tropical and temperate components of a single Japonica type, the Indica, Japonica and Javanica terms have been used by many rice workers to refer to three morphological types (Chang, 1976; Glaszmann, 1987; Khush, 1997). In recent decades, the classification of five major groups of rice cultivars identified by Garris et al. (2005) have been widely accepted, which provided a clearer description of the population structure and organization of rice germplasm. However, different subdivisions of rice cultivars have also been proposed (see reviews in Oka, 1988 and Khush, 1997) and implicated in recent molecular studies (for example, Ni et al., 2002; Zhang et al., 2009, 2011b; Ebana et al., 2010; Jin et al., 2010). Importantly, the ashina and rayada rice (groups III and IV of Glaszmann, 1987) have not been included in most previous studies such as Garris et al. (2005) and Huang et al. (2012), and few accessions from China were sampled in the analyses.
In this study, we investigated a panel of global rice cultivars that consisted of all previously recognized groups and included sufficient samples from China using multiple approaches including model-based grouping, NJ tree and PCA. We confirmed the five distinct groups recognized by Garris et al. (2005), that is, indica, aus, aromatic, temperate japonica and tropical japonica, corresponding to Glaszmann's (1987) groups I, II, V and VI (temperate japonica and tropical japonica), respectively. We also found that Glaszmann's (1987) group III (ashina) was mixed with group II (aus) and not a distinct group. Importantly, we demonstrated that Glaszmann's (1987) group IV (rayada) was unique in genealogy and maintained a high level of genetic diversity although it grows only in India and Bangladesh (Glaszmann, 1987). The rayada cultivars comprise a very particular rice sown in November–December and harvested as much as 12 months later. This group is able to tolerate cold and flooding, is photoperiod sensitive and lacks secondary dormancy (Glaszmann, 1987; Vaughan et al., 2008). Having these unusual characteristics and being genetically distinct, rayada rice should be considered as a new (the sixth) major group of rice germplasm. This rice group is not only an important genetic resource for rice breeding, but also provides materials for investigating rice domestication and adaptation in the complex environments of deepwater areas.
Three major groups present in Chinese rice germplasm
As one of the centres of rice domestication and the largest country of rice production (Normile, 1997; Liu et al., 2007), China maintains a huge amount of rice germplasm, with 53 263 rice landraces and 5633 modern varieties in the Chinese National Crop Genebank (Han and Cao, 2005). Although great efforts have been made to study Chinese rice germplasm, varied numbers of subgroups under two subspecies have been reported (Ting, 1957; Cheng, 1985; Zhang et al., 2009, 2011b; Huang et al., 2010; Jin et al., 2010; Li et al., 2010). For example, Jin et al. (2010) used 100 genome-wide SSRs to investigate the population genetic structure of 416 Chinese rice accessions and grouped them into seven groups, with six groups belonging to Indica and only one to Japonica. By re-sequencing 517 rice landraces, however, Huang et al. (2010) found that both subspecies contained three subgroups. This disaccord might arise from the employment of different genetic markers and analytical methods as well as different samples used in the studies. More importantly, all previous studies on Chinese rice germplasm focused exclusively on Chinese accessions. Without inclusion of collections from areas outside China, these studies were unable to make clear the number of cultivar groups and their genetic affinities in terms of the widely recognized systems proposed by Glaszmann (1987) and Garris et al. (2005).
Here we demonstrated that out of six groups identified for the global rice germplasm, only three major groups (indica, temperate japonica and tropical japonica) were found in China, with indica being the largest group. Although 13 cultivars that were sampled mainly from western China clustered with aus and aromatic on the genealogical tree, model-based STRUCTURE analyses indicated that they were all admixed accessions among groups and of limited significance. It is worthwhile mentioning that Chinese cultivars exhibited higher diversity than that of global samples for temperate japonica but slightly lower diversity than global samples for tropical japonica, probably reflecting the higher proportion of temperate japonica in Chinese rice germplasm (63.6%).
Diversity and differentiation of Chinese rice cultivars
Chinese farmers have traditionally recognized two major groups of rice cultivars, Hsien and Keng, based on grain types and sensitivity to temperature and photoperiod (Ting, 1957; Cheng, 1985). These two groups actually correspond to the two subspecies Indica and Japonica (Glaszmann, 1987) and have been widely accepted in China (Ting, 1957; Cheng, 1985; Zhang et al., 2009). According to six diagnostic characters, Cheng (1985) further classified Chinese rice into Hsein, Hsein-cline, Keng-cline and Keng, whereas Ting (1957) developed a five-level taxonomic system for Chinese germplasm, in which rice cultivars were divided into Hsien and Keng subspecies, two seasonal types (early/medium and late sown), two drought-tolerant (or watery regime) types (lowland and upland) and two endosperm types (waxy and non-waxy) and other cultivars. In addition to the Hsien and Keng classification, the other four types of divisions were mainly based on ecological or habitat features and were not supported by subsequent investigations, particularly molecular approaches (Zhang et al., 2009, 2011a). By screening 3024 Chinese landraces, for example, Zhang et al. (2009) found clear differentiation within two subspecies, that is, the subdivision among seasonal types within Indica and differentiation between drought-tolerant types within Japonica. They attributed such intra-subspecies differentiation to their different growth environments and cropping systems.
In this study, we found that all seasonal, drought-tolerant and endosperm types of cultivars occurred within each of three major groups. Within either temperate japonica or tropical japonica, almost all cultivar types are present (Supplementary Table S4). Furthermore, within indica, the three subgroups (indica-I, indica II and indica-III) are not correlated with differentiation of cultivar types (Supplementary Table S4). These observations do not support the findings of Zhang et al. (2009) that claimed seasonal differentiation in Indica and drought-tolerant differentiation in Japonica. Therefore, it is most likely that differentiation of cultivar types arose multiple times to adapt to local environments under artificial selection.
Interestingly, we observed that the number of alleles per locus (Na) was significantly lower in Chinese core collection than in the total Chinese germplasm at the group levels although the core collection retained very high percentage of total genetic diversity (over 98%). This finding suggested that numerous rare alleles might not be captured in the core collection. In recent decades, many elite genes or alleles have been discovered from landraces and utilized in rice breeding, including the semi-dwarf genes (Monna et al., 2002; Sasaki et al., 2002), resistant/tolerant genes to diseases and pests, and genes for improving grain yield and quality (Fukuoka et al., 2009; Ishimaru et al., 2013). It is likely, therefore, that three minor groups of rice germplasm (aus, aromatic and rayada) that have a limited presence in Chinese germplasm may be important genetic resources for future rice genetics and breeding in China given their unique genetic and phenotypic characteristics (Glaszmann, 1987; Khush, 1997).
Data archiving
Data were deposited in the Dryad repository: 10.5061/dryad.m8jj7. dryad.55425.
Acknowledgments
We thank Mr Daolong Ming (China National Rice Research Institute, China) for assistance in growing samples in greenhouse and to Richard Abbott for reading and improving the manuscript. We also thank the International Rice Research Institute (Los Banos, Philippines) for providing seed samples. This work was supported by the National Natural Science Foundation of China (91231201 and 31070195) and the Basic Research Budget of China National Rice Research Institute (2009RG001-3).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Caicedo AL, Williamson SH, Hernandez RD, Boyko A, Fledel-Alon A, York TL, et al. Genome-wide patterns of nucleotide polymorphism in domesticated rice. PLoS Genet. 2007;3:1745–1756. doi: 10.1371/journal.pgen.0030163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty R, Jin L. DNA Fingerprinting: State of the Science. Birkhauser Verlag, Basel, Switzerland; 1993. A unified approach to study hypervariable polymorphisms: statistical considerations of determining relatedness and population distances; pp. 153–175. [DOI] [PubMed] [Google Scholar]
- Chang TT. The origin, evolution, cultivation, dissemination, and diversification of Asian and African rices. Euphytica. 1976;25:425–441. [Google Scholar]
- Cheng KS. A statistical evaluation of the classification of rice cultivars into hsien and keng subspecies. Rice Genet Newsl. 1985;2:46–48. [Google Scholar]
- de Oliveira Borba TC, Brondani RP, Rangel PH, Brondani C. Microsatellite marker-mediated analysis of the EMBRAPA Rice Core Collection genetic diversity. Genetica. 2009;137:293–304. doi: 10.1007/s10709-009-9380-0. [DOI] [PubMed] [Google Scholar]
- Ebana K, Yonemaru J, Fukuoka S, Iwata H, Kanamori H, Namiki N, et al. Genetic structure revealed by a whole-genome single-nucleotide polymorphism survey of diverse accessions of cultivated Asian rice (Oryza sativa L.) Breeding Sci. 2010;60:390–397. [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuoka S, Saka N, Koga H, Ono K, Shimizu T, Ebana K, et al. Loss of function of a proline-containing protein confers durable disease resistance in rice. Science. 2009;325:998–1001. doi: 10.1126/science.1175550. [DOI] [PubMed] [Google Scholar]
- Garris AJ, Tai TH, Coburn J, Kresovich S, McCouch S. Genetic structure and diversity in Oryza sativa L. Genetics. 2005;169:1631–1638. doi: 10.1534/genetics.104.035642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaszmann JC. Isozymes and classification of Asian rice varieties. Theor Appl Genet. 1987;74:21–30. doi: 10.1007/BF00290078. [DOI] [PubMed] [Google Scholar]
- Goudet J. FSTAT, Version 2.9.3, A Program to Estimate and Test Gene Diversities and Fixation Indices. Lausanne University: Lausanne, Switzerland; 2001. [Google Scholar]
- Han LZ, Cao GL. Status of collection, conservation and propagation of rice germplasm in China. J Plant Genet Res. 2005;6:359–364. [Google Scholar]
- Huang XH, Wei XH, Sang T, Zhao QA, Feng Q, Zhao Y, et al. Genome-wide association studies of 14 agronomic traits in rice landraces. Nat Genet. 2010;42:961–976. doi: 10.1038/ng.695. [DOI] [PubMed] [Google Scholar]
- Huang XH, Zhao Y, Wei XH, Li CY, Wang A, Zhao Q, et al. Genome-wide association study of flowering time and grain yield traits in a worldwide collection of rice germplasm. Nat Genet. 2012;44:32–53. doi: 10.1038/ng.1018. [DOI] [PubMed] [Google Scholar]
- Ishimaru K, Hirotsu N, Madoka Y, Murakami N, Hara N, Onodera H, et al. Loss of function of the IAA-glucose hydrolase gene TGW6 enhances rice grain weight and increases yield. Nat Genet. 2013;45:707–711. doi: 10.1038/ng.2612. [DOI] [PubMed] [Google Scholar]
- Jin L, Lu Y, Xiao P, Sun M, Corke H, Bao JS. Genetic diversity and population structure of a diverse set of rice germplasm for association mapping. Theor Appl Genet. 2010;121:475–487. doi: 10.1007/s00122-010-1324-7. [DOI] [PubMed] [Google Scholar]
- Kato S, Kosaka H, Hara S. On the affinity of rice varieties as shown by fertility of hybrid plants. Agric Kyushu Univ. 1928;3:132–147. [Google Scholar]
- Khush GS. Origin, dispersal, cultivation and variation of rice. Plant Mol Biol. 1997;35:25–34. [PubMed] [Google Scholar]
- Li X, Yan W, Agrama H, Hu B, Jia L, Jia M, et al. Genotypic and phenotypic characterization of genetic differentiation and diversity in the USDA rice mini-core collection. Genetica. 2010;138:1221–1230. doi: 10.1007/s10709-010-9521-5. [DOI] [PubMed] [Google Scholar]
- Liu KJ, Muse SV. PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics. 2005;21:2128–2129. doi: 10.1093/bioinformatics/bti282. [DOI] [PubMed] [Google Scholar]
- Liu L, Lee GA, Jiang LP, Zhang JZ. The earliest rice domestication in China. Antiquity. 2007;81:313. [Google Scholar]
- Matsuo T. Genecological studies on cultivated rice. Bull Nat Inst Agr Sci Series. 1952;D3:1–111. [Google Scholar]
- McCouch SR, Temnykh L, Xu YB, Lobos KB, Clare K, et al. Development and mapping of 2240 new SSR markers for rice (Oryza sative L.) DNA Res. 2002;9:199–207. doi: 10.1093/dnares/9.6.199. [DOI] [PubMed] [Google Scholar]
- Monna L, Kitazawa N, Yoshino R, Suzuki J, Masuda H, Maehara Y, et al. Positional cloning of rice semidwarfing gene, sd-1: rice ‘Green revolution gene' encodes a mutant enzyme involved in gibberellin synthesis. DNA Res. 2002;9:11–17. doi: 10.1093/dnares/9.1.11. [DOI] [PubMed] [Google Scholar]
- Morinaga T, Kuriyama H. Intermediate type of rice in the subcontinent of India and Java. Jpn J Breed. 1958;7:253–259. [Google Scholar]
- Ni J, Colowit PM, Mackill DJ. Evaluation of genetic diversity in rice subspecies using microsatellite markers. Crop Sci. 2002;42:601–607. [Google Scholar]
- Normile D. Yangtze seen as earliest rice site. Science. 1997;275:309. [Google Scholar]
- Oka HI. Intervarietal variation and classification of cultivated rice. Indian J Genet Pl Br. 1958;18:79–89. [Google Scholar]
- Oka HI. Origin of Cultivated Rice. Japan Scientific Societies Press, Tokyo, Japan; 1988. [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohlf F. NTSYS-pc: Numerical Taxonomy and Multivariate Analysis System, Version 2.00. Exeter Software:: Setauket, NY, USA; 1997. [Google Scholar]
- Sang T, Ge S. Genetics and phylogenetics of rice domestication. Curr Opin Genet Dev. 2007;17:533–538. doi: 10.1016/j.gde.2007.09.005. [DOI] [PubMed] [Google Scholar]
- Sasaki A, Ashikari M, Ueguchi-Tanaka M, Itoh H, Nishimura A, Swapan D, et al. Green revolution: a mutant gibberellin-synthesis gene in rice—new insight into the rice variant that helped to avert famine over thirty years ago. Nature. 2002;416:701–702. doi: 10.1038/416701a. [DOI] [PubMed] [Google Scholar]
- Second G. Origin of the genetic diversity of cultivated rice (Oryza spp.): study of the polymorphism scored at 40 isozyme loci. Jpn J genet. 1982;57:25–57. [Google Scholar]
- Shimamoto K, Kyozuka J. Rice as a model for comparative genomics of plants. Annu Rev Plant Biol. 2002;53:399–419. doi: 10.1146/annurev.arplant.53.092401.134447. [DOI] [PubMed] [Google Scholar]
- Sweeney M, McCouch S. The complex history of the domestication of rice. Ann Bot-London. 2007;100:951–957. doi: 10.1093/aob/mcm128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting Y. The origin and evolution of cultivated rice in China. Acta Agron Sin. 1957;8:243–260. [Google Scholar]
- Vaughan DA, Lu BR, Tomooka N. The evolving story of rice evolution. Plant Sci. 2008;174:394–408. [Google Scholar]
- Wei XH, Tang SX, Yu HY, Jiang YZ, Qiu ZG. Studies on methods of developing a core collection for China traditional japonica rice. Chinese J Rice Sci. 2000;14:237–240. [Google Scholar]
- Wing RA, Ammiraju JSS, Luo MZX, Kim H, Yu YS, Kudrna D, et al. The Oryza Map Alignment Project: the golden path to unlocking the genetic potential of wild rice species. Plant Mol Biol. 2005;59:53–62. doi: 10.1007/s11103-004-6237-x. [DOI] [PubMed] [Google Scholar]
- Zhang DL, Zhang HL, Wang MX, Sun JL, Qi YW, Wang F, et al. Genetic structure and differentiation of Oryza sativa L. in China revealed by microsatellites. Theor Appl Genet. 2009;119:1105–1117. doi: 10.1007/s00122-009-1112-4. [DOI] [PubMed] [Google Scholar]
- Zhang HL, Zhang DL, Wang MX, Sun JL, Qi YW, Li JJ, et al. A core collection and mini core collection of Oryza sativa L. in China. Theor Appl Genet. 2011;122:49–61. doi: 10.1007/s00122-010-1421-7. [DOI] [PubMed] [Google Scholar]
- Zhang P, Li J, Li X, Liu X, Zhao X, Lu Y. Population structure and genetic diversity in a rice core collection (Oryza sativa L.) investigated with SSR markers. PLoS One. 2011;6:e27565. doi: 10.1371/journal.pone.0027565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang QF, Li JY, Xue YB, Han B, Deng XW. Rice 2020: a call for an international coordinated effort in rice functional genomics. Mol Plant. 2008;1:715–719. doi: 10.1093/mp/ssn043. [DOI] [PubMed] [Google Scholar]
- Zhao KY, Wright M, Kimball J, Eizenga G, McClung A, Kovach M, et al. Genomic diversity and introgression in O. sativa reveal the impact of domestication and breeding on the rice genome. PLoS One. 2010;5:e10780. doi: 10.1371/journal.pone.0010780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng KL, Subudhi PK, Domingo J, Magpantay G, Huang N. Rapid DNA isolation for marker assisted selection in rice breeding. Rice Genet Newsl. 1995;12:255–258. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.