Abstract
Changes in chromosome number and structure are important contributors to adaptation, speciation and macroevolution. In flowering plants, polyploidy and subsequent reductions in chromosome number by fusion are major sources of chromosomal evolution, but chromosome number increase by fission has been relatively unexplored. Here, we use comparative linkage mapping with gene-based markers to reconstruct chromosomal synteny within the model flowering plant genus Mimulus (monkeyflowers). Two sections of the genus with haploid numbers ⩾14 have been inferred to be relatively recent polyploids because they are phylogenetically nested within numerous taxa with low base numbers (n=8–10). We combined multiple data sets to build integrated genetic maps of the M. guttatus species complex (section Simiolus, n=14) and the M. lewisii group (section Erythranthe; n=8), and then aligned the two integrated maps using >100 shared markers. We observed strong segmental synteny between M. lewisii and M. guttatus maps, with essentially 1-to-1 correspondence across each of 16 chromosomal blocks. Assuming that the M. lewisii (and widespread) base number of 8 is ancestral, reconstruction of 14 M. guttatus chromosomes requires at least eight fission events (likely shared by Simiolus and sister section Paradanthus (n=16)), plus two fusion events. This apparent burst of fission in the yellow monkeyflower lineages raises new questions about mechanisms and consequences of chromosomal fission in plants. Our comparative maps also provide insight into the origins of a chromosome exhibiting centromere-associated female meiotic drive and create a framework for transferring M. guttatus genome resources across the entire genus.
Keywords: polyploidy, chromosomal rearrangement, chromosome number, speciation, karyotypic evolution, comparative map
Introduction
Changes in chromosome number and structure are important contributors to adaptation, speciation and macroevolutionary diversity (Levin, 2002). Closely related taxa often differ by a least a few structural rearrangements, and more distantly related taxa within genera often differ in chromosome number as well. In plants, polyploidy is an important source of chromosomal change, and doublings or near-doublings in chromosome number within families are often assumed to result from tetraploidy events (Jones, 1998; Wood et al., 2009). Nonetheless, changes in chromosome number between sister lineages may also occur by chromosome fission and fusion (Schubert and Lysak, 2011). However, both traditional cytogenetic studies (Baldwin and Wessa, 2000) and genomic analyses (Luo et al., 2009) suggest that dysploid (large steps) or aneuploid (small steps) decrease in chromosome number is more common than chromosome number increase by fission, with the exception of a few holocentric lineages in which fission may be less deleterious (Chung et al., 2012). Chromosomal fission can be difficult to distinguish from polyploidy when gaps in chromosome number are large, however, and assumptions about the prevalence of polyploidy in plants may bias simple chromosome-counting approaches to inferring the direction of karyotypic change (Mayrose et al., 2010; Cusimano et al., 2012). Therefore, directly identifying patterns of synteny between closely related taxa with very different chromosome numbers is an important first step toward investigating the causes and consequences of major karyotypic change.
Here, we investigate patterns of chromosomal evolution within the flowering plant genus Mimulus (Phrymaceae, formerly Scrophulariaceae). With >150 ecologically diverse species and numerous cross-compatible species complexes suitable for genetic investigation, Mimulus has long been a model system for understanding plant adaptation and speciation (Wu et al., 2008). The genus, which has its center of diversity and likely origin in western North America, has traditionally been divided into seven sections. Two sections, Simiolus and Paradanthus, consist of primarily yellow-flowered taxa and have n=14 and 16, respectively, as the common haploid chromosome numbers (Beardsley et al., 2004). Most of the remaining Mimulus sections, including the section Erythranthe, have a common haploid chromosome number of 8 (n=9–10 in a few cases). Paradanthus appears polyphyletic, with one clade sister to Simiolus and the other to Erythranthe (Beardsley et al., 2003, 2004). However, these three sections together form a monophyletic group contained within the rest of Mimulus (Beardsley et al., 2004), and share a most recent common ancestor about 20 million years ago (Mya) (Nie et al., 2006). Because phylogenetic reconstructions strongly suggests that n=8 is ancestral in Mimulus, the ∼2-fold difference in chromosome number between Erythranthe species (and most other taxa) and the yellow monkeyflowers has been inferred to represent a whole-genome duplication event specific to Simiolus and its sister clade of yellow Paradanthus (Figure 4 of Beardsley et al., 2004).
In this study, we compare integrated genetic maps of the Mimulus guttatus (yellow monkeyflower) species complex (section Simiolus; 2n=28) and the Mimulus lewisii species group (section Erythranthe; 2n=16) to infer chromosomal synteny. Comparative linkage mapping is a useful tool for the investigation of patterns of genome evolution, including chromosomal rearrangements, and also facilitates transfer of genetic tools and information among related but reproductively incompatible taxa (for examples, Burke et al., 2004; Wu and Tanksley, 2010). Both groups of monkeyflowers have been model systems for understanding speciation for nearly five decades and were among the first wild plants investigated with linkage and quantitative trait loci mapping approaches (Bradshaw et al., 1995; Fishman et al., 2001). The two species complexes remain models for understanding both chromosomal (Fishman and Saunders, 2008; Scoville et al., 2009; Lowry and Willis, 2010; Fishman et al., 2013) and genic contributions to the maintenance of fitness variation and the development of species barriers. More recently, whole-genome sequence from M. guttatus (http://www.Phytozome.net/) has facilitated candidate gene, fine mapping and positional cloning approaches to investigate the molecular basis of population differentiation and species divergence in Mimulus. Recent work has, for example, used the M. guttatus genome sequence as a guide for dissecting floral pigmentation loci in other Mimulus (Cooley et al., 2011; Streisfeld et al., 2013; Yuan et al., 2013). However, maximal use of M. guttatus genomic resources for research across the genus requires integration of genetic maps from other taxa with the genetic and physical maps of M. guttatus. The comparative maps presented in this study extend that connection genome wide to the M. cardinalis group, and also set the stage for investigating the molecular mechanisms and evolutionary consequences of major genomic restructuring in this model genus.
Materials and methods
Markers
A set of gene-based exon-primed intron-containing markers based on M. guttatus, cDNA sequences (MgSTS markers, for M. guttatus sequence-tagged site) were used for Mimulus map construction, integration and comparison. MgSTS markers were designed to be both broadly useful across the genus and highly polymorphic within and among species. Similar to COS markers of the Solanaceae (Wu and Tanksley, 2010) and Asteraceae (Chapman et al., 2007), MgSTS primers sets were designed from cDNA library of M. guttatus (IM62 inbred line) using a pipeline with three major features (Bouck and Vision, 2007). First, cDNA sequences were blasted against Arabidopsis and other dicot cDNA databases, and putatively low copy number genes were retained (criteria: first hit <e−20, no more than three other hits within e−10 window). Second, intron positions were inferred from comparisons of Arabidopsis genomic and cDNA sequences, assuming conservation of intron position. Third, primers were designed from the M. guttatus exon sequence to amplify one or more potential intron-containing regions, with each primer designed in a region of high amino-acid conservation with their 3′ ends anchored on second positions. Primer information for MgSTS markers (n=855) and sequences of corresponding cDNAs can be found at mimulusevolution.org. For mapping, primer sets were tested in the parental lines used to generate each mapping population, and fragment length polymorphisms scorable by capillary electrophoresis were identified. Multiplexes of three to six markers (forward primers 5′ fluorescently labeled) were amplified using a standard touchdown PCR protocol. Additional information on the MgSTS genotyping methods can be found elsewhere (Fishman et al., 2008, 2013).
Because the M. guttatus whole-genome sequence was not available at the time of MgSTS marker design, primer sequences were not initially screened for uniqueness. However, the physical position(s) of sequences corresponding to each MgSTS primer can now be determined by BLAST to the 7 × draft genome assembly of M. guttatus (V1.1; http://www.Phytozome.org/), and 50 markers (for examples, MgSTS.605 and MgSTS.598) were identified as identical by physical position. These pairs were collapsed to a single MgSTS number (whatever was most common across all maps) for the maps presented here and the other marker name deleted if it has also been mapped. Other markers amplified multiple segregating loci in a given cross- or amplified markers that mapped to distinct locations in two crosses within a group; these pairs could generally be confirmed as paralogs by BLAST (that is, equal match of both primers to two separate physical scaffolds) and have been designated by letter suffixes (a–d). It is also possible that some markers without such suffixes represent different loci in different maps, as only polymorphic markers could be mapped in a given cross.
Integrated M. guttatus map
For the integrated M. guttatus map (GIM, for guttatus integrated map), we concatenated linkage data from four F2-based mapping populations. With a few exceptions (see below), we used as our framework the combined Iron Mountain (CIM) map, which involves plants derived from a single well-studied M. guttatus population (Iron Mountain, OR, USA) and has the highest marker coverage (14 linkage groups, 274 markers, 1453 cM). This map is itself a consensus map based on three F2 hybrid maps, each created by crossing representatives of high and low outbred selection lines for flower size (Lee, 2009). The other three mapping populations all had the IM62 inbred line from Iron Mountain as one parent, with either a divergent ecotype or closely related species as the other, so the markers and maps are highly comparable. The second map (DIM; 184 markers, 1391 cM) was based on recombinant inbred lines derived from F2 hybrids of IM62 and the DUN10 inbred line, representing the coastal perennial ecotype of M. guttatus (Lowry et al., 2009; Lowry and Willis, 2010). The final two maps (F2N 2006 and F2N 2009 on mimulusevolution.org; here NIM1 and NIM2) are both based on an inbred line cross of Sherar's Falls (SF) M. nasutus × IM62 M. guttatus. M. nasutus is the most widespread of several highly self-fertilizing small-flowered species closely related to M. guttatus. The NIM1 map used a subset (N=276) of an initial larger mapping population genotyped for quantitative trait locus and transmission ratio distortion mapping (Fishman et al., 2001) and included 141 MgSTS markers, 12 SSR markers and 75 AFLPs (Fishman et al., 2008). The AFLPs were not included in map concatenation, and so terminal AFLPs were dropped from aligned NIM1 map (192 markers, shortened length=1328 cM). The NIM2 mapping population was grown and genotyped at Duke University for quantitative trait loci mapping of physiological traits (C Wu and JH Willis, unpublished data) and consists of 223 markers spanning 1406 cM. These four maps plus maps based on reciprocal IM62 M. guttatus × SF M. nasutus nearly isogenic line (NIL) populations (see below) are aligned in Supplementary Figure S1.
We first attempted to concatenate the M. guttatus maps using the map integration function in Joinmap 4.0 software (Van Ooijen, 2006), in which each of the base maps had been constructed. However, the maps produced with this approach often diverged substantially from the original, and generally parallel (Supplementary Figure S1), base maps in either order or intermarker distances. This discrepancy was likely because the Joinmap map integration protocol requires use of the regression-mapping algorithm, where recombination frequencies are averaged across maps for shared markers. However, recombination rates vary between maps just by chance, and each of the base maps also exhibits at least one region of suppressed recombination relative to others due to inversions that distinguish the parental lines (Lowry et al., 2009; Lee, 2009). Such systematic differences in recombination violate the assumptions necessary for automatic concatenation.
Therefore, we present a concatenated M. guttatus map (GIM; Supplementary Figure S2) constructed by designating a putatively collinear and relatively dense framework map for each linkage group (CIM for all but LG6, LG7 and LG11) and manually placing unshared markers from each map in the positions dictated by shared flanking markers. That is, if MgSTS.822 was placed in the center of the ∼9 cM interval between MgSTS.477 and MgSTS.306 in the DIM map (Supplementary Figure S1; LG4), but was absent from that interval in the CIM map, it was inserted into the CIM map in the appropriate relative position. This process was repeated iteratively, starting with markers present on multiple secondary maps and progressing to singletons. Because the number of shared markers was high, which implies that we often had multiple sources of positional information for markers not present in the framework group, placement was generally a straightforward process. In addition to the four primary maps, we also placed markers based on their locations in two NIL populations created by backcrossing reciprocal F1 hybrids to each parental line for four generations. These BN4 (SF × IM62 F1, M. nasutus recurrent parent, N=188) and BG4 (IM62 × SF F1, M. guttatus recurrent parent, N=192) populations were genotyped at MgSTS-, SSR- and gene-based markers designed following the MgSTS pipeline (232 and 194 markers each, respectively; Supplementary Figure S1). Markers in the NIL populations were readily grouped and ordered in Joinmap, although we cannot reasonably assume the Mendelian segregation and equal recombination in all generations required for accurate distance estimation (Fishman and Willis, 2005). A total of 43 markers were added from the two NIL maps (29 shared, 19 unique to BN4 and 1 unique to BG4), with the placement of singletons verified by physical linkage (based on primer BLAST) to a mapped marker on the same scaffold of the IM62 M. guttatus reference genome (V1.1; http://www.Phytozome.net/). For completeness, a handful of markers were also added from published fine-mapping studies of particular genomic regions (Barr and Fishman, 2010).
Integrated M. lewisii map
The M. lewisii integrated map was constructed by concatenating two maps based on inbred line crosses of M. parishii × M. lewisii (LP) and M. lewisii × M. cardinalis (LC), which are described in detail elsewhere (Fishman et al., 2013). The LEW inbred line was shared by both mapping populations and a high proportion of markers were shared between the LP and LC maps. However, marker distances and orders in both interspecific maps were affected by the presence of putative rearrangements, including both translocations and inversions, with an estimated minimum of five regions of recombination suppression in the LC map and two in the LP map (Fishman et al., 2013). Thus, the concatenated M. lewisii group map (linkage groups designated as LPC) was constructed using the relatively collinear LP map as base, with markers from the LC map added when they could be unambiguously assigned as tightly linked to a shared marker.
Map comparison
We aligned the integrated M. guttatus (GIM) and M. lewisii group (LPC) maps using markers shared by both genetic maps or, for LPC markers without a match, physical linkage (as indicated by colocalization on the same V1.1 M. guttatus genome scaffold; http://www.Phytozome.org/) between a LPC-mapped marker and a separate GIM marker. The V1.1 genome assembly consists of many small (maximum length ∼5 Mb) scaffolds, and thus such pairs were generally within 0.5 Mb. In a few cases, we could not make any links, and in a few others, we could not confidently determine which member of a duplicated marker pair on GIM matched the single locus mapped under that name in the LPC map, or vice versa; in the latter case, we made both links.
Results
Integrated M. guttatus map
The integrated M. guttatus map consists of 480 markers spanning 1504 cM on 14 linkage groups (Supplementary Figure S2). The total length is somewhat greater than each of the contributing maps because we chose a non-suppressed base map where possible for each chromosome containing a known polymorphic inversion. In addition to the three putative or confirmed inversions that suppress recombination on LG8 in DIM (Lowry and Willis, 2010), LG6 in CIM (Lee, 2009) and LG10 in NIM1 and NIM2 (Lee, 2009), there is local discrepancy of DIM vs CIM/NIM on LG5 as well as a few other regions (Supplementary Figure S1) that may indicate previously undetected differences in chromosome structure. The marker density in these areas is too low for further inferences at this time, but higher-resolution mapping with sequence-based markers may reveal additional chromosomal polymorphism within this diverse species complex. In addition, all of our mapping populations were polymorphic for a structural variant of LG11 associated with female meiotic drive (Fishman and Saunders, 2008), and the orders and marker distances on that linkage group reflect suppressed recombination relative to M. guttatus maps not segregating for the driving D haplotype (L Fishman, unpublished data).
Integrated M. lewisii map
The integrated M. lewisii map consists of 151 markers spanning 627 cM on seven linkage groups corresponding to eight chromosomes (Supplementary Figure S3). The two M. lewisii group maps differed in linkage group number and length because of putative inversions and translocations that suppress recombination in interspecific hybrids (Fishman et al., 2013). However, because >80% of markers were shared and the M. lewisii × M. parishii base map had high marker density placement of additional LC-only markers on the joint map was straightforward. Neither map had the expected eight linkage groups, because of an inferred M. lewisii-specific reciprocal translocation that joins two chromosomes into a single linkage group in both sets of interspecific hybrids (Fishman et al., 2013). Several markers from the cluster corresponding to LG10 on LP1+8 and LC1+8 (MgSTS.355, MgSTS.463), as well as one marker from LG12 (MgSTS.327), were completely unlinked from LG9 and LG14 markers in conspecific M. cardinalis (Fishman et al., 2013). An intraspecific M. lewisii map, which was constructed using gene-based MlSTS markers whose homologs can be located on the M. guttatus physical map by BLAST and thus physically connected to MgSTS markers in our maps, did not include any markers representing M. guttatus LG10 (Pince, 2009). However, a small number of markers each with homologs on LG12, LG9 and LG14 mapped to a single linkage group in that study. In addition, mapping of markers from across this region in M. cardinalis × M. parishii F2 hybrids, which should not segregate for the putative M. lewisii-specific translocation involving LPC 1+8, recovers two distinct (unlinked) groups (A Stathos and L Fishman, unpublished data). Taken together, these studies suggest that M. lewisii has chromosomes corresponding roughly to LG12+LG9+LG14 and LG10 and that M. cardinalis and M. parishii have chromosomes corresponding roughly to LG10+LG12 and LG9+LG14a, and that the M. lewisii arrangement is derived. However, because we did not have sufficient informative marker coverage to characterize the full extent of intrasection rearrangement and derive an order for any given species, we present LPC1+8 here as a single linkage group.
Synteny mapping
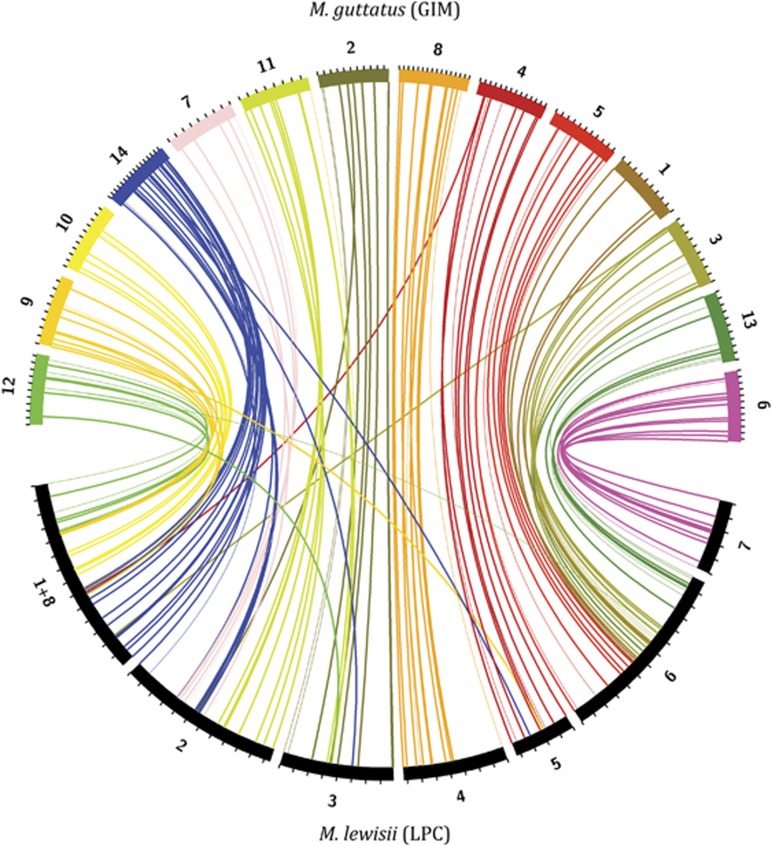
Alignment of the M. guttatus (GIM) and M. lewisii (LPC) integrated maps using 112 shared marker pairs (plus 29 genetic–physical pairs) revealed a complex history of fission, fusion and conservation resulting in major change in chromosome number (Figure 1). We found no evidence of recent within-genus polyploidy, as the vast majority of MgSTS primers amplified single polymorphic loci in a given cross and those that amplified multiple, presumably paralogous, loci showed no strong pattern of colocalization in the M. guttatus maps (Supplementary Figure S1). We did observe a few sets of paralogous markers potentially indicative of recent duplication (and potentially transposition) in M. guttatus, for example, a tightly linked group of markers on LG4 (e228a, e243a, e242a) all had partners clustered on LG9. However, markers from across LG4 and LG9 did not generally co-occur in the LPC-integrated map. Most importantly, we did not see intermingling of markers from multiple M. guttatus (GIM) groups in each M. lewisii (LPC) group (Figure 1), as might be expected if each LPC group corresponded to two homeologous M. guttatus groups derived since the split between the sections. Instead, we see a clear pattern of segmental synteny, with a few groups (LG6=LPC7, LG8=LPC4, LG4=LPC5) essentially unchanged at a large scale, and the rest exhibiting fission and/or fusion of large contiguous chromosomal blocks.
Figure 1.
Synteny between concatenated M. guttatus (GIM 1–14; colors) and M. lewisii (LPC 1+8–7; black) linkage maps. Lines connect either a single marker genetically mapped in both groups (heavy) or an LPC marker physically linked to a second GIM marker (light), and are colored by the M. guttatus group. Tick marks show 10 cM intervals. The LPC groups are shown to scale, but the GIM groups are equalized for clarity. The figure was constructed using Circos software (Krzywinski et al., 2009).
We identified 16 conserved chromosomal segments corresponding to 12 entire M. guttatus chromosomes plus two pairs of partial chromosomes (LG14a and LG14b and LG11a and LG11b). Because most Mimulus outside of Simiolus and sister section Paradanthus have base chromosome numbers of ∼8, we present a working hypothesis for increase in chromosome number from 8 to 14 (via n=16, as found in the Simiolus-sister clade Paradanthus). If we assume an ancestral M. cardinalis-like arrangement of LCP1+8 (LC1a=LG9+LG14a and LC1b=LG10+LG12), this hypothesis requires a minimum of eight ‘fission' events to separate LPC1+8 into LG10, LG12, LG9 and LG14a, LPC2 into LG14b, LG7 and LG11a, LPC3 into LG2 and LG11b and LPC6 into LG5, LG1, LG3 and LG13, plus two ‘fusions' (or reciprocal translocations plus losses of the minor products) to join LG14 and LG11. This is a conservative estimate of the number of karyotypic changes occurring between M. guttatus and M. lewisii groups, as it includes only large-scale fission–fusion events and does not include intrachromosome inversions or translocations. Given that three of the four relatively stable chromosomes exhibit a major inversion/translocation polymorphism within one or both species complexes, it is likely that such events have also occurred on a broader evolutionary scale.
It is also theoretically possible that there was a polyploidization event within Mimulus along the branch leading to Paradanthus (polyphyletic, mostly n=16), Simiolus (monophyletic, mostly n=14) and Erythranthe (monophyletic, n=8 or 9) (Beardsley et al., 2003). In this case, the lower chromosome numbers of Erythranthe would be secondarily derived by descending aneuploidy and homoplasic with the other n=8 taxa. However, analyses of expressed sequence tag libraries from M. lewisii and M. guttatus indicate that these taxa share a most recent whole-genome duplication event about 46–70 Mya (Clarke, 2012), which pre-dates the estimated origins of both the genus Mimulus (35 Mya) and the family Phrymaceae (40 Mya) (Beardsley et al., 2004; Nie et al., 2006). It is also theoretically possible that Paradanthus and Simiolus retain ancestral high chromosome numbers from this ancient polyploidization, and that all Mimulus clades with lower chromosome numbers have separately undergone reduction in chromosome number by fusion (in a minimum of four distinct lineages; Beardsley et al., 2004). In this case, the eight haploid chromosomes of Erythranthe taxa would result from recent (<20 Mya) fusion, and would again not correspond to the chromosomes of other low-number taxa. Further comparative mapping, genomic and cytogenetic analyses of diverse Mimulus species will be necessary to definitively rule out these latter possibilities. However, a burst of fission in the yellow monkeyflowers, rather than recent polyploidy and fusion in the Erythranthe group or multiple convergent fusions in different lineages, appears to be the most likely scenario given our comparative map data. In either case, we can certainly rule out a polyploidy event specific to yellow monkeyflowers previously hypothesized on the basis of chromosome number evolution alone.
Map length of segments
We are cautious about making direct intersection comparisons of map length because both M. lewisii maps were based on heterospecific crosses, and may exhibit undetected recombination suppression in addition to the five major rearrangements inferred from intrasection comparative mapping (Pince, 2009). However, it is apparent that the inferred increase in chromosome number in the M. guttatus group has resulted in substantial increases in intrachromosome recombination as well as the independent segregation of genes on new chromosomes (Figure 1). Most notably, all four markers from M. guttatus LG1 (spanning 100 cM) occupy a region of just <20 cM near the center of LPC6, indicating a >5-fold increase in recombination among genes in this region after fission. Similarly, the two M. lewisii groups that appear fully collinear with their corresponding M. guttatus groups and are not known to differ by rearrangements within the M. lewisii group (LPC7=LG6 and LPC5=LG4) are each less than half as long as the syntenic region spanned by the same markers in M. guttatus. This suggests either that undetected M. lewisii-specific inversions suppress recombination in both the LC and LP maps of these groups or that patterns of recombination have fundamentally changed across the section in concert with the change in chromosome number, perhaps due to changes in the location and size of centromeric regions.
Discussion
Changes in chromosome number are an important correlate of species diversification, and can be a direct cause of reproductive isolation. Reconstructing the mutational events that lead to chromosomal divergence is the key step in understanding the evolutionary processes that generate species and shape larger patterns of genome evolution. We infer that fission (and, less commonly, fusion), rather than whole genome duplication, accounts for a near-doubling of chromosome numbers in the lineage leading to the yellow monkeyflowers (M. guttatus species complex), a model system for ecological genetics and genomics. Despite this major karyotypic reorganization, the comparative linkage maps used to make this inference also demonstrate strong segmental synteny between M. guttatus group and M. lewisii group chromosomes, facilitating investigations of the genetic and molecular basis of phenotypic variation in the latter group and genus-wide. Furthermore, our results raise new questions about the mutational mechanisms and evolutionary causes of chromosomal number increase by fission, which may be more common in plants than has been generally appreciated.
In vertebrates, centric fission (whether simple or involving preduplication of centromeres Perry et al., 2004) is thought to be an important mechanism of karyotypic diversification (Ruiz-Herrera et al., 2011). In fact, a general trend of chromosome number increase by centric fission within some animal lineages led to the proposal of the ‘minimum interaction hypothesis', which posited that such fissions are universally favored because they reduce the probability of potentially costly reciprocal translocations (Imai et al., 1986). More recently, Pardo-Manuel de Villena and Sapienza (2001a) provided an elegant mechanism for mammalian chromosomal evolution by fission and fusion based on direct selection on chromosomal structure. They proposed that centromeric competition during asymmetric female meiosis (female meiotic drive; Pardo-Manuel de Villena and Sapienza, 2001b) may explain the observed bimodal pattern of karyotypes (all acrocentric or all metacentric, few mixed karyotypes) across mammals. Specifically, they argued that the underlying polarity of female meiosis in a given lineage may consistently favor either a pair of acrocentrics (two centromeres) or the homologous metacentric (one centromere) in heterokaryotypes leading to biased fixation probabilities for the products of chromosomal fission or fusion, respectively. A consistent transmission bias for either acrocentrics or metacentrics would result in similar karyotypic structures within a lineage, and periodic shifts in spindle polarity would then result in the mammal-wide bimodal pattern.
In plants, chromosomal fusion (Schubert and Lysak, 2011; Lysak and Schubert, 2012) has been widely inferred as a cause for chromosome number reductions following ancestral polyploidization in model taxa such as Arabidopsis and maize (Lysak et al., 2006; Wang and Bennetzen, 2012). In contrast, fission has received relatively little attention as a mechanism of chromosome number change in plants. Fission–fusion cycles have been observed in the common bean (Schubert et al., 1995) and there is no obvious mutational reason why fissions should not contribute to karyotypic evolution in plants as well as in mammals. Non-centric fission (that is, breakage of a chromosome into two pieces, only one of which includes a centromere) can carry obvious deleterious effects, but centric fissions should be less intrinsically deleterious, and no more so in plants than in mammals. In addition, chromosome breakage can lead to the recruitment of kinetochore proteins to new DNA sequences/locations and the epigenetic creation of functional neocentromeres (Topp et al., 2009; Fu et al., 2013). Such centromere repositioning events, accompanied by a shift in the centromere-associated tandem DNA repeat, appear to have been important in chromosome number evolution within the cucurbits (Han et al., 2009). Nonetheless, polyploidy has been concluded to be the major source of chromosome number increases in plants (Jones, 1998), except in a few odd lineages such as Zamia (Olson and Gorelick, 2011), slipper orchids (Cox et al., 1998) and sedges (Chung et al., 2012). In sedges, which have holocentric chromosomes, fission may be common in part because the lack of localized centromeres allows breakage anywhere along a chromosome with relatively low risks of meiotic dysfunction (Chung et al., 2012). However, increasing evidence of the dynamic and epigenetic nature of centromeres indicates that conservation of centromere position may not be a strong constraint on chromosome evolution, and thus suggests that chromosome increase by fission may be common even in non-holocentric taxa.
Because previous inferences of fission as a dominant mechanism of structural evolution within a particular lineage have generally been made on the basis of relationships between genome size and chromosome number, they often could not reconstruct individual evolutionary events (but see Han et al., 2009). Our results for Mimulus suggest that wholesale chromosomal fission may be important even in taxa without incremental series of chromosome numbers, and provide an opportunity to investigate its mechanisms and consequences for plant evolution. In particular, our findings raise the question of whether the selfish processes proposed to drive mammalian karyotypic evolution (Pardo-Manuel de Villena and Sapienza, 2001a) may also shape plant genome structure. With just one transition, we cannot reach broad conclusions, but it appears that there has been a consistent bias toward fission, rather than equal amounts of fission and fusion, along the lineage leading to M. guttatus. However, it also appears that the chromosome number doubling we see does not result exclusively from a single burst of centric fissions, as some ancestral chromosomes must have undergone multiple rounds of fission (and subsequent fusion) to generate the current M. guttatus karyotype. Comparative genetic mapping in related taxa, as well as cytogenetic and physical mapping, will be necessary to reconstruct that process of karyotypic evolution fully. For example, we can ask whether the 16 syntenic segments we observe correspond to the chromosomes of the M. moschatus group (n=16), which would indicate that the two apparent M. guttatus fusions (forming LG11 and LG14, respectively) were relatively recent and potentially unique to section Simiolus.
Reconstructing karyotypic evolution in Mimulus is important not just to capture broad patterns of genomic evolution but also to understand the evolutionary mechanisms driving chromosomal change. Fissional doubling in chromosome number requires concurrent increase in centromere number, either by doubling/splitting of existing centromeres (centric fission) or by the recruitment of novel genomic locations and/or sequences to centromeric function. As the chromosome-level genome assembly of the M. guttatus genome is analyzed, and additional diverse Mimulus are sequenced, we have the opportunity to examine explicitly the process of centromere and genomic sequence evolution in concert with chromosome number increase. For example, our comparative mapping predicts that M. guttatus LG1 should be unusually gene poor (as it corresponds to a very small portion at the center of one putative ancestral chromosome) and potentially acrocentric, whereas LG4, LG6 and LG8 should be metacentric and symmetrically gene-rich, having remained stable in gross structure for at least the 20 million years inferred to separate the M. guttatus and M. lewisii groups.
More specifically, our comparative maps may also provide clues to the origins of an unusual, putatively centromeric, female meiotic drive system in M. guttatus. In this system, a variant M. guttatus LG11 with abnormally large arrays of the primary centromere-specific DNA repeat (Cent728) exhibits near 100% transmission advantage against heterospecific M. nasutus chromosomes and also drives relatively weakly (∼60:40 bias) against non-driving homologs within M. guttatus. Fitness costs of drive homozygosity maintain intrapopulation polymorphism of two large non-recombining haplotypes spanning a large portion of LG11 (driving D haplotype frequency=∼35% in the focal Oregon population; Fishman and Saunders, 2008) and generate high levels of fitness variation due to genetic associations with male sterility. Recent cytogenetic screens suggest that Cent728 may be present in very low copy numbers across M. lewisii chromosomes, but is neither common nor centromere-associated (YZ Dong and L Fishman, unpublished data). Thus, understanding the establishment of new centromere locations and DNA associations during karyotypic turnover may be important for understanding the origins of meiotic drive, and vice versa. Interestingly, M. guttatus LG11 is one of the few chromosomes to have been derived by both fission and fusion. Specifically, the drive-associated genetic region (including markers MgSTS.87–MgSTS.471) corresponds to a small portion of M. lewisii LPC3, which otherwise corresponds to M. guttatus LG2, whereas the rest of LG11 corresponds to the bottom of M. lewisii LPC2. Thus, it is intriguingly possible that the fission and fusion events leading to the formation of LG11 predisposed that chromosome to further rearrangements, centromere repeat expansion and meiotic drive.
Finally, as genomic sequence becomes increasingly available from other Mimulus species (and until physically contiguous and closed genome sequences become far cheaper), our comparative genetic maps provide a template for investigating both chromosomal and genic evolution across this diverse model genus. Recent work suggests that chromosomal rearrangements have important roles in the evolution of reproductive isolation between sister species (Fishman et al., 2013) and in the maintenance of within-species (Lowry and Willis, 2010) and even within-population (Fishman and Saunders, 2008; Lee, 2009; Scoville et al., 2009) phenotypic variation in Mimulus. Linking the processes that drive recent chromosomal evolution with the larger-scale patterns revealed by genomic and genetic map comparisons holds great promise. More immediately, our maps provide a useful framework for investigations of the genetics of adaptation and speciation. Gene-based MgSTS markers allowed comparative mapping across approximately 20 million years of divergence within Mimulus, and are a resource for population and evolutionary genetics across the diversity of the genus. Recent studies have successfully used the M. guttatus physical map to target candidate genes involved in flower color divergence in M. aurantiacus and M. lewisii–M. cardinalis group, respectively (Streisfeld et al., 2013; Yuan et al., 2013), and MgSTS-anchored genetic maps will continue to provide an important link between physical maps and quantitative trait loci across the genus.
Data archiving
Data available from the Dryad Digital Repository: 10.5061/dryad.4bh26.
Acknowledgments
We thank SR Miller for assistance in drawing Figure 1 in Circos, A Stathos for assistance with mapping in the M. lewisii group, A Sweigart for assistance with marker testing, NIL construction and mapping in the M. guttatus complex, TJ Vision for MgSTS marker pipeline development, T Clarke for discussions of the duplication history of Mimulus genomes, P Beardsley for consultation on Mimulus phylogenetics and the DOE Joint Genome Institute for access to draft M. guttatus genome assemblies. Funding for Mimulus marker development and genetic mapping was provided by Grants NSF EF-0328326 to JHW, LF and others, DEB-0316786, DEB-0846089 and DEB-0918902 to LF and NIH GM073990 and NSF EF-0723814 to JHW.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Baldwin BG, Wessa BL. Origin and relationships of the tarweed-silversword lineage (Compositae-Madiinae) Am J Bot. 2000;87:1890–1908. [PubMed] [Google Scholar]
- Barr CM, Fishman L. The nuclear component of a cytonuclear hybrid incompatibility in Mimulus maps to a cluster of pentatricopeptide repeat genes. Genetics. 2010;184:455–465. doi: 10.1534/genetics.109.108175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beardsley PM, Schoenig SE, Whittall JB, Olmstead RG. Patterns of evolution in western North American. Mimulus Am J Bot. 2004;91:474–489. doi: 10.3732/ajb.91.3.474. [DOI] [PubMed] [Google Scholar]
- Beardsley PM, Yen A, Olmstead RG. AFLP phylogeny of Mimulus section Erythranthe and the evolution of hummingbird pollination. Evolution. 2003;57:1397–1410. doi: 10.1111/j.0014-3820.2003.tb00347.x. [DOI] [PubMed] [Google Scholar]
- Bouck A, Vision TJ. The molecular ecologist's guide to expressed sequence tags. Mol Ecol. 2007;16:907–924. doi: 10.1111/j.1365-294X.2006.03195.x. [DOI] [PubMed] [Google Scholar]
- Bradshaw HDJ, Wilbert SM, Otto KG, Schemske DW. Genetic mapping of floral traits associated with reproductive isolation in monkeyflowers (Mimulus) Nature. 1995;376:762–765. [Google Scholar]
- Burke JM, Lai Z, Salmaso M, Nakazato T, Tang S, Heesacker A, et al. Comparative mapping and rapid karyotypic evolution in the genus Helianthus. Genetics. 2004;167:449–457. doi: 10.1534/genetics.167.1.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman MA, Chang J, Weisman D, Kesseli RV, Burke JM. Universal markers for comparative mapping and phylogenetic analysis in the Asteraceae (Compositae) Theor Appl Genet. 2007;115:747–755. doi: 10.1007/s00122-007-0605-2. [DOI] [PubMed] [Google Scholar]
- Clarke T.2012Independence of Paleopolyploidy from Approximate-doubling of Chromosome Number in the Genome of Mimulus guttatusPlant & Animal Genome XX:San Diego, CA, USA [Google Scholar]
- Chung K-S, Hipp AL, Roalson EH. Chromosome number evolves independently of genome size in a clade with nonlocalized centromeres (Carex: Cyperaceae) Evolution. 2012;66:2708–2722. doi: 10.1111/j.1558-5646.2012.01624.x. [DOI] [PubMed] [Google Scholar]
- Cooley AM, Modliszewski JL, Rommel ML, Willis JH. Gene duplication in Mimulus underlies parallel floral evolution via independent trans-regulatory changes. Curr Biol. 2011;21:700–704. doi: 10.1016/j.cub.2011.03.028. [DOI] [PubMed] [Google Scholar]
- Cox AV, Abdelnour GJ, Bennett MD, Leitch IJ. Genome size and karyotype evolution in the slipper orchids (Cypripedioideae: Orchidaceae) Am J Bot. 1998;85:681–687. [PubMed] [Google Scholar]
- Cusimano N, Sousa A, Renner SS. Maximum likelihood inference implies a high, not a low, ancestral haploid chromosome number in Araceae, with a critique of the bias introduced by ‘x'. Ann Bot. 2012;109:681–692. doi: 10.1093/aob/mcr302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishman L, Saunders A. Centromere-associated female meiotic drive entails male fitness costs in monkeyflowers. Science. 2008;322:1559–1562. doi: 10.1126/science.1161406. [DOI] [PubMed] [Google Scholar]
- Fishman L, Willis JH. A novel meiotic drive locus almost completely distorts segregation in Mimulus (monkeyflower) hybrids. Genetics. 2005;169:347–353. doi: 10.1534/genetics.104.032789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishman L, Aagaard J, Tuthill JC. Toward the evolutionary genomics of gametophytic divergence: patterns of transmission ratio distortion in monkeyflower (Mimulus) hybrids reveal a complex genetic basis for conspecific pollen precedence. Evolution. 2008;12:2958–2970. doi: 10.1111/j.1558-5646.2008.00475.x. [DOI] [PubMed] [Google Scholar]
- Fishman L, Kelly AJ, Morgan E, Willis JH. A genetic map in the Mimulus guttatus species complex reveals transmission ratio distortion due to heterospecific interactions. Genetics. 2001;159:1701–1716. doi: 10.1093/genetics/159.4.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishman L, Stathos A, Beardsley PM, Williams CF, Hill JP. Chromosomal rearrangements and the genetics of reproductive barriers in Mimulus (monkeyflowers) Evolution. 2013;67:2547–2560. doi: 10.1111/evo.12154. [DOI] [PubMed] [Google Scholar]
- Fu S, Lv Z, Gao Z, Wu H, Pang J, Zhang B, et al. De novo centromere formation on a chromosome fragment in maize. Proc Natl Acad Sci USA. 2013;110:6033–6036. doi: 10.1073/pnas.1303944110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Zhang Z, Liu C, Liu J, Huang S, Jiang J, et al. Centromere repositioning in cucurbit species: implication of the genomic impact from centromere activation and inactivation. Proc Natl Acad Sci USA. 2009;106:14937–14941. doi: 10.1073/pnas.0904833106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai HT, Maruyama T, Gojobori T, Inoue Y, Crozier RH. Theoretical bases for karyotype evolution. 1. The minimum-interaction hypothesis. Am Nat. 1986;128:900–920. [Google Scholar]
- Jones K. Robertsonian fusion and centric fission in karyotype evolution of higher plants. Bot Rev. 1998;64:273–289. [Google Scholar]
- Krzywinski MI, Schein JE, Birol I, Connors J, Gascoyne R, Horsman D, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. Genetics Analysis of Standing Variation for Floral Morphology and Fitness Components in a Natural Population of Mimulus guttatus (Common Monkeyflower) Duke University: Durham, NC, USA; 2009. [Google Scholar]
- Levin D. The Role of Chromosomal Change in Plant Evolution. Oxford University Press: New York, NY, USA; 2002. [Google Scholar]
- Lowry DB, Willis JH. A widespread chromosomal inversion polymorphism contributes to a major life-history transition, local adaptation, and reproductive isolation. PLoS Biol. 2010;8:e1000500. doi: 10.1371/journal.pbio.1000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry DB, Hall MC, Salt DE, Willis JH. Genetic and physiological basis of adaptive salt tolerance divergence between coastal and inland Mimulus guttatus. New Phytol. 2009;183:776–788. doi: 10.1111/j.1469-8137.2009.02901.x. [DOI] [PubMed] [Google Scholar]
- Luo MC, Deal KR, Akhunov ED, Akhunova AR, Anderson OD, Anderson JA, et al. Genome comparisons reveal a dominant mechanism of chromosome number reduction in grasses and accelerated genome evolution in Triticeae. Proc Natl Acad Sci USA. 2009;106:15780–15785. doi: 10.1073/pnas.0908195106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysak MA, Schubert I.2012Mechanisms of chromosomal rearrangementsIn Greilhuber J, Dolezel J, Wendel JF, (eds) Plant Genome DiversityVol. 2Springer: Vienna, Austria; 137–147. [Google Scholar]
- Lysak MA, Berr A, Pecinka A, Schmidt R, McBreen K, Schubert I. Mechanisms of chromosome number reduction in Arabidopsis thaliana and related Brassicaceae species. Proc Natl Acad Sci USA. 2006;103:5224–5229. doi: 10.1073/pnas.0510791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayrose I, Barker MS, Otto SP. Probabilistic models of chromosome number evolution and the inference of polyploidy. System Biol. 2010;59:132–144. doi: 10.1093/sysbio/syp083. [DOI] [PubMed] [Google Scholar]
- Nie Z-L, Sun H, Beardsley PM, Olmstead RG, Wen J. Evolution of biogeographic disjunction between eastern Asia and eastern North America in Phryma (Phrymaceae) Am J Bot. 2006;93:1343–1356. doi: 10.3732/ajb.93.9.1343. [DOI] [PubMed] [Google Scholar]
- Olson K, Gorelick R. Chromosomal fission accounts for small-scale radiations in Zamia (Zamiaceae; Cycadales) Bot J Linn Soc. 2011;165:168–185. [Google Scholar]
- Pardo-Manuel de Villena F, Sapienza C. Female meiosis drives karyotypic evolution in mammals. Genetics. 2001;159:1179–1189. doi: 10.1093/genetics/159.3.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo-Manuel de Villena F, Sapienza C. Nonrandom segregation during meiosis: the unfairness of females. Mamm Genome. 2001;12:331–339. doi: 10.1007/s003350040003. [DOI] [PubMed] [Google Scholar]
- Perry J, Slater HR, Choo KHA. Centric fission—simple and complex mechanisms. Chromosome Res. 2004;12:627–640. doi: 10.1023/B:CHRO.0000036594.38997.59. [DOI] [PubMed] [Google Scholar]
- Pince C. Evolution and Genetics of Floral Form in Mimulus. University of Washington: Seattle, WA, USA; 2009. [Google Scholar]
- Ruiz-Herrera A, Farre M, Robinson TJ. Molecular cytogenetic and genomic insights into chromosomal evolution. Heredity. 2011;108:28–36. doi: 10.1038/hdy.2011.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert I, Lysak MA. Interpretation of karyotype evolution should consider chromosome structural constraints. Trends Genet. 2011;27:207–216. doi: 10.1016/j.tig.2011.03.004. [DOI] [PubMed] [Google Scholar]
- Schubert I, Rieger R, Fuchs J. Alteration of basic chromosome number by fusion–fission cycles. Genome. 1995;38:1289–1292. doi: 10.1139/g95-170. [DOI] [PubMed] [Google Scholar]
- Scoville A, Lee YW, Willis JH, Kelly JK. Contribution of chromosomal polymorphisms to the G-matrix of Mimulus guttatus. New Phytol. 2009;183:803–815. doi: 10.1111/j.1469-8137.2009.02947.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streisfeld MA, Young WN, Sobel JM. Divergent selection drives genetic differentiation in an R2R3-MYB transcription factor that contributes to incipient speciation in Mimulus aurantiacus. PLoS Genet. 2013;9:e1003385. doi: 10.1371/journal.pgen.1003385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topp CN, Okagaki RJ, Melo JR, Kynast RG, Phillips RL, Dawe RK. Identification of a maize neocentromere in an Oat–Maize addition line. Cytogenet Genome Res. 2009;124:228–238. doi: 10.1159/000218128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ooijen J. JoinMap 4: Software for the Calculation of Genetic Linkage Maps in Experimental Populations. Kyazma BV: Wageningen, Netherlands; 2006. [Google Scholar]
- Wang H, Bennetzen JL. Centromere retention and loss during the descent of maize from a tetraploid ancestor. Proc Natl Acad Sci USA. 2012;109:21004–21009. doi: 10.1073/pnas.1218668109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood TE, Takebayashi N, Barker MS, Mayrose I, Greenspoon PB, Rieseberg LH. The frequency of polyploid speciation in vascular plants. Proc Natl Acad Sci USA. 2009;106:13875–13879. doi: 10.1073/pnas.0811575106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CA, Lowry DB, Cooley AM, Wright KM, Lee YW, Willis JH. Mimulus is an emerging model system for the integration of ecological and genomic studies. Heredity. 2008;100:220–230. doi: 10.1038/sj.hdy.6801018. [DOI] [PubMed] [Google Scholar]
- Wu F, Tanksley SD. Chromosomal evolution in the plant family Solanaceae. BMC Genom. 2010;11:182. doi: 10.1186/1471-2164-11-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan YW, Sagawa JM, Young RC, Christensen BJ, Bradshaw HDJ. Genetic dissection of a major anthocyanin QTL contributing to pollinator-mediated reproductive isolation between sister species of Mimulus. Genetics. 2013;194:255–263. doi: 10.1534/genetics.112.146852. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.