Abstract
Tuberculosis (TB) caused by Mycobacterium bovis is a re-emerging disease of livestock that is of major economic importance worldwide, as well as being a zoonotic risk. There is significant heritability for host resistance to bovine TB (bTB) in dairy cattle. To identify resistance loci for bTB, we undertook a genome-wide association study in female Holstein–Friesian cattle with 592 cases and 559 age-matched controls from case herds. Cases and controls were categorised into distinct phenotypes: skin test and lesion positive vs skin test negative on multiple occasions, respectively. These animals were genotyped with the Illumina BovineHD 700K BeadChip. Genome-wide rapid association using linear and logistic mixed models and regression (GRAMMAR), regional heritability mapping (RHM) and haplotype-sharing analysis identified two novel resistance loci that attained chromosome-wise significance, protein tyrosine phosphatase receptor T (PTPRT; P=4.8 × 10−7) and myosin IIIB (MYO3B; P=5.4 × 10−6). We estimated that 21% of the phenotypic variance in TB resistance could be explained by all of the informative single-nucleotide polymorphisms, of which the region encompassing the PTPRT gene accounted for 6.2% of the variance and a further 3.6% was associated with a putative copy number variant in MYO3B. The results from this study add to our understanding of variation in host control of infection and suggest that genetic marker-based selection for resistance to bTB has the potential to make a significant contribution to bTB control.
Keywords: genome-wide association study, bovine tuberculosis, novel resistance loci
Introduction
Bovine tuberculosis (bTB), caused by Mycobacterium bovis, is a chronic respiratory disease characterised by granulomas in affected tissues. This disease has substantial animal health and welfare consequences, as well as a serious economic impact in the Great Britain (GB), Northern Ireland (NI) and Republic of Ireland (ROI), none of which have bTB-free status despite more than five decades of compulsory skin testing and slaughter. The costs to governments in these countries were £227 million (£152 million in GB, £23 million in NI and £52 million in the ROI) in the 2010/2011 period alone (Abernethy et al., 2013). In addition, bTB is increasing across the European Union in both countries officially free of bTB (OTF) and in non-OTF countries (Schiller et al., 2011) and remains a significant risk in other OTF countries (Humblet et al., 2009). On a global scale, this zoonotic pathogen is estimated to cause 10–15% of human TB cases in the developing world (Michel et al., 2010) and is considered to be the fourth most significant livestock disease in terms of impact on human health and economics in developing countries, including risks to other livestock and wildlife (Perry et al., 2002). Furthermore, there is evidence that subclinical bTB has a negative impact on productivity in dairy cows, increasing the costs incurred by the dairy industry (Boland et al., 2010). As many parts of the world have no active surveillance programmes and limited epidemiological studies, the prevalence and impact of bTB worldwide is likely to be underestimated (Grace et al., 2012).
The risk of bTB to cattle is dependent on host, pathogen and environmental factors and there is a broad spectrum of outcomes to infection with M. bovis (Thoen et al., 2006), which are thought to be similar to the effects of the related pathogen M. tuberculosis (Waters et al., 2010) in humans. The wide range of responses is partly due to the complexity of host–pathogen interactions across time involving both the innate and adaptive immune systems. This complexity limits efficacy of the current diagnostic tests (De la Rua-Domenech et al., 2006), which in turn leads to difficulties in disease control. Control measures in the United Kingdom and ROI involve statutory screening of all cattle herds using the single intradermal comparative tuberculin test (SICTT), followed by culling and movement restrictions when skin test-positive cattle are detected. In addition, abattoir surveillance is carried out by specialist meat inspectors on all SICCT test reactor cattle looking for evidence of suspect tuberculous lesions in defined organs and body sites. All cattle destined for the human food chain are subjected to carcase inspection.
Given the multifaceted issues surrounding bTB, it is likely that control and eradication will require additional control strategies. A potentially powerful approach would be the exploitation of genetic variation in host resistance to bTB. The aim would be to selectively breed cattle that are genetically more resistant to infection with M. bovis, complementing other control approaches. Substantial evidence suggests between-host genetic variation in resistance to TB exists in many species, including humans, mice, deer and cattle (reviewed by Allen et al., 2010). In particular, quantitative genetic studies in herds in GB and ROI have shown convincingly that significant heritability for host resistance exists in dairy cattle herds (Bermingham et al., 2009, 2010; Brotherstone et al., 2010). Recently, several studies have begun to address the identification of genetic loci associated with bTB resistance. Polymorphisms in candidate genes, SLC11A1 in African Zebu cattle (Kadarmideen et al., 2011) and (TLR1) in Chinese Holsteins (Sun et al., 2012), have been significantly associated with bTB outcome. Two genomic regions identified by the microsatellite markers INRA111 and BMS2753 were also reported to be associated with SICTT reactor status in UK cattle (Driscoll et al., 2011). However, this study used a methodology via association tests based on microsatellite markers in a mixed breed population, which may not give robust results. Finally, a recent genome-wide association study (GWAS) in Holsteins in ROI identified a genomic region on Bos taurus chromosome 22 containing the taurine transporter gene SLC6A6, which was suggestively associated with bTB resistance, although it only explained a small proportion of the total variance due to genetic factors (Finlay et al., 2012).
Thus, host genetic control of resistance to bTB in cattle is likely to be complex and involve many loci of varying effect. Furthermore, each study to date has had limitations in terms of the number of animals studied and the genomic tools deployed. Therefore, to more robustly explore the genetic basis of TB, we performed a GWAS in naturally exposed Holstein–Friesian cattle from 146 dairy herds in Northern NI, using both more animals and more powerful genomic tools than previous studies. The aim was to demonstrate the existence of genetic variation in the risk of bTB and to identify specific genomic regions contributing to this variation.
Materials and methods
Phenotypes and case–control definitions
Our data were collected from commercial Holstein–Friesian cattle in NI. Phenotypes used to define bTB status, and hence to group animals into cases or controls, were derived from data collected as part of the NI bTB control programme. Specifically, SICTT results and post-mortem abattoir inspection data were made available from the Animal and Public Health Information System (Abernethy et al., 2006) of the NI Department of Agriculture and Rural Development.
The SICCT is the primary screening test in the bTB surveillance and control programme in GB and Ireland (De la Rua-Domenech et al., 2006). M. bovis-PPD antigen is injected into the neck of the animal and after 72 h the size of reaction to M. bovis-PPD is compared with the reaction similarly induced by M. avium-PPD, to correct for potential exposure to other non-tuberculous Mycobacterium spp. For our purposes, differences in reaction between M. bovis and M. avium antigens of >4, 1–4 and <1 mm were deemed positive, inconclusive and negative, respectively. All cattle testing positive to the SICTT are slaughtered and undergo standard meat inspection, including organ and lymph node inspection and palpation. Lesions from SICTT-positive cattle undergo histopathology and, from SICTT-positive cattle in which no lesions are observed, M. bovis culture is performed on specified tissues. Confirmation of M. bovis infection is determined through a combination of histopathology, bacterial culture and genotyping (Skuce et al., 2010). The statutory bTB control programme definition of an infected animal is based on the presence of a positive SICTT and/or confirmed bacterial culture from diseased tissue. Diagnostic sensitivity of all of these procedures is variable. For example, cattle in the early stages of infection with positive SICTT diagnostic test results may not disclose lesions at slaughter. Furthermore, cattle with advanced TB may be anergic to the SICTT. Thus, as described by Allen et al. (2010), a spectrum of phenotypes may be observed. To ensure that the cases in our study consisted of truly infected animals, we defined the case phenotype as an animal that was positive to both the SICTT and abattoir inspection, that is, animals that had exhibited an immune response to M. bovis PPD as well as evidence of pathology in the form of granuloma, lesions at slaughter, positive culture and molecular confirmation of M. bovis. Controls were defined as female cattle that were raised contemporaneously with cases but had negative SICTT results before sampling, and on every test (two or more occasions, mean=10 tests) after sampling, and had at least an equal opportunity of being exposed to the pathogen as had their bTB case herd mates. Thus, as far as was feasible, we minimised errors in phenotype assignment to improve the power of the study to detect true associations.
Sampling cases and controls and definition of experimental animals
Blood samples from 2975 tuberculin-positive cattle were sampled at slaughter between August 2008 and June 2009. Of these animals, 822 had infection confirmed through histopathology, bacterial culture and genotyping, and hence were defined as potential cases.
To ensure accurate phenotype definition, and hence increase experimental power, it is desirable for controls to have had a high probability of exposure and both groups to come from epidemiologically comparable groups (Allen et al., 2010). To achieve this, longitudinal epidemiological data for herds, from which case cattle were sampled, were made available from the APHIS database. The data comprised herd- and animal-level demographic data and animal movement records, together with disease data: SICTT results, abattoir inspection data, laboratory M. bovis culture and histopathology records. These data were then used to define episodes of infection (Bermingham et al., 2009) in case herds. We defined an episode as herd restrictions initiated by two SICTT-positive cattle and terminated by two consecutive clear herd tests. In total, 1355 cattle (685 cases, 670 controls) from 178 case herds were grouped within defined episodes. Furthermore, cattle that were moved into the herd during or within 6 weeks before the beginning of an episode were excluded, as they may have been infected before entry to the herd. Age-matched control animals were sampled from 165 episodes within 146 case herds, avoiding herd episodes with two or fewer cases in an attempt to increase the probability of exposure to infection. There were generally close to equal numbers of cases and controls per farm, except for a bias for more controls from farms with a higher prevalence to increase the probability of exposure to infection.
These longitudinal data were also used retrospectively to identify and remove control cattle that subsequently exhibited a positive SICTT result as well as negative animals that disclosed a lesion at slaughter. Male cattle were also removed as they have transient residence in dairy production systems, and hence likely to experience different M. bovis infection pressures. In total, 1223 female cattle (629 cases, 594 controls) from 146 case herds were selected for DNA extraction and genotyping.
SNP chip genotyping
All DNA extraction and genotyping was conducted by the ARK-Genomics laboratory at The Roslin Institute (http://www.ark-genomics.org). Genomic DNA was extracted from blood samples using the Promega Maxwell 16 Blood DNA Purification Kit (Promega, Madison, WI, USA) and DNA recovery from each sample was quantified using Invitrogen's Molecular Probes QuantitPicoGreen ds DNA Assay Kit (Invitrogen, Carlsbad, CA, USA) and measured on a Thermo Labsystems Fluoroskan Ascent (Labotal Scientific Equipment, Abu-Gosh, Israel). The extracted DNA samples were then normalised to 50 ng μl−1. Samples that failed to meet the 50 ng μl−1 minimum DNA concentration recommended for Illumina genotyping were removed.
The BovineHD Genotyping BeadChip (Illumina Inc., San Diego, CA, USA) was then used to genotype the female cattle in the study, of which 628 cases and 591 controls were genotyped. The BovineHD BeadChip assays 727 252 single-nucleotide polymorphism (SNP) markers with a median interval of 3 kb between SNPs (Illumina Inc.; 2011 BovineHD Genotyping BeadChip Data Sheet: DNA Analysis. http://www.illumina.com/Documents/products/datasheets/datasheet_bovineHD.pdf). The BeadChips were scanned using iScan (Illumina Inc.) and analysed using GenomeStudio software version 2011.1 (Illumina Inc.). The genotypes in this study were inferred based on forward (positive) strand output from GenomeStudio. Stringent quality control parameters were applied at the sample and SNP level to ensure reliability of results. The minimal acceptable Gentrain call (GC) score for individual typings was 0.60. Loci with a minor allelic frequency of <5% or with call rates <90% were excluded. Cattle with average GC scores below 0.65 or call rate of <90% were excluded. Finally, four pairs of samples that appeared to be duplicates from the same animal, as indicated by their high concordance for SNP identity by state, were removed. The final data set comprised 1151 animals (592 cases, 559 controls) genotyped at 617 010 loci. The SNP positions within a chromosome were mapped to the bovine genome assembly (Bos taurus UMD 3.0).
Statistical analyses
The genome-wide association analysis (GWA) was performed using the genome-wide rapid association using mixed model and regression (GRAMMAR; Aulchenko et al., 2007) approach, this being a two-step approximation to full mixed-model analyses. Pedigree information was not available for these animals, so the pedigree relationship matrix was replaced with a marker-based relationship matrix, that is, the identity-by-state matrix (G) adjusts for average allele sharing or relatedness among sampled cattle and hence removed genetic stratification. The identity-by-state coefficients between pairs of animals i and j were estimated from genotype data as follows:
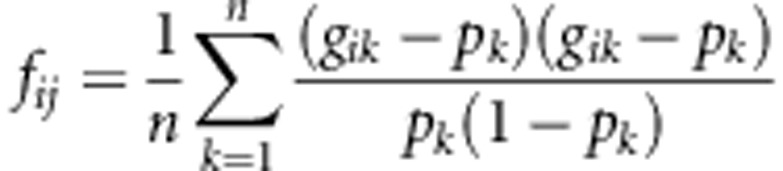 |
where gik is the genotype of the ith animal at the kth SNP (coded as 0, 1/2 and 1, for minor allele homozygote, heterozygote and common homozygote, respectively), pk is the frequency of the major allele and n is the number of SNPs used for kinship estimation in the statistical package GenABEL (Aulchenko et al., 2007). The genomic relationship matrix was then calculated as twice the kinship matrix.
Multidimensional scaling procedures were used to assess population stratification in GenABEL. We performed a multidimensional scaling analysis on the 1151 × 1151 matrix of genome-wide identity-by-state pairwise distances (G), using the multidimensional scaling plot option in conjunction with clustering. The first principal component values were plotted against the second principal component values to identify potential clustering among the cases and controls. However, no apparent clustering of cases and controls was observed (Supplementary Figure 1) nor was there any observable difference between animals classified as Holstein or Friesian (results not shown). We concluded that there was no apparent substructure differentiating the case and control samples; hence, fitting the genomic relationship matrix should be sufficient to account for genetic structure in the population.
In the first step of the GRAMMAR approach, we fitted the following mixed linear animal model to the data:
where y represents binary bTB status, μ is the overall mean,
b is the vector of fixed effects, a is the additive genetic effect,
matrices X and Z are incidence matrices and e is a vector
containing residuals, in the statistical package ASReml (Gilmour
et al., 2009). The covariance structure of the additive genetic
effects was accounted for by fitting the genomic relationship matrix G, as
described above. The binary trait was treated as a continuous trait as often done in
genetic and GWA studies (Bermingham et al., 2009;
Minozzi et al., 2012). Fixed effects were
chosen to account for risks of bTB, these being breed (Holstein vs Friesian), age at,
season of, year of and reason for episode-initiating SICTT. As well as providing
residual values for step two, this model also yields an estimated heritability for bTB
resistance. The whole genomic heritability (h2) was estimated from
the variance components from Model (1) using the formula
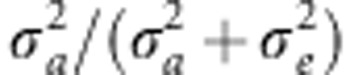 (Nagamine et al., 2012), where
σ2a is the additive genetic variance estimated
using the G matrix and σ2e is the residual
variance.
(Nagamine et al., 2012), where
σ2a is the additive genetic variance estimated
using the G matrix and σ2e is the residual
variance.
In the second step of the GRAMMAR approach, single SNP associations were performed using the residuals from the mixed model as the phenotype; these residuals capture much of the SNP effect and are independent of familial structure. The residuals were regressed on each SNP in turn using GenABEL, assuming an additive effect of the number of alleles on the trait. There was little evidence of any general inflation of the test statistics (Supplementary Figure 2). Nevertheless, the genome-wide degree of inflation (λ) was calculated to test and correct for any hidden substructure that may cause an inflation of significant results. The λ was less than one across all analyses, indicating that genomic control for substructure (in which test statistics are scaled by 1/λ) was not required in subsequent analyses. Appropriate significance levels, after correction for multiple testing, at the chromosome- and genome-wide level were estimated using the permutation procedure (1151 permutations, as there were 1151 animals) available in the GenABEL package. Associations were deemed statistically significant at the 5% empirical significance cutoff.
An alternative means of identifying genomic regions associated with the phenotype, known as RHM (Nagamine et al., 2012), was also investigated. RHM is a variance components approach to map genomic regions influencing complex traits, which combines information across SNPS. Each chromosome is divided into windows of a predefined number of SNPs, and the additive variance attributable to each window is estimated and compared with the null hypothesis of no variance in that window (i.e., Model (1), above). The full mixed model was as follows:
where the notation and fitted effects are the same as Model (1), and r is regional genomic additive genetic effect.
The whole genomic relationship matrix G was estimated using all SNPs and the regional genomic additive effect was estimated from a regional genomic relationship matrix constructed from adjacent SNPs from each region. In our baseline analysis, window sizes of 100 or 50 SNPs were used to construct a regional relationship matrix and the window was shifted every 50 or 25 SNPs, respectively. In total, 11 767 and 22 758 windows were tested across the 29 bovine autosomal chromosomes.
The regional heritability (h2r) was estimated from the
variance components from Model (2) using the formula:
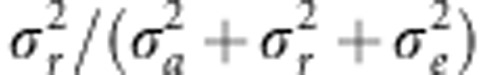 (Nagamine et al., 2012), where
σ2r is the region-specific additive genetic
variance. We used the likelihood ratio test statistic
(LRT)=−2ln(L0/L1) to test for
the presence of regional variance against the null hypothesis of no regional variance at
each window, where L0 and L1 represent the
respective likelihood values under the reduced (1) and full (2) models, respectively.
Bonferroni correction was used to adjust P-values for multiple testing at the
chromosome and genome-wide level, based on the number of windows tested. The LRT was
judged to be significant at the 5% significance level.
(Nagamine et al., 2012), where
σ2r is the region-specific additive genetic
variance. We used the likelihood ratio test statistic
(LRT)=−2ln(L0/L1) to test for
the presence of regional variance against the null hypothesis of no regional variance at
each window, where L0 and L1 represent the
respective likelihood values under the reduced (1) and full (2) models, respectively.
Bonferroni correction was used to adjust P-values for multiple testing at the
chromosome and genome-wide level, based on the number of windows tested. The LRT was
judged to be significant at the 5% significance level.
To obtain unbiased estimates of the allele substitution and genotypic effects of, and the variance (σ2SNP) explained by SNPs found to be significant in the GRAMMAR model, these SNPs were included in full mixed-model analyses (Model (1)), separately and simultaneously as a covariate (i.e., regression on allele number), as an additional fixed effect (i.e., three categories being the two homozygous classes and heterozygous) or as a random effect (i.e., assuming SNP∼N [0, σ2 SNP]), respectively. Further, a logistic animal linear mixed model was fitted in ASReml to estimate the odds ratios of significant bTB-associated risk alleles. These analyses were all conducted in ASReml.
Linkage disequilibrium (LD) blocks were inferred in regions of association in Haploview using the Gabriel confidence interval method (Barrett et al., 2005). The Gabriel protocol has an upper D′ confidence interval bound of 0.98, a lower D′ confidence interval bound of 0.70 and with 5% of informative markers required to be in strong LD. Haplotypes, specified in terms of the forward strand, were phased in PLINK (Purcell et al., 2007). The random effect of phased haplotypes were fitted in Model (1) using the software ASReml.
Statistical significance of the fixed effect of genotype and haplotype was determined using the F-test, with individual allele substitution effects evaluated using a t-test. The LRT was performed to evaluate the random effects of genotype/haplotypes on bTB status. All tests were judged significant at the nominal 5% significance level.
To investigate experimental power, the non-centrality parameter (assuming no phenotype misclassification, and perfect LD with the causal variant) (Yang et al., 2010) was calculated for SNPs contributing different levels of phenotypic variance. The false-negative rate (β) was then calculated using non-central χ2 distribution function in R package normal distribution, with threshold corresponding to the genome-wide significance. Further, we then assumed an expected r2=0.5 between SNP and causative mutation. Power was then calculated as 1−β. Finally, for the most significant results found, we investigated differential no-call rates within the case and control groups, or across the genotypes using the PLINK missingness-by-phenotype and non-random missingness routines, respectively (Purcell et al., 2007).
Identification of candidate genes
Genomic regions where significant results were obtained were further explored to attempt to identify candidate genes underlying the loci. The location of SNPs and gene annotations were based on the UMD_3.1 assembly (http://www.ncbi.nlm.nih.gov/genome/82?project_id=33843). Transcripts and annotation were downloaded on the UCSC Genome Browser (http://genome.ucsc.edu) and aligned with human, mouse, rat and zebra fish RefSeq mRNA sequences using BLAT.
Results
Overview of results
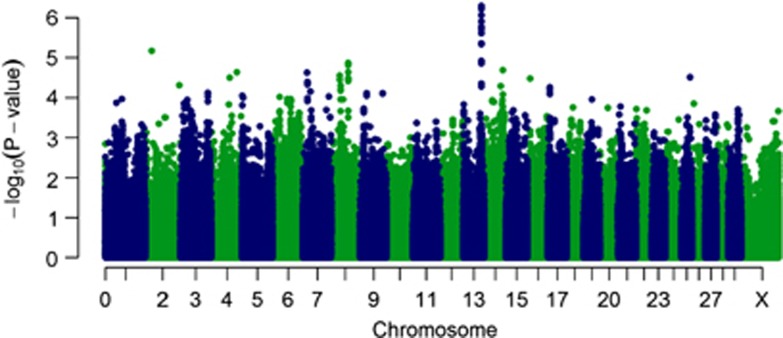
The heritability of bTB resistance in this study was estimated at 21.0% (95% confidence interval: 8.6–33.4). Power analysis (under the assumption of perfect case–control classification and perfect linkage disequilibrium with the causal variant) has demonstrated that this study is moderate (⩾0.80) to well-powered (⩾0.99) to detect variants contributing ⩾0.4% and 1.0% of phenotypic variance, respectively, at the genome-wide level. The GWAS analysis identified SNPs with significant association with resistance to bTB on chromosomes 2 and 13. A notable deviation of the observed significance level from that expected by chance was observed, suggestive of the presence of true associations (Supplementary Figure 2). The Manhattan plot of observed P-values (Figure 1) revealed a set of seven closely located SNPs (rs42494357, rs110465273, rs42494342, rs109809949, rs109042660, rs137562332, rs132841890; Table 1 and Figure 2) between 71.78 and 71.79 Mb on chromosome 13 and a single SNP (rs136617760; Table 1 and Supplementary Figure 3) at 25.90 Mb on chromosome 2, which were statistically significant at the chromosome-wide level (empirical P-value from permutation testing <0.05). When fitted in a full mixed model, the SNP (rs136617760) on chromosome 2 explained 4.0% (χ2=12.86; P-value 3.4 × 10−04), and the GG haplotype on chromosome 13 (see below) explained 6.6% (χ2=21.21; P-value 4.3 × 10−06) of phenotypic variance. When fitted simultaneously, we estimated that 18.8% of the variance in TB resistance can be explained by polygenic effects other than the two significant regions, 6.2% can be explained by the GG haplotype on chromosome 13 and a further 3.6% by rs136617760 on chromosome 2 (χ2=33.46; P-value 7.3 × 10−09).
Figure 1.
Manhattan plots showing P-values of association for each SNP, expressed as −log 10 (P-value). Manhattan plot showing P-values from the GRAMMAR approach. The y axis shows the –log 10 P-values of 617 010 SNPs, and the x axis shows the chromosomal positions.
Table 1. Results from the additive and genotype models of significantly associated variants in the GWAS.
| SNP |
Additive model
|
Genotype model
|
||||||
|---|---|---|---|---|---|---|---|---|
| P-valuea |
Allele substitution effect
|
P-valuea |
Heterozygous non-ref. effect
|
Homozygous non-ref. effect
|
||||
| b(95% CI)b | OR(95% CI) c | b(95% CI)b | OR(95% CI) c | b(95% CI)b | OR(95% CI) c | |||
| Rs136617760 | 6.8 × 10−06 | −0.10(−0.15, −0.05) | 0.64(0.51–0.81) | 4.5 × 10−05 | −0.20(−0.35, −0.05) | 0.41(0.20–0.86) | −0.19(−0.30, −0.09) | 0.36(0.21–0.62) |
| Rs42494357 | 8.6 × 10−07 | −0.13(−0.18, −0.08) | 0.58(0.46–0.73) | 1.5 × 10−05 | −0.13(−020, −0.07) | 0.57(0.43–0.75) | −0.26(−0.39, −0.12) | 0.35(0.19–0.65) |
| Rs110465273 | 6.1 × 10−07 | −0.13(−0.18, −0.08) | 0.58(0.46–0.73) | 1.5 × 10−05 | −0.13(−020, −0.07) | 0.57(0.43–0.75) | −0.26(−0.39, −0.12) | 0.36(0.19–0.69) |
| Rs42494342 | 5.9 × 10−07 | −0.13(−0.18, −0.08) | 0.58(0.46–0.73) | 1.5 × 10−05 | −0.13(−020, −0.07) | 0.52(0.39–0.71) | −0.26(−0.39, −0.12) | 0.35(0.19–0.65) |
| Rs109809949 | 1.2 × 10−06 | −0.13(−0.18, −0.08) | 0.58(0.46–0.73) | 2.8 × 10−05 | −0.13(−0.19, −0.07) | 0.53(0.39–0.71) | −0.25(−0.39, −0.12) | 0.37(0.19–0.71) |
| Rs109042660 | 5.2 × 10−07 | −0.13(−0.18, −0.08) | 0.58(0.46–0.73) | 1.3 × 10−05 | −0.13(−0.20, −0.07) | 0.52(0.38–0.70) | −0.26(−0.39, −0.12) | 0.37(0.19–0.71) |
| Rs137562332 | 6.1 × 10−07 | −0.13(−0.18, −0.08) | 0.58(0.46–0.73) | 1.5 × 10−05 | −0.13(−020, −0.07) | 0.57(0.43–0.75) | −0.26(−0.39, −0.12) | 0.35(0.19–0.65) |
| Rs132841890 | 5.9 × 10−07 | −0.13(−0.18, −0.08) | 0.58(0.46–0.73) | 1.4 × 10−05 | −0.13(−0.19, −0.06) | 0.58(0.44–0.77) | −0.27(−040, −0.13) | 0.33(0.18–0.61) |
Abbreviations: CI, confidence interval; gBLUP, genomic best linear unbiased prediction; GWAS, genome-wide association study; non-ref., non-reference (minor) allele on Illumina BovineHD BeadChip; OR, odds ratio; P-value, probability of obtaining the observed effect size (b), assuming that the null hypothesis is true; SNP, single-nucleotide polymorphism GenBank accession number.
P-values from genome-wide rapid association using linear and logistic mixed models and regression in GenABEL.
Effect size estimated from gBLUP using a normal link function.
OR estimated from gBLUP using a logit link function.
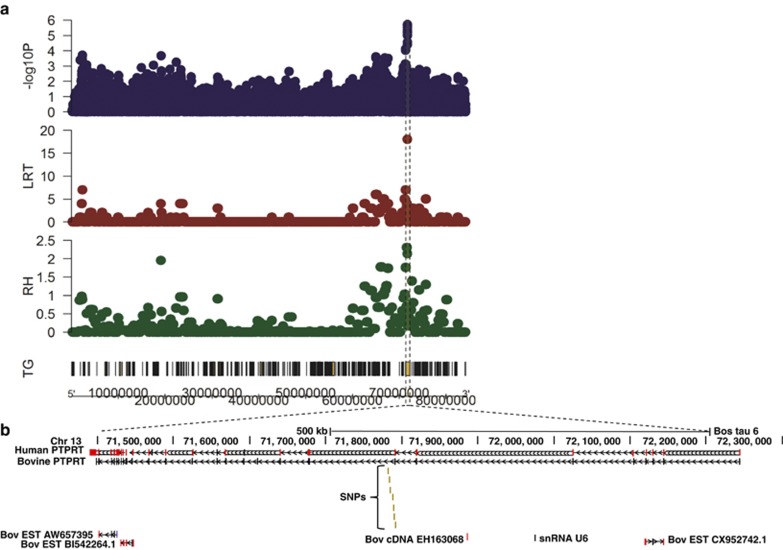
Figure 2.
Regional association plot of chromosome 13 (a) and UCSC browser image (b) of the region surrounding significant SNPs on BTA 13 (UMD_3.1, Btau 6.1). The y axis shows the –log 10 P-values from GWA and the estimated regional heritabilities (RH) and corresponding values for the LRT of the regional heritability from the RHM approaches for tuberculosis resistance, and transcribed genes (TG) on chromosome 13. The x axis shows the chromosomal positions. The shaded area highlights the region of association in PTPRT. The label SNPs refers to rs132841890, rs42494357, rs110465273, rs42494342, rs109809949, rs134449285 and rs137562332; the SNPs are shown in relation to the following sequences: bovine PTPRT predicted transcript XM_002692322.1 (bovine PTPR); RefSeq (NM_133170.3) mRNA transcript 1 (human PTPRT); three bovine EST sequences, one bovine cDNA clone and a bovine U6 spliceosomal RNA (snRNA U6) also map to this region as shown.
Chromosome 13 associations
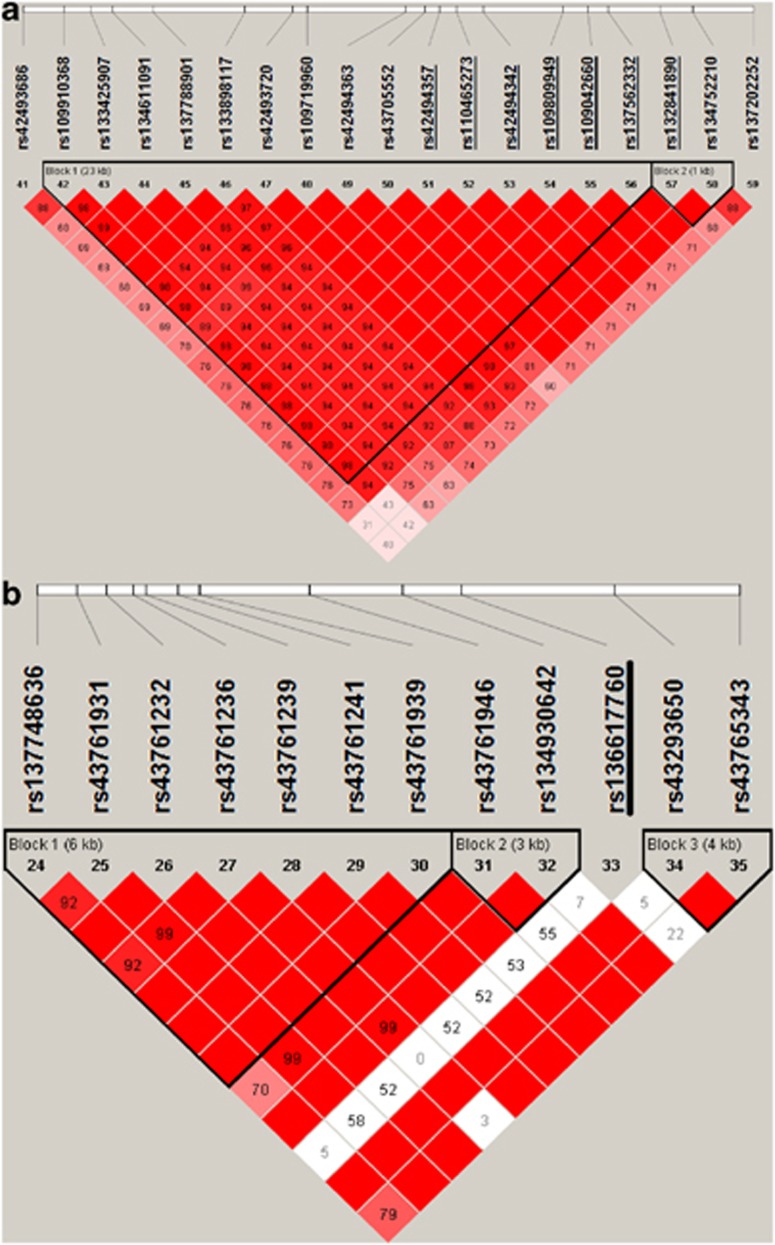
The seven SNPs in the region of association on chromosome 13 had high call rates approaching 100% (Table 2) and were in Hardy–Weinberg equilibrium (HWE) (P>0.05; Table 2). The SNPs were also in strong LD (Figure 3a), and close together (<10 kb apart; Table 2), thus making it currently impossible to distinguish them in terms of their effect on the infection phenotype (Table 1). The major allele A of the top hit rs110465273 was the allele associated with greater risk in this population (F-test 29.09; P-value 6.1 × 10−7), with an additive model providing the best fit (Table 1). Two haplotype blocks were found in the region of association on chromosome 13 (Figure 3a). The first haplotype block 1 containing 15 SNPs—rs109910368 to rs137562332 (Figure 3a)—was significantly associated with resistance to bTB (F-test 23.28; P-value 1.6 × 10−6). Three frequent haplotypes (5′-GCAGAGGAAGAGAGG-3′, 5′-ACAGAGGAAGAGAGG-3′ and 5′-AAGAGAAGGAGAGAA-3′ haplotypes 1, 2 and 3, respectively) were predicted with frequencies of 0.42, 0.32 and 0.21, respectively. The additive model provided the best fit for haplotype 3. Cattle carrying two copies of this haplotype had lower risk of bTB infection than those carrying one (Table 3). The second haplotype block 2 containing two SNPs—rs132841890 and rs134752210 (Figure 3a)—was also significantly associated with resistance to bTB (F-test 25.63; P-value 4.8 × 10−7). Three frequent haplotypes (AA, AG and GG) were predicted with frequencies of 0.42, 0.35 and 0.23, respectively. The additive model also provided the better fit for the GG haplotype. Cattle carrying two copies of this haplotype had lower risk than those carrying one (Table 3). The phased data of two haplotypes was highly confounded; the effect of haplotype 3 (F-test 0.04; P-value 8.4 × 10−1) was reduced to near zero when it was fitted simultaneously with the GG haplotype (F-test 25.61; P-value 4.9 × 10−7). The phased GG haplotype data was also confounded with the top hit rs110465273 and reduced the allele substitution effect to near zero (F-test 0.01; P-value 9.2 × 10−1) when fitted together in a single analysis.
Table 2. Summary of genotype data from significantly associated variants in the GWAS.
| SNP | Chr. | Position (bp) | N | Ref./non-ref. | MAF | MAF ca | MAF co | HWE | HWE ca | HWE co |
|---|---|---|---|---|---|---|---|---|---|---|
| Rs136617760 | 2 | 25 899 036 | 1058 | C/A | 0.11 | 0.07 | 0.15 | 7.5 × 10−93 | 2.2 × 10−39 | 2.0 × 10−52 |
| Rs42494357 | 13 | 71 782 488 | 1151 | G/A | 0.22 | 0.18 | 0.27 | 9.0 × 10−01 | 8.2 × 10−01 | 4.2 × 10−01 |
| Rs110465273 | 13 | 71 783 216 | 1151 | A/G | 0.22 | 0.18 | 0.27 | 9.0 × 10−01 | 8.2 × 10−01 | 4.2 × 10−01 |
| Rs42494342 | 13 | 71 784 332 | 1147 | G/A | 0.22 | 0.18 | 0.27 | 9.7 × 10−01 | 7.8 × 10−01 | 4.6 × 10−01 |
| Rs109809949 | 13 | 71 787 722 | 1148 | A/G | 0.22 | 0.18 | 0.26 | 10.0 × 10−01 | 8.0 × 10−01 | 5.2 × 10−01 |
| Rs109042660 | 13 | 71 788 784 | 1150 | G/A | 0.22 | 0.18 | 0.27 | 9.0 × 10−01 | 8.2 × 10−01 | 4.1 × 10−01 |
| Rs137562332 | 13 | 71 789 620 | 1151 | G/A | 0.22 | 0.18 | 0.27 | 9.0 × 10−01 | 8.2 × 10−01 | 4.2 × 10−01 |
| Rs132841890 | 13 | 71 791 844 | 1151 | A/G | 0.23 | 0.18 | 0.27 | 9.3 × 10−01 | 9.8 × 10−01 | 5.5 × 10−01 |
Abbreviations: bp, base pairs; ca, cases; Chr., chromosome; co, controls; GWAS, genome-wide association study; HWE, probability that the SNP is in Hardy–Weinberg equilibrium; MAF, minor allele frequency; N, number of genotyped samples; non-ref., non-reference (minor) allele on BeadChip; Position, Illumina BovineHD BeadChip SNP map position on the chromosome; Ref., reference (major) allele on BeadChip; SNP, single-nucleotide polymorphism GenBank accession number.
Figure 3.
LD maps of the candidate regions. Pairwise SNP LD maps of (a) chromosomes 13 and (b) 2 containing the most significantly associated variants identified in our GWAS using the GRAMMAR approach (SNP accession identifiers underlined). LD is based on D′ (coefficient of LD), and maps are drawn based on the genotype data of all case and control samples. The D ′ values are shown inside each diamond. Red diamonds without a number represent D ′=1. The black triangles indicate the LD blocks identified by Haploview using Gabriel's method (Gabriel et al., 2002).
Table 3. Results from the additive and haplotype models of two haplotype blocks in the region of association on chromosome 13.
| Haplotype block |
Additive model
|
Haplotype model
|
||||||
|---|---|---|---|---|---|---|---|---|
| P-valuea |
Haplotype substitution effect
|
P-valuea |
Heterozygous non-ref. effect
|
Homozygous non-ref. effect
|
||||
| b(95% CI)b | OR(95% CI) c | b(95% CI)b | OR(95% CI) c | b(95% CI)b | OR(95% CI) c | |||
| 1 | 1.6 × 10−06 | −0.13(−0.18, −0.08) | 0.58(0.46, 0.73) | 9.6 × 10−06 | −0.12(−0.19, −0.06) | 0.59(0.45, 0.78) | −0.27(−0.41, −0.13) | 0.32(0.17, 0.63) |
| 2 | 4.8 × 10−07 | −0.13(−0.18, −0.08) | 0.58(0.46, 0.72) | 3.1 × 10−06 | −0.12(−0.19, −0.07) | 0.58(0.44, 0.76) | −0.27(−0.41, −0.14) | 0.33(0.18, 0.61) |
Abbreviations: CI, confidence interval; gBLUP, genomic best linear unbiased prediction; non-ref., non-reference (minor) haplotype on Illumina BovineHD BeadChip; OR, odds ratio; P-value, probability of obtaining the observed effect size (b), assuming that the null hypothesis is true.
P-values from genome-wide rapid association using linear and logistic mixed models and regression in GenABEL.
Effect size estimated from gBLUP using a normal link function.
OR estimated from gBLUP using a logit link function.
The SNP rs132841890 in the GG haplotype block is close to the 3′ end of intron 7 of the gene for bovine PTPRT, a gene that is highly conserved between eutherian mammalian species and is in a region of high synteny (Figure 2). The other six significant SNPs are in the 3′ end of haplotype block 1 and are close to the beginning of intron 8 of this gene (Figures 2 and 3a).
Chromosome 2 associations
The SNP (rs136617760) on chromosome 2 had a moderate call rate of 92% (Table 2). On examination, the SNP displayed no differential missing rates between cases and controls (P>0.05) nor non-random genotyping failure (P>0.05). However, the SNP was not in HWE in either the cases or controls (P<0.0001; Table 2) and had low LD with adjacent SNPs (D′<0.50; Figure 3b). The major C allele for rs136617760 was associated with the greater risk of bTB in this population, with the dominance model providing the best fit (Table 1). The SNP falls within intron 32 of the gene for bovine myosin IIIB (MYO3B) (Supplementary Figure 3), a gene that is highly conserved across species and is found within a region of high synteny. Further inspection of the cluster plot for rs136617760 revealed that in addition to the 93 samples that were not called, there was an excess of B alleles (alternative base forms at a specific genomic position), as well as a number of multimodal clusters (Supplementary Figure 4). The two SNPs flanking rs136617760 (rs134930642 and rs43293650) are also within the same intron 32 of MYO3B (Supplementary Figure 3), yet they are in the adjacent haplotype blocks (Figure 3b); neither of them showed any association with bTB (results not shown). The cluster plots of these flanking SNPs displayed normal genotype clusters (data not shown).
Regional heritability mapping
The RHM approach did not uncover new regions that explain additional trait variation beyond those identified from GWAS. However, this approach also identified the region of association on chromosome 13 (Figure 2), which was significant at the chromosome-wide level (χ2=18.05; P-value 1.0 × 10−04). The estimated heritability for bTB in this region was 2.5%. The region of association on chromosome 2 (Supplementary Figure 3) was not identified by RHM. The size of the SNP window size did not greatly influence the performance of RHM in this study.
To further explore the association within MYO3B and PTPRT, we estimated the phenotypic variance explained by the two genes using regional gene-wise kinship matrices. No significant additive variance was detected using the 134 SNPs within MYO3B present on the SNP chip (LRT 0.11; P-value 7.5 × 10−1), indicating that the associated phenotypic variance is generated by a single SNP (Supplementary Figure 3). However, 303 SNPs in the PRPRT gene explained 1.5% (LRT 7.23; P-value 7.2 × 10−03). Nevertheless, when any single SNP within the region of association was fitted simultaneously as a random covariate in the analysis, the regional heritability estimate was reduced to 0.0%. The GG haplotype explained 6.6% of phenotypic variance (LRT 21.12; P-value 4.3 × 10−06). This indicated that the phenotypic variance within PTPRT is generated by the region of association identified by GWAS (Figure 2).
Discussion
A GWAS, regional heritability and haplotype-sharing analyses have identified two potential novel loci containing candidate genes for resistance to bTB in cattle. In addition, a substantial proportion of variation in resistance to bTB (21%) is determined by a sum of all the SNP genotypes, indicating that the trait is polygenic. Further support for a genomic basis to host resistance to bTB in Holstein–Friesian dairy cattle has recently been reported from the ROI (Finlay et al., 2012). In cattle populations, there is a high level of relatedness, particularly Holstein–Friesian dairy cattle where the effective population size is very small, which may inflate the rate of false-positive associations between the trait and the markers (Minozzi et al., 2012). In this study however, we used a polygenic model that included the genomic relationship matrix that should have removed any statistical anomalies arising because of population structure. In principle, population admixture could reduce the experimental power. However, multidimensional scaling did not reveal any apparent differential clustering of Holstein and Friesian cows. Under the assumption that we have a population of cows from the same breed, and no phenotype misclassification our power analysis demonstrated that this study is sufficiently powered to detect genome-wide significant variants that contribute moderate to high levels of phenotypic variance. Nevertheless, the previously reported association with resistance to bTB in the locus containing the taurine transporter gene SLC6A6 (Finlay et al., 2012) was not replicated in our study. Although there is a possibility that the significant results of either study may include false positives, there can be many reasons why significant associations do not replicate across populations. For example, even if the same quantitative trait loci is segregating in both populations, different allele frequencies of either the marker or causative mutation, or different linkage phases between marker and quantitative trait loci, can lead to different results.
The case–control study design typically entails the identification of cases (affected individuals) and controls (i.e., unaffected individuals) that match the cases with regard to environment and genetic background (Allen et al., 2010). Thus, to estimate the influence of the host genotype on the outcome of bTB infection, the cases and controls in this study were assigned using stringent criteria. As described in humans, a spectrum of phenotypes is observed following exposure to M. bovis, ranging from no apparent infection to asymptomatic carriage (cattle with positive SICTT, but no evidence of pathology or M. bovis culture) through to progressive disease (cattle with a positive skin test that also exhibit evidence of pathology and M. bovis culture) (Allen et al., 2010). This is partly due to the time lag between exposure and emergence of clinical signs, and the immunological anergy that develops in late-stage bTB infection (De la Rua-Domenech et al., 2006), but the range of phenotypes is also a result of host, pathogen and environmental factors, including limitations of the tests. However, our requirement for cases to be positive both to the SICTT, abattoir inspection and molecular confirmation of M. bovis infection removed ambiguity over the case definition, particularly as the SICTT and abattoir tests are known to have high specificity (>99% De la Rua-Domenech et al., 2006). Nevertheless, it is important to note that in an attempt to maximise the power in this study, we have excluded many potentially interesting phenotypes from the case spectrum. In particular, cattle that become infected (SICTT positive) but never go on to develop disease-associated pathology (abattoir negative), or infected cattle that rapidly develop progressive disease (i.e., potentially abattoir positive) and become anergic to the SICTT, may become a latent source of infection in the herd. An investigation of their genotypes could reveal further insights into associations with bTB.
Control definition is more challenging. The imperfect test sensitivity of the SICTT diagnostic tool used in this study results in the non-detection of around 10% of infected cattle (Clegg et al., 2010). The second diagnostic tool, post-mortem inspection is even less sensitive, with only around 47% of M. bovis infected cattle detected (Corner et al., 1990). These relatively low sensitivities mean that classifying the control samples as exposed to M. bovis yet not infected is inherently more difficult. Therefore, to increase the probability that the controls represented resistant animals, all controls were sampled from bTB-restricted case herds favouring higher prevalence herds in an attempt to increase their probability of exposure to infection (Bishop et al., 2012). Furthermore, individual cattle were only included if they also had two or more consecutive negative SICTT diagnoses. In addition, where feasible, prospective SICTT and abattoir inspection results were used to remove control cattle that were subsequently diagnosed as bTB positive. Although it is still possible that some of the SICTT-negative cattle were, or subsequently became, infected (i.e., should have been reclassified as cases), this would only have the effect of reducing the power of our study and would not lead to false-positive associations. In fact, in many case–control designs used in humans, controls are simply a random sample of individuals (Craddock et al., 2010). It must be stated, however, that the control definition that would have maximised power in this study is SICTT- and abattoir-negative cattle. It was not logistically feasible in the given time frame of this study, but as more data become available it may be possible to assign prospectively abattoir results to controls and perform analysis on such a data set.
This study identified seven additive SNPs in high LD on BTA 13 that were significantly associated with resistance to bTB. Haplotype-sharing analysis showed strong support for two SNP block of LD from 71 791 844 to71 793 206 bp at the 3′ end of the seventh intron in PTPRT. PTPRT belongs to the type IIB PTPμ-like subfamily of receptor protein tyrosine phosphatases, which are essential for the regulation of signalling pathways in which they dephosphorylate tyrosine residues on a variety of targets. So far, no variants of PTPRT have been associated with infectious disease susceptibility. However, PTPRT mutations are commonly associated with cancer (Wang et al., 2004) and SNP variants with type 2 diabetes (Hayes et al., 2007), systemic lupus erythematosus (Armstrong et al., 2009) and rheumatoid arthritis (Harley et al., 2008). The functional importance of many receptor protein tyrosine phosphatases is still being unravelled (Tonks, 2006).
A single dominant SNP on BTA 2 was also significantly associated with resistance to bTB. However, it is possible that this singleton association may represent a false association owing to genotyping artefacts or chance. Indeed, the region was not confirmed by either RHM or haplotype-sharing analysis. However, our results may be explicable by the genotype pattern results for this SNP; multimodal genotype clusters for this SNP were observed both in the present study as well as an independent Australian Holstein–Friesian cattle population (Dr Ben Hayes, personal communication; Supplementary Figure 5). Such features can be characteristic of a copy number variant (McCarthy et al., 2008). Further investigation would be required to confirm the hypothesis that this locus represents a copy number variant.
In any case, the putative regions of association on BTA 13 and BTA 2 may be due to single quantitative trait loci in LD with the regions of the SNPs, or they may indicate the presence of more than one quantitative trait loci in these regions. The possibility that these associations are due to chance should also be considered, and thus functional validation and replication of these findings in independent populations is critical.
The results of this study are consistent with a polygenic model, with 18.8% of variation in resistance to bTB captured by SNPs other than those associated with PTPRT and MYO3B, and 6.2% and 3.6% by the regions of associations in the PTPRT and MYO3B genes, respectively. The polygenic component reported in this study is almost identical to the heritability estimates of 18% reported for resistance to bTB in two recent quantitative genetic studies in Irish and British Holstein–Friesian cow populations (Bermingham et al., 2009; Brotherstone et al., 2010). These results indicate that genetic improvement in bTB resistance is possible, most likely through genomic selection methods originally suggested by Meuwissen et al. (2001), rather than relying on individual SNPs, whose frequency in the population as a whole is still unknown. In principle, this is possible as the dairy industry in the United Kingdom has implemented genomic selection for many traits. However, further research is required to determine optimal selection methods for resistance to bTB that are feasible in practice. Genetic and phenotypic correlations of this trait with other economically important traits also need to be quantified so that bTB resistance can be weighted appropriately within the Holstein–Friesian dairy cattle breeding objectives.
Data archiving
Data available from the Dryad Digital Repository: 10.5061/dryad.519bm.
Acknowledgments
We gratefully acknowledge the financial support of the Biotechnology and Biological Sciences Research Council, through CEDFAS initiative grants BB/E018335/1 and 2, the Roslin Institute Strategic Programme Grant and support received from a European Union Framework 7 Project Grant (No: KBBE-211602-MACROSYS); Richard Talbot, David Morrice and their colleagues from ARK-Genomics, Roslin Institute, Edinburgh, UK for the timely and efficient DNA extraction and genotyping, and excellent technical support throughout the project; Department of Agriculture and Rural Development for access to Animal and Public Health Information System data; the support provided by Will Barker and Department of Agriculture and Rural Development Veterinary Service, staff at WD Meats and Veterinary Sciences Division Farm Staff who delivered all the abattoir case sampling and farm control sampling; herd keepers who gave permission to sample their control cattle; and VSD-AFBI specialist histology/pathology and bacteriology staff involved in TB case confirmation.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Abernethy D, Denny G, Menzies F, McGuckian P, Honhold N, Roberts A. The Northern Ireland programme for the control and eradication of Mycobacterium bovis. Vet Microbiol. 2006;112:231–237. doi: 10.1016/j.vetmic.2005.11.023. [DOI] [PubMed] [Google Scholar]
- Abernethy DA, Upton P, Higgins IM, McGrath G, Goodchild AV, Rolfe SJ, et al. Bovine tuberculosis trends in the UK and the Republic of Ireland, 1995–2010. Vet Rec. 2013;172:312. doi: 10.1136/vr.100969. [DOI] [PubMed] [Google Scholar]
- Allen A, Minozzi G, Glass E, Skuce R, McDowell S, Woolliams J, et al. Bovine tuberculosis: the genetic basis of host susceptibility. Proc R Soc Ser B. 2010;277:2737. doi: 10.1098/rspb.2010.0830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong DL, Reiff A, Myones BL, Quismorio FP, Klein-Gitelman M, McCurdy D, et al. Identification of new SLE-associated genes with a two-step Bayesian study design. Genes Immun. 2009;10:446–456. doi: 10.1038/gene.2009.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aulchenko YS, Ripke S, Isaacs A, van Duijn CM. GenABEL: an R library for genome-wide association analysis. Bioinformatics. 2007;23:1294–1296. doi: 10.1093/bioinformatics/btm108. [DOI] [PubMed] [Google Scholar]
- Barrett J, Fry B, Maller J, Daly M. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Bermingham M, More S, Good M, Cromie A, Higgins I, Brotherstone S, et al. Genetics of tuberculosis in Irish Holstein–Friesian dairy herds. J Dairy Sci. 2009;92:3447. doi: 10.3168/jds.2008-1848. [DOI] [PubMed] [Google Scholar]
- Bermingham ML, More SJ, Good M, Cromie AR, Higgins IM, Berry DP. Genetic correlations between measures of Mycobacterium bovis infection and economically important traits in Irish Holstein–Friesian dairy cows. J Dairy Sci. 2010;93:5413–5422. doi: 10.3168/jds.2009-2925. [DOI] [PubMed] [Google Scholar]
- Bishop SC, Doeschl-Wilson AB, Woolliams JA. Uses and implications of field disease data for livestock genomic and genetics studies. Front Genet. 2012;3:114. doi: 10.3389/fgene.2012.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland F, Kelly G, Good M, More S. Bovine tuberculosis and milk production in infected dairy herds in Ireland. Prev Vet Med. 2010;93:153–161. doi: 10.1016/j.prevetmed.2009.09.021. [DOI] [PubMed] [Google Scholar]
- Brotherstone S, White I, Coffey M, Downs S, Mitchell A, Clifton-Hadley R, et al. Evidence of genetic resistance of cattle to infection with Mycobacterium bovis. J Dairy Sci. 2010;93:1234–1242. doi: 10.3168/jds.2009-2609. [DOI] [PubMed] [Google Scholar]
- Clegg T, Duignan A, Whelan C, Gormley E, Good M, Clarke J, et al. 2010. In: More S, Collins D (eds) The Centre for Veterinary Epidemiology and Risk Analysis The TB Diagnostics and Immunology Research Centre The Badger Vaccine Project Biennial Report, 2008–09 Veterinary Sciences Centre, University College Dublin: Dublin, UK [Google Scholar]
- Corner L, Melville L, McCubbin K, Small K, McCormick B, Wood P, et al. Efficiency of inspection procedures for the detection of tuberculous lesions in cattle. Austr Vet J. 1990;67:389–392. doi: 10.1111/j.1751-0813.1990.tb03020.x. [DOI] [PubMed] [Google Scholar]
- Craddock N, Hurles ME, Cardin N, Pearson RD, Plagnol V, Robson S, et al. Genome-wide association study of CNVs in 16 000 cases of eight common diseases and 3000 shared controls. Nature. 2010;464:713–720. doi: 10.1038/nature08979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De la Rua-Domenech R, Goodchild A, Vordermeier H, Hewinson R, Christiansen K, Clifton-Hadley R. Ante mortem diagnosis of tuberculosis in cattle: a review of the tuberculin tests, [gamma]-interferon assay and other ancillary diagnostic techniques. Res Vet Science. 2006;81:190–210. doi: 10.1016/j.rvsc.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Driscoll EE, Hoffman JI, Green LE, Medley GF, Amos W. A preliminary study of genetic factors that influence susceptibility to bovine tuberculosis in the british cattle herd. PLoS One. 2011;6:e18806. doi: 10.1371/journal.pone.0018806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay EK, Berry DP, Wickham B, Gormley EP, Bradley DG. A genome wide association scan of bovine tuberculosis susceptibility in Holstein–Friesian dairy cattle. PLoS One. 2012;7:e30545. doi: 10.1371/journal.pone.0030545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- Gilmour AR, Gogel B, Cullis B, Thompson R, Butler D. ASReml user Guide Release 3.0. VSN International Ltd: Hemel Hempstead, UK; 2009. [Google Scholar]
- Grace D, Gilbert J, Randolph T, Kang'ethe E. The multiple burdens of zoonotic disease and an ecohealth approach to their assessment. Trop Anim Health Prod. 2012;44:67–73. doi: 10.1007/s11250-012-0209-y. [DOI] [PubMed] [Google Scholar]
- Harley JB, Alarcón-Riquelme ME, Criswell LA, Jacob CO, Kimberly RP, Moser KL, et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet. 2008;40:204–210. doi: 10.1038/ng.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes MG, Pluzhnikov A, Miyake K, Sun Y, Ng MCY, Roe CA, et al. Identification of type 2 diabetes genes in Mexican Americans through genome-wide association studies. Diabetes. 2007;56:3033–3044. doi: 10.2337/db07-0482. [DOI] [PubMed] [Google Scholar]
- Humblet M-F, Boschiroli ML, Saegerman C. Classification of worldwide bovine tuberculosis risk factors in cattle: a stratified approach. Vet Res. 2009;40:50–50. doi: 10.1051/vetres/2009033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadarmideen HN, Ali AA, Thomson PC, Müller B, Zinsstag J. Polymorphisms of the SLC11A1 gene and resistance to bovine tuberculosis in African Zebu cattle. Anim Genet. 2011;42:656–658. doi: 10.1111/j.1365-2052.2011.02203.x. [DOI] [PubMed] [Google Scholar]
- McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JPA, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- Meuwissen THE, Hayes BJ, Goddard ME. Prediction of total genetic value using genome-wide dense marker maps. Genetics. 2001;157:1819–1829. doi: 10.1093/genetics/157.4.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel AL, Müller B, van Helden PD. Mycobacterium bovis at the animal–human interface: a problem, or not. Vet Microbiol. 2010;140:371–381. doi: 10.1016/j.vetmic.2009.08.029. [DOI] [PubMed] [Google Scholar]
- Minozzi G, Williams JL, Stella A, Strozzi F, Luini M, Settles ML, et al. Meta-analysis of two genome-wide association studies of bovine paratuberculosis. PLoS One. 2012;7:e32578. doi: 10.1371/journal.pone.0032578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagamine Y, Pong-Wong R, Navarro P, Vitart V, Hayward C, Rudan I, et al. Localising loci underlying complex trait variation using regional genomic relationship mapping. PLoS One. 2012;7:e46501. doi: 10.1371/journal.pone.0046501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry BD, Randolph TF, JJ McDermott, KR Sones, PK Thornton. Investing in Animal Health Research to Alleviate Poverty. ILRI (International Livestock Research Institute): Nairobi, Kenya; 2002. [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiller I, RayWaters W, Vordermeier HM, Jemmi T, Welsh M, Keck N, et al. Bovine tuberculosis in Europe from the perspective of an officially tuberculosis free country: trade, surveillance and diagnostics. Vet Microbiol. 2011;151:153–159. doi: 10.1016/j.vetmic.2011.02.039. [DOI] [PubMed] [Google Scholar]
- Skuce RA, Mallon TR, McCormick CM, McBride SH, Clarke G, Thompson A, et al. Mycobacterium bovis genotypes in Northern Ireland: herd-level surveillance (2003 to 2008) Vet Rec. 2010;167:684–689. doi: 10.1136/vr.c5108. [DOI] [PubMed] [Google Scholar]
- Sun L, Song Y, Riaz H, Yang H, Hua G, Guo A, et al. Polymorphisms in toll-like receptor 1 and 9 genes and their association with tuberculosis susceptibility in Chinese Holstein cattle. Vet Immunol Immunopathol. 2012;147:195–201. doi: 10.1016/j.vetimm.2012.04.016. [DOI] [PubMed] [Google Scholar]
- Thoen CO, Steele JH, Gilsdorf MJ. Mycobacterium bovis Infection in Animals and Humans. Wiley-Blackwell; 2006. [Google Scholar]
- Tonks NK. Protein tyrosine phosphatases: from genes, to function, to disease. Nat Rev Mol Cell Biol. 2006;7:833–846. doi: 10.1038/nrm2039. [DOI] [PubMed] [Google Scholar]
- Wang Z, Shen D, Parsons DW, Bardelli A, Sager J, Szabo S, et al. Mutational analysis of the tyrosine phosphatome in colorectal cancers. Science. 2004;304:1164–1166. doi: 10.1126/science.1096096. [DOI] [PubMed] [Google Scholar]
- Waters W, Palmer MV, Thacker TC, Davis WC, Sreevatsan S, Coussens P, et al. Tuberculosis immunity: opportunities from studies with cattle. Clin Dev Immunol. 2010;2011:768542. doi: 10.1155/2011/768542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Wray NR, Visscher PM. Comparing apples and oranges: equating the power of case–control and quantitative trait association studies. Genet Epidemiol. 2010;34:254–257. doi: 10.1002/gepi.20456. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.