Abstract
External ear hole closure in LG/J mice represents a model of regenerative response. It is accompanied by the formation of a blastema-like structure and the re-growth of multiple tissues, including cartilage. The ability to regenerate tissue is heritable. An F34 advanced intercross line of mice (Wustl:LG,SM-G34) was generated to identify genomic loci involved in ear hole closure over a 30-day healing period. We mapped 19 quantitative trait loci (QTL) for ear hole closure. Individual gene effects are relatively small (0.08 mm), and most loci have co-dominant effects with phenotypically intermediate heterozygotes. QTL support regions were limited to a median size of 2 Mb containing a median of 19 genes. Positional candidate genes were evaluated using differential transcript expression between LG/J and SM/J healing tissue, function analysis and bioinformatic analysis of single-nucleotide polymorphisms in and around positional candidate genes of interest. Analysis of the set of 34 positional candidate genes and those displaying expression differences revealed over-representation of genes involved in cell cycle regulation/DNA damage, cell migration and adhesion, developmentally related genes and metabolism. This indicates that the healing phenotype in LG/J mice involves multiple physiological mechanisms.
Keywords: wound healing, tissue regeneration, QTL, LG/J, SM/J
Introduction
The ability of mammals to mount a regenerative response by closing holes made in ear pinnae was first reported in rabbits (Vorontsova and Liosner, 1960; Joseph and Dyson 1966; Grimes and Goss, 1970). Such ear hole closure is accompanied by the formation of a regeneration blastema-like structure around the injury site and the growth and replacement of the underlying cartilage. These features suggest that, in addition to invertebrates and various vertebrate groups including salamanders, mammals are capable of appendage and tissue regeneration. In 1998, it was reported that MRL/MpJ (MRL) mice, like the rabbit, could also close through-and-through external ear holes with perfect replacement of histological architecture and cartilage in a scarless manner (Joseph and Dyson, 1966; Clark et al., 1998). Since that time, there have been many reports showing that multiple tissue types in MRL mice display healing responses. These include heart myocardium (Leferovich et al., 2001; Haris Naseem et al., 2007; Alfaro et al., 2008), digits (Chadwick et al., 2007; Gourevitch et al., 2009), articular cartilage (Fitzgerald et al., 2008; Rai et al., 2012), intra-articular fractures (Ward et al., 2008), corneal epithelium (Ueno et al., 2005), CNS stem cells and tissue (Hampton et al., 2004; Baker et al., 2006; Balu et al., 2009; Thuret et al., 2009), central and peripheral nerves (Buckley et al., 2011; Thuret et al., 2012) and myometrial healing (Buhimschi et al., 2010). Further, dorsal skin wounds repaired with skin transplants also show enhanced healing without scarring in MRL (Tolba et al., 2010).
To identify genes associated with multigenic ear hole closure, linkage mapping studies have been conducted in a number of different MRL F2 intercrosses, including MRL × C57BL/6 (B6) (McBrearty et al., 1998; Blankenhorn et al., 2003), MRL × CAST/EiJ (Heber-Katz et al., 2004; Yu et al., 2005) and MRL × SJL/J (Masinde et al., 2001). MRL mice were originally bred from a series of crosses of four mouse strains (Murphy and Roths, 1978), AKR/J, C3H/HeDi, C57BL/6J, 65 and two final backcrosses with LG/J (LG). Only one of these parental lines, LG, which contributed 75% of the MRL genome, has been shown to have a regenerative ear hole response (Kench et al., 1999; Li et al., 2010). Recently, a linkage study using LG/J × SM/J (SM) F2 intercrosses identified multiple quantitative trait loci (QTL) that regulate wound healing with regeneration (Blankenhorn et al., 2009). Further analysis of LG,SM QTL show that when compared with the QTL identified in MRL crosses, there are both shared and unique loci (Cheverud et al., 2012).
In the current study, we use an advanced intercross line (AIL) and a linkage mapping strategy originally described by Darvasi and Soller (1995) to narrow the wide support intervals obtained from F2 intercross analysis by enhancing the frequency of recombination between loci. We examined ear hole closure in the Wustl:LG,SM-G34 AIL experimental population. We identified 19 QTL, with support intervals ranging from 0.66 to 7.14 Mb in length. Genes in these regions were analyzed for differential expression in parental strain healing tissue, and both coding and non-coding single-nucleotide polymorphisms (SNPs) were evaluated using genomic sequences for the LG/J and SM/J strains. Candidate genes, their expression patterns and function analysis was interrogated in these QTL and support previous findings showing that metabolism, cell cycle/DNA damage, cell adhesion and migration and developmental processes are likely candidates for regeneration.
Materials and methods
Samples
The animals studied in this experiment are from the F34 generation of the LG/J by SM/J AIL (Wustl:LG,SM-G34). This line was originally formed from an F2 Intercross of the LG/J and SM/J mouse inbred strains obtained from The Jackson Laboratory (Bar Harbor, ME, USA). LG/J mice were generated by a selection experiment for greater body weight at 60 days (Goodale 1938; Wilson et al., 1971) while the SM/J mice were selected for lesser body weight at 60 days (MacArthur, 1944). The original stock that produced LG/J mice is unknown (Goodale, 1938). The base population from which the SM/J strain was derived was formed by a seven-way cross of various inbred strains (MacArthur, 1944). Both strains were accessioned by the Jackson Laboratories in the mid-twentieth century and have been maintained by brother–sister mating ever since (Chai, 1956). The SM/J strain is maintained by forced heterozygosity at the agouti locus on mouse chromosome 2. Attempts to fix the strain for either the non-agouti (a) allele or the whitebellied agouti allele (Aw) have ended in strain failure (Hrbek et al., 2006).
The AI line was formed by first mating 10 SM males with 10 LG females to produce 51 F1 hybrid animals. The F1 hybrids were intercrossed to produce a F2 hybrid generation where each individual is a genetically unique recombination of the parental genotypes (Kramer et al., 1998; Vaughn et al., 1999). Two hundred pairs of F2 hybrids were randomly mated to form a F3 generation of 1600 individuals in 200 full-sibships (Kramer et al., 1998; Fawcett et al., 2008). Two random sets of 50 mating pairs were chosen to start two different AI lines as a control for an inbreeding experiment (Cheverud et al., 1999). A male and a female were randomly selected from each F3 sibship to use for breeding purposes. These animals were randomly mated except that sib-mating was avoided. Limiting each litter to providing only two breeders from the previous generation doubles the effective population size relative to the census size and dramatically reduces inbreeding over time (Falconer and Mackay, 1996). The two AI lines were combined at the F8 generation after which the population has been maintained using at least 75 mating pairs per generation. All animal care and handling procedures conformed to IACUC guidelines.
The F34 population comprised 1169 animals in 137 full-sib families with an average litter size of 8.5 pups per family. Animals were reared by their mothers for 3 weeks after which half of each litter was weaned onto a high- or low-fat diet (Cheverud et al., 2004). The diets are isocaloric, but the low-fat diet has 15% of its calories from fat while the high-fat diet has 42% of its calories from fat. At 5 weeks of age, each animal had a 2-mm hole punched in both right and left ears. The diameter of the hole was recorded 30 days after hole punch for each animal. Animals were anesthetized both for the hole punch procedure and to read the punch. The healing phenotype for each animal is the average of its healing on the left and right sides. Torn or damaged holes were treated as missing data, and in these cases, only the intact side is used for that animal.
Genotypes
Both the F33 parents and F34 offspring were genotyped for 3072 polymorphic SNPs selected from the Oxford/CTC SNP set (http://www.well.ox.ac.uk/mouse/INBREDS/). Of these, 2842 autosomal SNPs are used in the present analysis. SNPs were scored with the Illumina GoldenGate Bead Array (Illumina, San Diego, CA, USA) by the Center for Inherited Disease Research (CIDR). Centimorgan distances were calculated using R/QTL (Broman and Sen, 2009). The map length of the F34 AI line population is approximately 17 times the length of the F2 map over short distances allowing much finer QTL mapping resolution. Marker positions in the F34 map are provided in Supplementary Table S1.
Analysis
We first examined the population for the effects of sex, diet and family membership on the level of wound healing using an analysis of variance model:
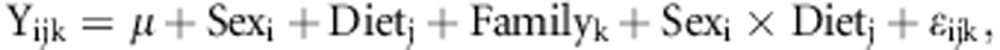 |
where sex is male or female, diet is high or low fat and family identifies the random effect of sibship membership. In this model, the family effect provides a measure of the heritability for the trait in question, pooled over sexes and diets.
Interval mapping model
QTL interval mapping followed the procedures described by Haley and Knott (1992). Using the genotypes at adjacent markers and the recombination rate between them, we imputed additive (Xai) and dominance (Xdi) genotype scores every 1 cM along the F34 marker map and inserted marker scores at their map positions. Additive genotype scores (Xai) at markers are +1, 0, and −1, and dominance genotype scores (Xdi) are 0, +1 and 0 for the LL, LS and SS genotypes respectively. We used a mixed model (SAS version 9.1; SAS Institute, Cary, NC, USA) with sex, diet and additive and dominance genotype scores:
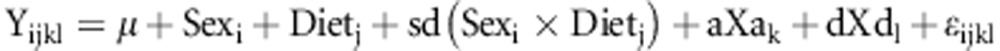 |
at each chromosomal position and included the random effects of family, family by sex, family by diet and family by sex by diet interactions in the model. Results were compared with a null model of no additive and dominance effects. The additive genotypic value (a) is half the difference between the two homozygotes, and the dominance genotypic value (d) is the deviation of the heterozygote from the mid-point of the two homozygotes using the marker scores described above. With this parameterization, a negative additive genotypic value indicates that the LG/J allele promotes healing (smaller hole) and a positive additive genotypic value indicates the SM/J allele promotes healing. The random effects of family and its interactions with sex and diet corrects for the inclusion of non-independent family members in the analysis. Probabilities were obtained from likelihood ratio tests where 2ln (likelihood) of the genetic model was compared with that of the null using a chi-squared test with two degrees of freedom. An additional model including two- and three-way interactions was also tested. The fixed effects are modeled by:
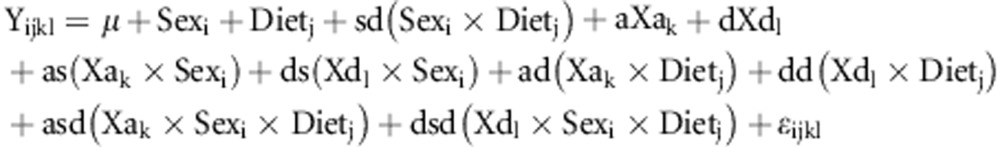 |
where regression coefficients for additive and dominance genotype scores are given as an ‘a' or ‘d' respectively, along with the interacting terms for sex (s), diet (d) or both sex and diet together (sd). Probabilities were transformed by –log10(probability) in order to present the results on a scale consistent with limit of detection (LOD) scores
To account for multiple testing after the mixed model analysis (which accounts for family structure as discussed above), we performed a Bonferroni correction using the number of independent tests as suggested by Li and Ji (2005) using the eigenvalues of the intermarker correlation matrix for each chromosome to obtain multiple tests' corrected thresholds for each chromosome and for the genome chromosomes range from 2.97 for chromosome 19 to 3.66 for chromosome 1. Using chromosome-wise thresholds, we expect one false-positive finding under the null model across as a whole. The whole-genome threshold is 4.72 while thresholds on individual the whole genome. Chen and Storey (2006) suggest this approach to balance false-positive and false-negative decisions. When multiple QTL mapped to the same region of a chromosome, we performed a two-QTL test to determine whether the two-QTL model had a significantly better fit than the best-fit single QTL model. In these circumstances, the parameter values presented are from the multiple QTL model. In addition to testing for the direct effects of QTLs, we also examined significant locations for interactions with sex, diet or both sex and diet. When the full or reduced model had a significant LOD score, we examined the LOD scores for the individual interaction terms. If they were significant at the point-wise level (LOD>1.3), we analyzed the sex- and/or diet cohorts separately to determine the meaning of the interaction term. In these instances, cohort-specific genotypic values are presented.
Gene expression and function analysis
Gene expression studies for ear hole closure in LG/J and SM/J mice were previously reported (Blankenhorn et al., 2009). RNA samples were prepared in triplicate from day 0 ear holes and from day 7 ear hole donuts from LG/J and SM/J female mice (3 mm circular blastemas formed around the hole punch). Tissue was derived from three mice of each strain for each time point. RNA was isolated using a standard Trizol extraction protocol. RNA quality was assessed by gel electrophoresis and nanodrop analysis and then amplified according to Illumina protocols. Three micro-array replicates per time point were used. Illumina raw intensities and detection P-values were obtained using the Illumina Bead Studio v3.0. Arrays were quantile normalized, and probes were removed from further analysis if their intensity was lower than the experimental background (detection P-value>0.05) in all samples or if the maximum ratio between any two samples was not at least 1.2. Microarray data analysis was performed using the Matlab v7.2 functions. All genes from the identified QTL regions were tested for differential expression between the LG and SM groups at day 0 or day 7 using a two-tailed t-test. False discovery rate was estimated using the procedure of Storey and Tibshirani (2003) and P-value<0.05 results that met the false discovery rate<10% threshold. Fold change for each gene was calculated as a ratio of median expressions of LG/J and SM/J when the expression in LG/J exceeds that in SM/J and as −SM/LG when SM expression exceeds that of LG. Maximum fold change among the day 0 and day 7 comparisons is reported.
A select list of 34 candidate genes was analyzed for enriched common functions using the Ingenuity Pathway Analysis software (Ingenuity Systems, Richmond, CA, USA). The significance threshold was set to P<0.001.
SNP analysis
For each QTL support interval, positional candidate genes were identified as residing in non-conserved 10 kb regions between LG/J and SM/J based on the density of SNPs between the two strains (Lawson et al., 2011). Genes falling within conserved regions of QTL do not contain SNPs. Genomic regions designated as conserved have a genome-wide SNP frequency of 0.01% while non-conserved regions have a frequency of 0.35%. Conserved and non-conserved regions between LG/J and SM/J cover 51.5% and 48.5% of the mouse genome, respectively.
Positional candidate genes in enriched common pathways and/or showing significant differential expression were evaluated for molecular variation. SNPs were classified by genomic region using the UCSC genome browser annotations (NCBI37/mm9 assembly; http://genome.ucsc.edu/) as residing 2500 bp upstream, in 5′ untranslated region (UTR), exons, introns, 3′UTR or 2500 bp downstream of the 3′UTR. Mammalian conservation scores for each SNP were obtained from the PhastCons30wayPlacental table downloaded from the UCSC browser. Highly conserved mammalian (HCM) SNPs (>0.90 conservation score) in non-coding DNA were evaluated for potential impact.
The potential functional significance of non-synonymous SNPs was evaluated using three algorithms, SIFT (Kumar et al., 2009), PolyPhen-2 (Adzhubei et al., 2010) and LRT (Chun and Fay, 2009). SIFT uses sequence homology and a median conservation score to measure protein conservation. PolyPhen-2 uses both structure and sequence-based predictive features to judge the functional importance of an amino-acid variant. LRT is a likelihood ratio test comparing the probability that a codon has evolved under a conserved model to its corresponding probability under a neutral model. The conserved model allows a codon to have evolved under negative selection while the neutral model assumes that the rate of non-synonymous and synonymous mutations are the same.
Our molecular analysis focused on non-synonymous SNPs and on HCM non-coding SNPs in candidate genes, because these SNPs are most likely to affect function and underlie QTL effects. However, it is possible that some functionally important SNPs are synonymous or are relatively lowly conserved among mammals. We chose to focus on the top or high-priority candidates based on the defined criteria when ranking SNPs for potential functional importance rather than securely eliminate any particular SNP.
Immunohistochemistry
To examine gene expression in hole-punched ear tissue, ear pinnae (two ears/strain/time point) were removed, and ear tissue was prepared in Prefer fixative, followed by immersion in 70% ethyl alcohol (ETOH), and then in 1X phosphate-buffered saline (PBS) each for 24 h. Paraffin embedding was followed by 5-micron sectioning of the tissue. Sectioned tissue was deparaffinized and dehydrated in preparation for antibody staining. Immunostaining was carried out by placing tissue on slides.
To examine gene expression in fibroblast-like cells derived from normal ear tissue from SM/J and LG/J mice, cells were grown on sterile coverslips for immunostaining. When 70–80% confluent and ready for staining, cells were washed in 1X PBS, fixed in methanol for 10 min and rewashed in PBS. Staining results are from three separate experiments.
Three genes, Wnt3a, Hif1a (hypoxia-inducible factor 1a) and Kif23, were examined by immunostaining of ear tissue and cultured cells. Antibody specific for WNT3a (R&D Biosystems, Minneapolis, MN, USA; Cat no. MAB1324; rat immunoglobulin G (IgG); 1:50); for KIF23 (Santa Cruz, Dallas, TX, USA; Cat no. sc-22793; rabbit polyclonal; 1:300) and for HIF1a (Abcam, Cambridge, MA, USA; Cat no. 2185; rabbit polyclonal; 1:100) was used. Tissue sections were incubated with 2% bovine serum albumin as a blocking buffer for 1 h at room temperature, then primary antibody was applied for 16 h at +4 °C. After washing in 1X PBS, secondary antibody was applied to tissue for 1 h room temperature. Goat anti-rat IgG (Molecular Probes, Invitrogen, Grand Island, NY, USA; Cat no. 11007; 1:200) was used with the WNT3a antibody; goat anti-rabbit IgG (Molecular Probes, Invitrogen; Cat no. A11036; 1:800) was used with the KIF23 antibody; and goat-anti rabbit IgG (Molecular Probes, Invitrogen; Cat no. A11008; 1:200) was used with the HIF1a antibody. Controls were carried out using secondary antibodies alone after bovine serum albumin blocking of tissue.
Mitotracker was used to determine the mitochondrial mass. Fixed LG/J and SM/J ear-derived fibroblasts were stained with mitotracker orange (Invitrogen, Cat no. M7510) and 12 cells/strain were analyzed. Fluorescence was used to determine a proxy measure of total mitochondrial content.
Image analysis was carried out by first photographing tissue samples at × 20 magnification using an Olympus AX70 fluorescent microscope (Olympus, Center Valley, PA, USA) and SPOT camera with bounded software. The antigen-specific white signal was then measured by the ImageJ v1.46f software, provided by the NIH (Bethesda, MD, USA), as the number of pixels in black-and-white tissue images. The number of positive pixels was plotted against the time points with s.e. bars determined from 2–4 tissue samples. Student's t-test was used to calculate P-values.
Western blotting analysis
Ear tissue samples (ear hole tissue donuts from three different mice) were homogenized in radioimmunoprecipitation assay buffer (50 mM Tris–HCl pH 7.6, containing 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 1 mM EDTA and 0.1% sodium dodecyl sulfate) with 1 mM phenylmethanesulfonylfluoride and a protease inhibitor cocktail (Sigma, St Louis MO, USA). Samples with equal amounts of protein (about 40 μg) were loaded onto a NuPAGE 4-12%Bis–Tris gradient gel (Life Technologies, Grand Island, NY, USA), electrophoresed and then electro-transferred onto a PVDF-FL membrane (Immobilon, Billerica, MA, USA). The membrane was subsequently blocked with the Odyssey blocking buffer (LI-COR, Lincoln, NE, USA), probed with primary antibodies (HIF1a (10006421, Cayman Chemical, Ann Arbor, MI, USA) or WNT3a (MAB1324, R&D Systems, Minneapolis, MN, USA)) overnight at 4 °C, then further incubated with Alexa Fluor-labeled secondary antibodies (IRDye 800CW goat-anti rat or IRDye 800CW goat-anti rabbit (LI-COR)) for 1 h and scanned using the Odyssey system (LI-COR).
Results
Genetics of wound healing revealed in the LG,SM AIL
The heritability of wound healing pooled across sex and diet cohorts is 49% (Probability=6.5 × 10−12), indicating that approximately half of the phenotypic variation is due to uncontrolled environmental and non-heritable genetic effects.
We mapped 19 QTLs for wound healing in the LG,SM AI line (see Table 1) distributed over chromosomes (chr) 1, 7, 9, 10, 11, 12, 13, 14 and 18. Several chromosomes, 1, 7, 9, 11, 13 and 18, had multiple QTLs where the two QTL model fit significantly better than the best one QTL model. Eight of the 19 QTL are significant at the genome-wide level while the remaining 11 are significant at the chromosome-wise level.
Table 1. Ear-healing QTLs in the Wustl:LG,SM-G34 advanced intercross line.
| QTL | LOD a | Chr | Effects b | Peak genome coordinates | Proximal interval genome coordinates | Distance interval genome coordinates | Interval length (bp) | Additive (mm) c % | Dominance (mm) c % | Additive (s.e.) | Dominance (s.e.) | Genes d | Non, split, cons e |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Q1.1 | 5.98 | 1 | a | 72 252 316 | 71 676 295 | 73 411 001 | 1 734 706 | 0.088 | 0.010 | 0.018 | 0.024 | 14 | 5, 5, 4 |
| Q1.2 | 5.18 | 1 | a, d | 134 931 146 | 133 749 487 | 137 468 445 | 3 718 958 | −0.066 | −0.044 | 0.019 | 0.023 | 68 | 40, 7, 21 |
| Q1.3 | 3.70 | 1 | a | 140 498 264 | 139 536 983 | 142 773 121 | 3 236 138 | −0.057 | −0.030 | 0.020 | 0.024 | 19 | 7, 4, 8 |
| Q7.1 | 4.63 | 7 | a, d | 70 741 322 | 66 490 265 | 72 631 840 | 6 141 575 | −0.050 | 0.053 | 0.019 | 0.023 | 31 | 22, 6, 3 |
| Q7.2 | 4.87 | 7 | a, d | 82 773 421 | 81 045 510 | 86 086 593 | 5 041 083 | 0.057 | −0.055 | 0.019 | 0.024 | 13 | 5, 5, 3 |
| Q7.3 | 4.97 | 7 | a | 87 330 462 | 86 950 051 | 88 125 934 | 1 175 883 | 0.084 | 0.025 | 0.019 | 0.029 | 27 | 21, 0, 6 |
| Q9.1 | 4.66 | 9 | a | 62 013 984 | 61 092 166 | 62 280 288 | 1 188 122 | −0.091 | 0.021 | 0.021 | 0.025 | 12 | 9, 0, 1 |
| Q9.2 | 3.79 | 9 | a | 95 077 076 | 93 516 401 | 96 892 086 | 3 375 685 | −0.079 | 0.000 | 0.023 | 0.028 | 20 | 20, 0, 0 |
| Q10.1 | 3.53 | 10 | d | 128 553 075 | 127 751 828 | 128 553 075 | 801 247 | 0.020 | 0.091 | 0.018 | 0.022 | 35 | 35, 0, 0 |
| Q11.1 | 6.74 | 11 | a | 58 818 355 | 57 795 593 | 59 802 286 | 2 006 693 | −0.116 | 0.028 | 0.022 | 0.027 | 50 | 50, 0, 0 |
| Q11.2 | 5.99 | 11 | a | 99 864 706 | 99 642 739 | 100 202 942 | 560 202 | −0.276 | −0.317 | 0.056 | 0.278 | 27 | 13, 1, 13 |
| Q11.3 | 5.49 | 11 | a | 101 318 869 | 101 126 956 | 101 510 781 | 383 825 | −0.105 | 0.001 | 0.023 | 0.033 | 18 | 12, 1, 5 |
| Q12.1 | 3.96 | 12 | a | 73 203 378 | 70 704 786 | 73 595 736 | 2 890 950 | −0.069 | −0.020 | 0.019 | 0.025 | 28 | 19, 7, 2 |
| Q13.1 d | 3.97 | 13 | a, d, dd, as | 70 313 984 | 69 468 028 | 71 022 095 | 1 554 067 | −0.091 | 0.085 | 0.026 | 0.030 | 10 | 9, 0, 0 |
| −0.099 | 0.135 | 0.039 | 0.044 | ||||||||||
| Q13.2 | 7.96 | 13 | a | 106 712 772 | 105 104 329 | 107 727 570 | 2 623 240 | 0.101 | 0.023 | 0.017 | 0.024 | 9 | 1, 0, 8 |
| Q14.1 | 3.40 | 14 | a, d | 49 832 566 | 47 748 393 | 50 848 141 | 3 099 748 | −0.035 | 0.060 | 0.018 | 0.022 | 19 | 17, 2, 0 |
| Q18.1 d | 3.65 | 18 | a, as, ds, asd | 38 525 762 | 37 267 434 | 39 270 239 | 2 002 805 | −0.045 | 0.002 | 0.016 | 0.021 | 75 | 33, 2, 40 |
| −0.093 | 0.068 | 0.030 | 0.042 | ||||||||||
| Q18.2 d | 3.73 | 18 | a, d, ad, dd | 42 341 135 | 40 489 461 | 42 861 162 | 2 371 701 | 0.016 | 0.066 | 0.029 | 0.024 | 13 | 3, 4, 6 |
| 0.053 | 0.072 | 0.038 | 0.032 | ||||||||||
| Q18.3 | 3.60 | 18 | a | 44 011 475 | 43 794 001 | 45 574 479 | 1 780 477 | −0.071 | 0.043 | 0.026 | 0.027 | 14 | 10, 1, 3 |
Abbreviations: LOD, limit of detection; QTL, quantitative trait loci.
LOD scores in bold are significant at the genome-wide level while others are significant at the chromosome-wise level.
Significant effects at locus: a=additive; d=dominance; second letter s=sex by genotype interaction; second letter d=diet by genotype interaction; sd=genotype by sex by diet interaction.
Millimeter percentage effects in boldface are significantly different from zero at the 5% level.
Multiple a and d values for sex/diet interactions.
Number of genes in non-conserved areas (non), divided between conserved and non-conserved areas (split), and in conserved areas (cons).
Two-thirds of the significant additive effects are negative, indicating that the LG/J allele promotes enhanced healing, although the SM/J allele promotes healing at four loci. Summed over all loci, the LG/J haplotype heals 1.64 mm (4.94 s.d.) better than the SM/J haplotype. Additive effects can also be considered in relation to the phenotypic s.d. by dividing the ‘a' genotypic value by the s.d. The absolute values of significant additive standardized effects range from 0.11 to 0.83 s.d., with an average effect of 0.26 s.d. The QTL with the largest effect size, Q11.2 at −0.276 mm, lies in a small physical interval, 560 kb, with a large amount of recombination (52 cM). The peak lies at the mid-point of this interval where the linkage disequilibrium with scored markers is relatively small. In such circumstances, effect sizes are likely to be inflated, and the result for this QTL should be taken as provisional. Although individual QTL effects are relatively small, collectively they produce a large difference in tissue regeneration between the LG/J and SM/J parental strains.
Of the 19 QTLs, 12 showed only additive effects. The LG/J allele was dominant to the SM/J allele once and the SM/J allele was dominant to the LG/J allele four times. In these five cases, the non-healing allele was dominant to the healing allele three times. There were two QTLs where the heterozygote healed worse than either homozygote. As seen with the additive effects, significant dominance effects were also relatively small, ranging from 0.13 to 0.27 s.d. with an average of 0.19 s.d.
Three of the 19 loci had genotype by sex or diet interactions, Q13.1, Q18.1 and Q18. (Heber-Katz et al., manuscript in preparation; Norgard et al., 2011). The effects of Q13.1 were specific to high-fat fed animals. Effects at Q18.2 were restricted to females, and at Q18.1, the additive effects were limited to the high-fat fed males while dominance effects were restricted to high-fat fed females.
The F34 AI generation and its expanded map make for much finer resolution of QTL locations than was possible in earlier studies of wound healing in F2 intercrosses. Support intervals defined in the present study are about 1 cM on the F2 scale. In terms of base pair lengths, support intervals range from 0.4 to 6 Mb, with a median interval size of 2 Mb and 70% of the intervals <3 Mb.
Candidate genes and genetic elements in the intervals
There were a total of 499 protein coding genes and annotated microRNAs in the QTL support intervals based on the MGI database (Eppig et al., 2012) (Supplementary Table S2). This gives a median of 19 and mean of 26 genes per QTL. When the 499 genes were intersected with conserved and non-conserved intervals of the LG/J and SM/J genomes, 331 genes were in non-conserved intervals, 123 were in conserved intervals and 45 were in straddled conserved and non-conserved intervals. Hence, 25% of the positional candidate genes can be considered unlikely sources of the QTL effects based on their lack of variation between the strains. Subtracting conserved intervals from these QTL reduces the median number of genes per interval to 13 with an average of 18.6.
The 34 positional candidate genes (Table 2), whose sequences were considered based on the criteria defined above (see Materials and methods), contained 137 exonic SNPs, 56% of which were HCM. One hundred and two of these are synonymous substitutions while 35 result in an amino-acid change (see Table 3). Of these 35 SNPs, 4 are predicted to have relatively severe functional effects based on SIFT (Kumar et al., 2009), PolyPhen (Adzhubei et al., 2010) or LRT (Chun and Fay, 2009). These include a Thr:624:Asn substitution (ACC in SM to AAC in LG at chr7: 87 647 213; (−) strand) in exon 7 in Blm; a His:722:Arg substitution (CAC in SM to CGC in LG at chr9: 95 771 930; (+) strand) in Atr's exon 10; an Arg:315:His substitution (CGC in SM to CAC in LG at chr10: 128 380 017; (+) strand) in exon 6 of Itga7; and an Ala:1120:Asp substitution (GCT in LG to GAT in SM at chr13: 70 874.574; (−) strand) in exon 13 of Adamts16.
Table 2. SNPs in positional candidate genes.
| QTL | Gene | X a |
Exons
|
Non-coding DNA
|
2500 upstream
|
5′UTR
|
Introns
|
3′UTR
|
2500 downstream
|
Total
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNPs | HCM | nsyn | syn | SNPs | HCM | SNPs | HCM | SNPs | HCM | SNPs | HCM | SNPs | HCM | SNPs | HCM | SNPs | HCM | |||
| Q1.1 | Fn1 | Y | 7 | 4 | 0 | 7 | 180 | 6 | 8 | 1 | 0 | 0 | 170 | 5 | 0 | 0 | 2 | 0 | 187 | 10 |
| Q1.1 | Smarcal1 | N | 0 | 0 | 0 | 0 | 29 | 0 | 0 | 0 | 1 | 0 | 26 | 0 | 2 | 0 | 0 | 0 | 29 | 0 |
| Q1.1 | Xrcc5 | N | 5 | 3 | 1 | 4 | 525 | 5 | 1 | 0 | 0 | 0 | 515 | 5 | 0 | 0 | 9 | 0 | 530 | 8 |
| Q1.2 | Lgr6 | N | 5 | 2 | 1 | 4 | 447 | 8 | 2 | 0 | 1 | 0 | 437 | 8 | 0 | 0 | 7 | 0 | 452 | 10 |
| Q1.2 | Ptprv | Y | 4 | 0 | 1 | 3 | 25 | 0 | 0 | 0 | 0 | 0 | 22 | 0 | 0 | 0 | 3 | 0 | 29 | 0 |
| Q1.3 | Ptprc | Y | 2 | 1 | 1 | 1 | 188 | 4 | 9 | 1 | 1 | 1 | 174 | 2 | 1 | 0 | 3 | 0 | 190 | 5 |
| Q7.1 | Fan1 | N | 0 | 0 | 0 | 0 | 9 | 0 | 3 | 0 | 1 | 0 | 4 | 0 | 0 | 0 | 1 | 0 | 9 | 0 |
| Q7.1 | Ndn | Y | 0 | 0 | 0 | 0 | 4 | 0 | 1 | 0 | 0 | 0 | — | — | 0 | 0 | 3 | 0 | 4 | 0 |
| Q7.3 | Blm | N | 8 | 2 | 4 | 4 | 127 | 1 | 2 | 0 | 0 | 0 | 124 | 1 | 0 | 0 | 1 | 0 | 135 | 3 |
| Q7.3 | Mesp2 | Y | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Q9.1 | Kif23 | Y | 5 | 4 | 4 | 1 | 91 | 1 | 4 | 0 | 0 | 0 | 56 | 0 | 3 | 1 | 28 | 0 | 96 | 5 |
| Q9.1 | Coro2b | Y | 0 | 0 | 0 | 0 | 568 | 8 | 3 | 0 | 0 | 0 | 522 | 5 | 22 | 3 | 21 | 0 | 568 | 8 |
| Q9.2 | Atr | N | 7 | 6 | 1 | 6 | 276 | 2 | 15 | 0 | 0 | 0 | 250 | 2 | 0 | 0 | 11 | 0 | 283 | 8 |
| Q9.2 | Rnf7 | Y | 1 | 1 | 0 | 1 | 112 | 6 | 21 | 0 | 0 | 0 | 68 | 5 | 1 | 1 | 22 | 0 | 113 | 7 |
| Q10.1 | Cdk2 | Y | 0 | 0 | 0 | 0 | 64 | 4 | 24 | 1 | 0 | 0 | 25 | 2 | 3 | 0 | 12 | 1 | 64 | 4 |
| Q10.1 | Cs | Y | 2 | 2 | 0 | 2 | 109 | 6 | 8 | 1 | 0 | 0 | 88 | 2 | 6 | 1 | 7 | 2 | 111 | 8 |
| Q10.1 | Itga7 | Y | 19 | 12 | 2 | 17 | 200 | 2 | 15 | 0 | 3 | 0 | 160 | 2 | 4 | 0 | 18 | 0 | 219 | 14 |
| Q10.1 | Myl6 | Y | 1 | 0 | 1 | 0 | 39 | 4 | 14 | 1 | 1 | 0 | 14 | 1 | 2 | 1 | 8 | 1 | 40 | 4 |
| Q10.1 | Obfc2b | Y | 4 | 3 | 0 | 4 | 58 | 2 | 13 | 1 | 1 | 0 | 31 | 0 | 1 | 0 | 12 | 1 | 62 | 5 |
| Q10.1 | Smarcc2 | N | 6 | 4 | 0 | 6 | 194 | 2 | 10 | 0 | 0 | 0 | 173 | 1 | 3 | 0 | 8 | 1 | 200 | 6 |
| Q11.1 | Hist3h2a | N | 1 | 1 | 0 | 1 | 26 | 2 | 18 | 1 | 0 | 0 | — | — | 1 | 1 | 7 | 0 | 27 | 3 |
| Q11.1 | Hist3h2ba | N | 0 | 0 | 0 | 0 | 10 | 0 | 2 | 0 | 1 | 0 | — | — | 1 | 0 | 6 | 0 | 10 | 0 |
| Q11.1 | Wnt3a | Y | 1 | 1 | 0 | 1 | 71 | 1 | 4 | 0 | 0 | 0 | 56 | 1 | 1 | 0 | 10 | 0 | 72 | 2 |
| Q11.2 | Krt15 | Y | 2 | 2 | 0 | 2 | 3 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 5 | 2 |
| Q11.3 | Becn1 | Y | 5 | 5 | 0 | 5 | 112 | 9 | 13 | 0 | 0 | 0 | 79 | 3 | 3 | 1 | 17 | 5 | 117 | 14 |
| Q11.3 | Brca1 | N | 5 | 3 | 5 | 0 | 24 | 0 | 0 | 0 | 0 | 0 | 22 | 0 | 1 | 0 | 1 | 0 | 29 | 3 |
| Q11.3 | Rdm1 | N | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 2 | 0 |
| Q13.1 | Adamts16 | Y | 27 | 6 | 10 | 17 | 1162 | 1 | 34 | 0 | 5 | 0 | 1071 | 1 | 13 | 0 | 39 | 0 | 1189 | 7 |
| Q13.2 | Adamts6 | N | 8 | 7 | 1 | 7 | 1224 | 66 | 7 | 2 | 1 | 0 | 1195 | 59 | 3 | 3 | 18 | 2 | 1232 | 73 |
| Q14.1 | Atg14 | N | 0 | 0 | 0 | 0 | 12 | 0 | 2 | 0 | 0 | 0 | 8 | 0 | 0 | 0 | 2 | 0 | 12 | 0 |
| Q18.1 | Diap1 | Y | 5 | 2 | 1 | 4 | 224 | 7 | 23 | 3 | 0 | 0 | 198 | 4 | 0 | 0 | 3 | 0 | 229 | 9 |
| Q18.1 | Fgf1 | Y | 1 | 1 | 0 | 1 | 217 | 4 | 2 | 0 | 1 | 0 | 171 | 1 | 34 | 3 | 9 | 0 | 218 | 5 |
| Q18.1 | Hdac3 | Y | 3 | 3 | 0 | 3 | 89 | 1 | 0 | 0 | 0 | 0 | 70 | 0 | 5 | 0 | 14 | 1 | 92 | 4 |
| Q18.3 | Mcc | Y | 3 | 2 | 2 | 1 | 1383 | 23 | 12 | 0 | 0 | 0 | 1327 | 19 | 21 | 3 | 23 | 1 | 1386 | 25 |
| Total | 21 | 137 | 77 | 35 | 102 | 7804 | 175 | 271 | 12 | 17 | 1 | 7059 | 129 | 131 | 18 | 326 | 15 | 7941 | 252 | |
Abbreviations: HCM, highly conserved in mammals; QTL, quantitative trait loci; SNP, single-nucleotide polymorphism; UTR, untranslated region.
Expression differences.
Table 3. Non-synonomous coding SNPs in positional candidate genes.
| Gene | QTL | Chr | Pos | Exon | Substitution | AA no. | Significant a |
|---|---|---|---|---|---|---|---|
| Xrcc5 | Q1.1 | 1 | 72 392 997 | 14 | Asn:His | 558 | |
| Lgr6 | Q1.2 | 1 | 137 001 653 | 1 | Ala:Gly | 22 | |
| Ptprv | Q1.2 | 1 | 137 020 322 | 9 | Val:Ala | 494 | |
| Ptprc | Q1.3 | 1 | 140 008 831 | 9 | Lys:Glu | 300 | |
| Blm | Q7.3 | 7 | 87 647 213 | 7 | Thr/Asn | 624 | SIFT |
| Blm | Q7.3 | 7 | 87 647 304 | 7 | Thr/Ala | 594 | |
| Blm | Q7.3 | 7 | 87 647 435 | 7 | Asn/Thr | 550 | |
| Blm | Q7.3 | 7 | 87 647 472 | 7 | Val/Met | 538 | |
| Kif23 | Q9.1 | 9 | 61 774 853 | 13 | Val:Ile | 464 | |
| Atr | Q9.2 | 9 | 95 771 930 | 10 | His:Arg | 722 | PolyPhen |
| Myl6 | Q10.1 | 10 | 127 929 109 | 5 | Gly:Trp | 145 | |
| Itga7 | Q10.1 | 10 | 128 380 017 | 6 | Arg:His | 315 | PolyPhen |
| Itga7 | Q10.1 | 10 | 128 382 560 | 13 | Thr:Pro | 613 | |
| Brca1 | Q11.3 | 11 | 101 371 241 | 13 | Glu:Lys | 1510 | |
| Brca1 | Q11.3 | 11 | 101 385 554 | 9 | Gly:Ser | 1023 | |
| Brca1 | Q11.3 | 11 | 101 385 593 | 9 | Val:Ile | 1010 | |
| Brca1 | Q11.3 | 11 | 101 385 823 | 9 | Cys:Leu | 933 | |
| Brca1 | Q11.3 | 11 | 101 386 328 | 9 | Ser:Pro | 765 | |
| Adamts16 | Q13.1 | 13 | 70 874 574 | 21 | Ala:Asp | 1120 | LRT |
| Adamts16 | Q13.1 | 13 | 70 874 586 | 21 | Pro:Leu | 1116 | |
| Adamts16 | Q13.1 | 13 | 70 874 611 | 21 | Gly:Ser | 1108 | |
| Adamts16 | Q13.1 | 13 | 70 877 487 | 20 | Gln:Glu | 1021 | |
| Adamts16 | Q13.1 | 13 | 70 892 160 | 18 | Ala:Val | 892 | |
| Adamts16 | Q13.1 | 13 | 70 936 853 | 5 | Arg:Pro | 274 | |
| Adamts16 | Q13.1 | 13 | 70 975 021 | 3 | Val:Leu | 134 | |
| Adamts16 | Q13.1 | 13 | 70 980 197 | 2 | Glu:Gly | 38 | |
| Adamts16 | Q13.1 | 13 | 70 980 224 | 2 | Leu:Arg | 29 | |
| Adamts6 | Q13.2 | 13 | 105 102 839 | 4 | Ser:Gln | 226 | |
| Diap1 | Q18.1 | 18 | 38 050 634 | 15 | Ser:Pro | 602 | |
| Mcc | Q18.3 | 18 | 44 919 042 | 2 | Ser:Asn | 53 |
Abbreviation: SNP, single-nucleotide polymorphism.
Non-synonomous coding SNPs rated as significant by different criteria.
There are 7804 SNPs found in the non-coding regions of these positional candidates, of which 175 are HCM. Only one of the 17 5′UTR SNPs is highly conserved while 14% of the 131 3′UTR SNPs are highly conserved. Introns, representing the most common class of non-coding sequence, had the most SNPs (7059) but only 2% of these were highly conserved. Of these highly conserved intronic SNPs, five were at intron–exon boundaries, one in Xrcc5, two in Becn1, two in Adamts16 and one in Adamts6.
Overall, based on our SNP classification criteria and differential expression results, we predict functional effects for exonic variants in Lgr6, Ptprv, Xrcc5, Blm, Atr, Itga7, Krt15, Adamts6, Adamts16, Myl6, Brca1, Diap1 and Mcc and for regulatory variants in Fn1, Rnf7, Cdk2, Cs, Itga7, Myl6, Obfc2b, Wnt3a, Becn1, Lars, Diap1, Kif23, Coro2b, Fgf1, Hdac3, Ptprc and Mcc.
Expression differences in genes with regulatory variation
Of the 499 genes (Supplementary Table S2), 152 were differentially expressed between the LG/J and SM/J parental strains (Supplementary Table S3). We focused on the 34 best candidate genes (Table 2, Supplementary Table S4) and carried out function enrichment analysis showing cell cycle, cytoskeleton and developmentally related genes as being the top categories (Table 4).
Table 4. Molecular processes significantly over-represented among 34 candidate genes found in QTL regions.
| Function | P-value | N | Molecules |
|---|---|---|---|
| Cell cycle/DNA damage | |||
| S phase | 6.9E−07 | 7 | Blm, Brca1, Cdk2, Fgf1, Fn1, Ptprc, Xrcc5 |
| M phase | 2.1E−06 | 6 | Atr, Becn1, Cdk2, Fgf1, Kif23, Xrcc5 |
| Arrest in G2/M phase transition | 2.3E−06 | 4 | Atr, Blm, Brca1, Cdk2 |
| Homologous recombination of DNA | 7E−06 | 3 | Blm, Brca1, Cdk2 |
| Double-stranded DNA break repair of cells | 6.3E−05 | 3 | Blm, Brca1, Xrcc5 |
| Quantity of centrosome | 6.3E−05 | 3 | Becn1, Cdk2, Kif23 |
| Cytoskeleton and motility | |||
| Cell movement of gonadal cell lines | 0.00015 | 3 | Diaph1, Fn1, Wnt3A |
| Growth failure | 0.00022 | 7 | Blm, Brca1, Diaph1, Fn1, Itga7, Smarcal1, Xrcc5 |
| Organismal death | 4.6E−05 | 15 | Atr, Becn1, Blm, Brca1, Cdk2, Fn1, Hdac3, Itga7, Mesp2, Myl6, Ndn, Ptprc, Smarcal1, Wnt3A, Xrcc5 |
| Developmental | |||
| Growth of neurites | 2.8E−05 | 7 | Cdk2, Fgf1, Fn1, Kif23, Mesp2, Ndn, Wnt3A |
| Differentiation of cells | 2.9E−06 | 15 | Becn1, Blm, Brca1, Cdk2, Diaph1, Fgf1, Fn1, Hdac3, Itga7, Mcc, Ndn, Ptprc, Ptprv, Wnt3A, Xrcc5 |
| Formation of embryonic tissue | 0.00025 | 5 | Brca1, Fgf1, Fn1, Mesp2, Wnt3A |
| Proliferation of cells | 7.4E−05 | 18 | Atr, Becn1, Blm, Brca1, Cdk2, Diaph1, Fgf1, Fn1, Hdac3, Itga7, Mcc, Ndn, Ptprc, Ptprv, Rnf7, Smarcal1, Wnt3A, Xrcc5 |
| Proliferation of epithelial cells | 9.9E−05 | 6 | Brca1, Cdk2, Fgf1, Fn1, Mcc, Rnf7 |
N=number of genes from the set found in the category.
Genes in bold have been examined for expression levels in regenerating tissues.
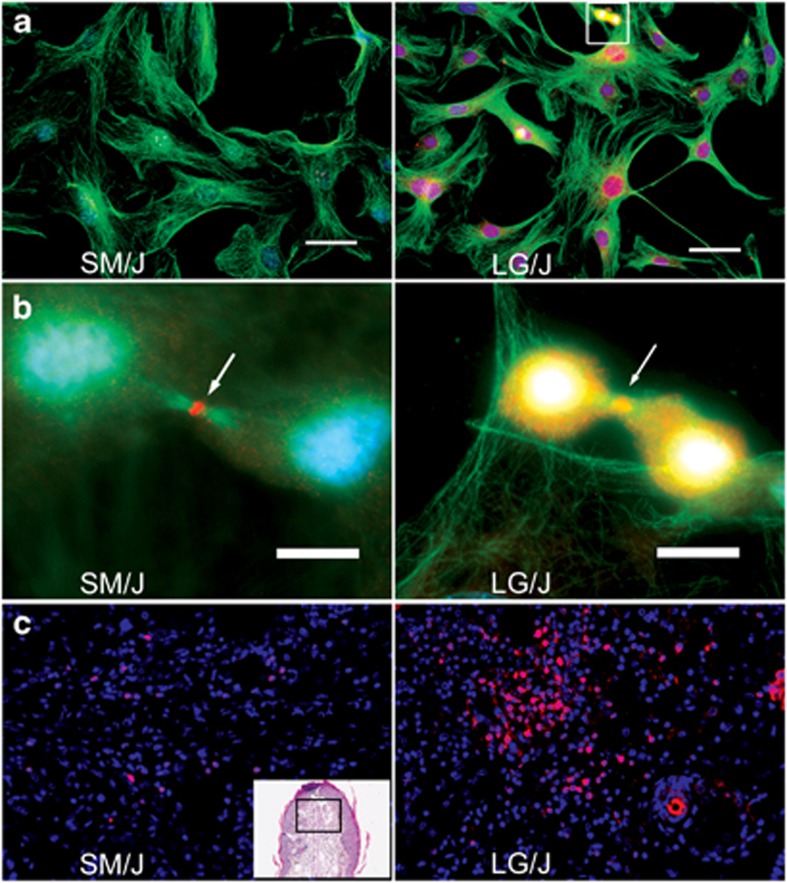
From the 34 gene set, we examined several candidate genes based on data from our previous studies, the identification of molecules known to be important in regeneration, and common function analysis from Table 4. Kif23 (Q9.1), a molecule involved in cytokinesis and developmental processes, is over-expressed in LG/J tissue at the mRNA level (expression ratio=+3.38). Using fibroblasts derived from normal ear tissue from LG/J and SM/J mice, we examined KIF23 protein expression by immunohistochemistry. Both LG/J and SM/J fibroblast populations show expression at the midzone during cytokinesis (Figure 1b); however, LG/J over-expresses this molecule throughout the process of daughter cell separation. Furthermore, only LG/J shows high levels of protein expression in the nucleus in interphase cells (Figure 1a). Thus, KIF23 has a different subcellular distribution in LG/J vs SM/J mice. KIF23 protein levels were also assessed in ear tissue (Figure 1c) where it was found in both epithelial and dermal cells with LG/J and SM/J healing ear tissue, but it had different patterns of expression. In LG/J ear tissue, KIF23 was high in the regeneration blastema and in the epidermis near hair follicles in LG/J, but it was not seen there in SM/J tissues (data not shown).
Figure 1.
KIF23 immunohistochemistry (IHC) in LG/J and SM/J ear cells and tissue. The upper two panels (a) show IHC of SM/J and LG/J fibroblasts with DAPI (blue, DNA), KIF23 (red), a-tubulin (green, microtubules) stains. Bars=50 μm. The middle two panels (b) show dividing cells ( × 100) and co-localization of green, blue, and red for microtubules, DNA, and KIF23, respectively. Arrows show the midzone. Here the bar=10 μm. The lower two panels (c) show KIF23 expression (red) in LG/J and SM/J hole-punched ear tissue (Day 7). The inset on the left is a low magnification H&E of a day 7 ear, and the boxed area is the stained region shown in both panels.
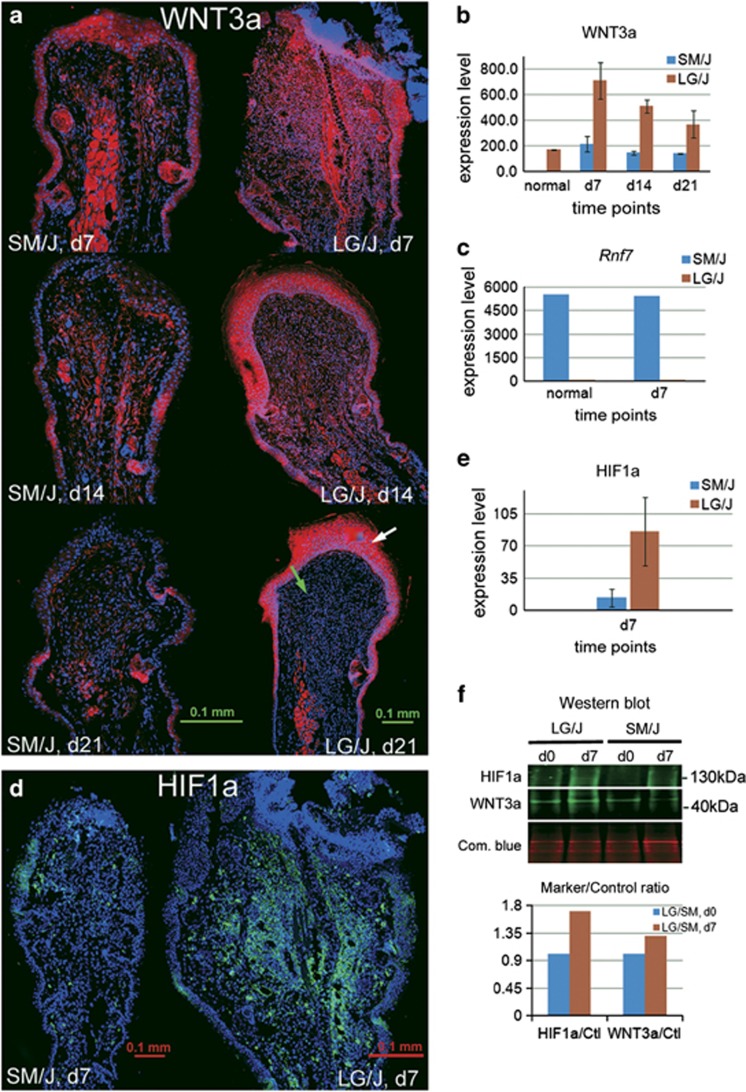
A second gene, Wnt3a (Q11.1), is involved in proliferation and regenerative healing and is also upregulated (expression ratio=+1.96) in LG/J. We examined WNT3a protein expression in LG/J and SM/J ear tissue throughout a 21-day post-injury period of healing by immunostaining (Figures 2a and b) and found a significant difference in level and location of expression. In the epidermis, WNT3a is high in LG/J from day 7 through day 21 of healing but only sparsely present on day 7 in SM/J; in the dermis, WNT3a is high in LG/J ear on days 7 and 14 (Figure 2a). This is supported by western blotting analysis (Figure 2f).
Figure 2.
WNT3a expression levels in LG/J and SM/J tissue on days 7, 14, and 21 post-ear hole injury (a). The exact localization and kinetics of staining for WNT3a (red) compared to DAPI (blue) is seen with a white arrow showing epidermis and a green arrow showing dermis. Quantitation of staining was carried out using ImageJ v1.46f software as seen in (b) for whole tissue. Rnf7 mRNA levels are shown in (c). HIF1a levels of immunostaining are seen in (d) and quantitation of HIF1a staining for the whole tissue is found in (e). Western blot analysis is presented in (f) with commassie blue as a loading control and below that is the ratio of the western analysis of specific band density/commassie blue band density as control. P-values for WNT3a are d7=0.08, d14=0.02; and d21=0.16; and for HIF1a=0.1995.
The mRNA expression level of the third gene, Rnf7 (Q9.2), shows a 54-fold reduction in LG/J compared with SM/J ear tissue (Figure 2c). This molecule acts as a HIF1a-specific E3 ligase, and the low level in LG/J should result in stabilization of HIF1a protein rather than its degradation. We examined HIF1a protein levels in LG/J and SM/J ear tissue on day 7 post injury and found higher levels in LG/J both by immunostaining (Figures 2d and e) and by western blotting analysis (Figure 2f). Further support for HIF stabilization is the activation of HIF1a target genes on day 7 after injury (Table 5) with a significant increase in Gapdh, Glut1, Leptin, Tgfb3, and Vegfb in LG/J over SM/J.
Table 5. HIF target gene expression in LG/J vs SM/J day 7 healing tissue.
| LG/SM | Day 7 ratio | P-value |
|---|---|---|
| Gapdh | 1.239 | 0.024 |
| Glut1 | 1.233 | 0.001 |
| lep | 3.143 | 0 |
| Tgfb3 | 1.654 | 0.012 |
| Vegfb | 1.506 | 0.006 |
Abbreviation: HIF, hypoxia-inducble factor.
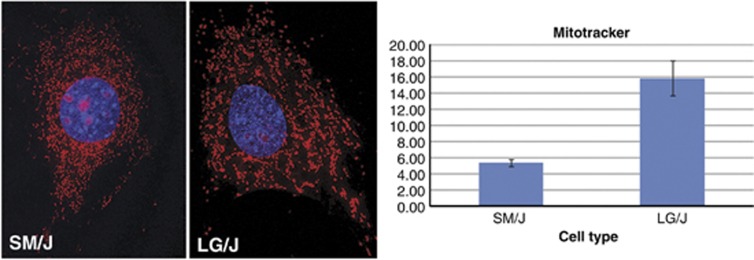
The fourth gene, Cs or citrate synthase (Q10.1), a mitochondrial structural gene, is strongly upregulated in LG/J (expression ratio=+5.51), As a second measure of mitochondrial content, we determined the level of mitotracker binding in LG/J and SM/J fibroblasts and found that LG/J showed higher levels of fluorescence (Figure 3), consistent with the high levels of Cs mRNA levels erxpressed in this tissue.
Figure 3.
In the two panels on the left are seen mitotracker staining of SM/J and LG/J ear-derived fibroblasts. Staining was quantified using the ImageJ v1.46f software as above and s.e. determined as seen in the right panel. P-value for mitotracker staining differences is 0.0032.
Discussion
The examination of LG/J and MRL/MpJ ear hole closure reflects mammalian epimorphic regeneration; conversely, the examination of control B6 and SM/J ear hole closure reflects mammalian wound repair. Over the past 15 years, mapping studies have attempted to identify candidate genes for these traits in very large genetic intervals. In the current study, real progress has been made in narrowing the quantitative trait intervals by use of the (LG,SM) F34 AIL. An important feature of the present work is that QTLs on chromosomes 9, 10, 11, 14 and 18 discovered in earlier studies (Blankenhorn et al., 2009; Cheverud et al., 2012) were confirmed in the AI line, lending further credence to their role in regenerative healing. New QTLs have been identified on chromosomes 1, 7, 12 and 13 in this study. Healing QTLs reported by Cheverud et al (2012) on chromosomes 2, 4, 6, 8 and 19 did not show up as significant at the 5% Bonferroni-corrected significance threshold in the present study. However, each of these QTL locations did pass a point-wise 5% threshold appropriate for testing an a priori hypothesis in the F34 generation given the earlier F2 results. Novel QTL found on chromosomes 1, 7 and 13 may have been missed in the F2 crosses, because positive and negative effects on the same chromosome would have cancelled each other out in the F2 high linkage disequilibrium environment.
It should be noted that the LG/J and SM/J mice have been selected for size differences and also differ in other ways, including their autoimmune responses and metabolic responses (Naviaux et al., 2009, for example). There are many events that are occurring between day 0 and day 7. There may be differences seen at the starting point (day 0), which could give one strain an advantage over another, but may not be operating by day 7. Wound healing/regeneration is a highly dynamic phenomenon and early events are quite important in setting the course of these two alternative healing pathways. Many of these events occur in the wound site between day 0 and day 7, so we examined gene expression for those two time points. These are discussed below.
Positional candidate genes for regenerative healing
Within our QTL support intervals, we selected groups of candidate genes based on data from our previous studies as well as a canonical pathway analysis. These include metabolism, cell cycle regulation, cell migration and adhesion and developmentally related molecules. We narrowed our analysis to more carefully examine four genes involved in each of these pathways/processes.
Metabolism
Differentially expressed genes within our intervals and involved in metabolism include Cs, Rnf7 and Becn1. We focused on this group because of previous studies showing the importance of aerobic glycolysis and mitochondrial variations in MRL ear hole regeneration (Sachadyn et al., 2008; Naviaux et al., 2009) and the role of glycolytic enzymes during amphibian regeneration (Schmidt, 1958, Rao et al., 2009). We examined Cs or citrate synthase, a Kreb cycle enzyme and structural mitochondrial protein found in the mitochondrial matrix. CS levels can be used to quantify the mitochondrial content of cells. MRL tissues contain more CS protein than B6 tissues (in liver 67% more; in heart 82% more) (Naviaux et al., 2009), consistent with the mRNA upregulation in LG/J vs SM/J (expression ratio=+5.51) and the increased levels of mitotracker labeling in LG/J we show here (Figure 3). This finding supports the notion that regenerating tissue contains increased numbers of mitochondria. Citrate synthase has six HCM non-coding SNPs spread across the gene, one in the 2500 bp upstream and two downstream, two in introns (2) and one in the 3′UTR, suggesting a regulatory mechanism controlling expression levels.
We also examined Rnf7 (SAG/ROC2/RBX2), which codes for an E3 ligase component and has a massive 54-fold reduction in LG/J compared with SM/J mice. Interest in this molecule relates to its involvement in HIF1a levels. HIF1a is a key positive regulator of glycolysis in response to hypoxia that transcriptionally activates a large number of genes in the glycolytic pathway with the effect of enhancing pyruvate conversion to lactate and suppressing conversion to acetyl CoA (Semenza, 2011). RNF7 is responsible for the ubiquination and then degradation of the HIF1a protein (Tan et al., 2007). With this severe lack of Rnf7 mRNA in LG/J, it is not unexpected that HIF1a would remain at high levels once transcribed and translated in LG/J mice. MRL mice has been found to show increased levels of glycolysis (Naviaux et al., 2009), and one possible explanation for this may be the HIF1a upregulation and reduced levels of breakdown in LG/J due to the absence of RNF7. Rnf7 has five HCM non-coding SNPs, four intronic and one in the 3′UTR, again suggesting a regulatory mechanism controlling expression levels.
Cell cycle regulation
Our previous study of ear hole closure showed that cells and tissue from MRL/MpJ and LG/J regenerative mice display an increased DNA damage response, increased p53 levels and G2/M arrest compared with the non-regenerative controls (Bedelbaeva et al., 2010, Heber-Katz et al., 2013). Furthermore, the G1 checkpoint regulatory molecule, p21Cip1/Waf1, was found in very reduced levels in regenerative ears and, when deleted in KO mice, led to a highly proliferative cellular response, a G2 bias, DNA damage and ear hole regeneration (Bedelbaeva et al., 2010). Analysis of our selected genes showed that 14 of our 19 QTLs are involved in cell cycle regulation or DNA damage and repair with Cdk2, Kif23, Mcc as cell cycle regulators; Brca1, Obfc2b (Hssb1), Atr as DNA damage checkpoint genes; Blm and Smarca1 involved in fork stability; Rdm1, Fan1, Xrcc5 involved in DNA repair and recombination; and Hdac3, H2a, H2ba and Smarcc2 being involved in chromatin remodeling with several differentially expressed between LG/J and SM/J healing tissues (Kif23, Rnf7, Cdk2, Obfc2b and Mcc).
The first gene we examined in this group, Kif23, is a motor molecule known to be involved in cytokinesis (Nislow et al., 1992; Raich et al., 1998; Neef et al., 2006), a process important in axolotl (Rao et al., 2009, Heber-Katz et al., 2013) and zebrafish regeneration (Poss et al., 2002, 2004). Kif23, also known as MKLP1, is part of a critical complex formed at the midzone or midbody and associates with CYK-4 in the central spindlin (Glotzer, 2005). By immunohistochemistry, both LG/J and SM/J fibroblast populations show expression at the midzone during cytokinesis (Figure 1b) though LG/J expresses this molecule throughout the process of daughter cell separation. Only LG/J shows high levels of protein expression in the nucleus of interphase cells (Figure 1c). Thus, KIF23 has a different subcellular distribution in LG/J vs SM/J strain animals. The cellular distribution of KIF23 in SM/J is the typical expression pattern. KIF23 has previously been shown in CHO1 cells to have the typical KIF23 transcript (MKLP1) (Nislow et al., 1992) and an additional, unique splice variant that includes exon 18, making it 100 amino acids longer than MKLP1 and allowing it to bind to actin in the cytoplasm (CHO-1) (Kuriyama et al., 2002). Kif23 (Figure 1a) has only one HCM non-coding SNP located in the 3′UTR, which may modify expression. It also has a single non-synonymous coding SNP in exon 13 (Val:464:Ile). Given the non-synonymous coding SNP, it is not known if there is a gain or loss of function of this gene.
The second gene, Rnf7, the E3 ligase discussed above (in ‘Metabolism' section), is also associated with the ubiquination and degradation of p27, a cell cycle checkpoint gene which inhibits S phase entry (Duan et al., 2001). Though our previous results showed that LG/J fibroblasts display increased DNA damage and arrest in G2 (Bedelbaeva et al., 2010), p27 disregulation was not explored.
Cell migration and adhesion
Cells involved in migration need to polarize and adhere to the extracellular matrix to move forward. Kif23 discussed above in relation to its role in cytokinesis (in ‘Cell cycle regulation' section) has another potential function. It was originally found to be important in body closure during embryogenesis in Caenorhabditis elegans with the formation of the digestive tract through its attachment to the external epidermis. Kif23 appears to function in epithelial polarity and epithelialization of embryos (Portereiko et al., 2004). This provides a novel function that could very well have a key role in mammalian wound closure. Protein levels of KIF23 were assessed by immunohistochemical staining of ear tissue (Figure 2d) where it is found in both epithelial and dermal cells. LG/J and SM/J healing ear tissue show different patterns of expression. In LG/J ear tissue, KIF23 shows high levels of expression in the regeneration blastema not seen in SM/J. Also, there is expression in the epidermis near hair follicles in LG/J not seen in SM/J (data not shown).
Developmentally related genes
The establishment of the role of developmental genes involved in the regenerative response is long standing (Stocum and Cameron, 2011), and we identified four genes: Wnt3a, Lgr6, Mesp2 and Fgf1, which are in healing QTLs and known to be important for developmental regulation.
Wnt3a is involved in the proliferation, differentiation and migration of mesenchymal stem cells in rats (Shang et al., 2007). In postnatal mice, canonical WNT signaling increases following wounding and functions with transforming growth factor-β in this context (Carre et al., 2010). Also, WNT proteins regulate the rate of satellite cell proliferation during adult skeletal muscle cell regeneration (Otto et al., 2008). WNT has been shown to have a key role in the initiation of amphibian limb (Kawakami et al., 2006; Yokoyama et al., 2007) and zebrafish fin regeneration (Goessling et al., 2009). Wnt3a has only one highly conserved intronic non-coding SNP in its sequence. As Wnt3a mRNA is upregulated in LG/J tissue, we examined WNT3a protein expression in LG/J and SM/J ear hole sections (Figure 2a) and found a highly significant difference in level and location of expression (Figure 2b). The WNT3a level is extremely high in the LG/J epidermis from day 7 through day 21 of healing but only sparsely present on day 7 in SM/J. It is also at high levels in the dermis on days 7 and 14. As WNT3a is a canonical WNT and signals through β-catenin, which enters the nucleus and activates proliferation through binding to the TCF/LEF transcription factors, this difference could, in part, account for the proliferative response in the LG/J ear leading to ear hole closure. Interestingly, the microRNA 344 family is found in our Q7.1 locus, is differentially expressed, and may increase canonical WNT signaling and affect Wnt3a expression (Qin et al., 2010). Another candidate gene, Lgr6, enhances Wnt/β-catenin, and thus WNT3a, signaling through increased LGR6 phosphorylation by high affinity binding to R-spondins 1–3 (Gong et al., 2012). Thus, we have identified at least three candidate genes involved with enhancing canonical WNT signaling in LG/J compared with SM/J supporting an important role for canonical wnts in regeneration.
Conclusion
This comprehensive survey of expression and molecular variation within strong QTL-based positional candidate genes identifies likely SNPs affecting variation in ear wound healing in LG, SM mice. Future studies will evaluate the role of high probability variants in genes and physiological pathways most closely related to wound healing, especially cell cycle and DNA repair, cell migration and adhesion, cell differentiation and proliferation, fibrosis and specific metabolic processes. Although this study focuses on ear tissue regeneration, it is possible that these QTLs are also involved in the regeneration of the diverse array of tissues in MRL/MpJ and LG/J discussed in the Introduction section. Rai et al. (2012) found a strong positive genetic correlation between ear hole and articular cartilage regeneration in a set of LGXSM Recombinant Inbred (RI) lines, indicating a common genetic basis for these two regenerative processes. Investigation into other regenerating tissues in the LGXSM RI lines may show that the QTLs and genes identified here operate more generally in the regeneration of a variety of tissues.
Data archiving
Gene expression microarray data for LG/SM mice has been deposited to GEO: GSE17168. The genomic SNPs for the LG/J and SM/J strain used in this study are deposited in dbSNP: under the handle ‘Cheverud'. DRYAD 10.5061/dryad.vc473: Genomic coordinates for SNPs in mapping LG,SM F34 AIL.
Acknowledgments
These studies were supported by a grant from the NIH, NIGMS GM073226, a grant from the Commonwealth of Pennsylvania, and an NCI Cancer Center Grant (P30 CA10815) to the Wistar Institute. These studies were also supported by the Genomics and Animal Wistar Core Facilities. Genotyping services were provided by the Center for Inherited Disease Research (CIDR). CIDR is funded through a federal contract from the National Institutes of Health to The Johns Hopkins University (contract number HHSN268200782096C). We thank Peter Burgers for his insights into the DNA replication and cell cycle functions of positional candidate genes in our QTLs.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Mat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfaro MP, Pagni M, Vincent A, Atkinson J, Hill MF, Cates J, et al. The Wnt modulator sFRP2 enhances mesenchymal stem cell engraftment, granulation tissue formation and myocardial repair. Proc Natl Acad Sci. 2008;105:18366–18371. doi: 10.1073/pnas.0803437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker KL, Daniels SB, Lennington JB, Lardaro T, Czap A, Notti RQ, et al. Neuroblast protuberances in the subventricular zone of the regenerative MRL/MpJ mouse. J Comp Neurol. 2006;498:747–761. doi: 10.1002/cne.21090. [DOI] [PubMed] [Google Scholar]
- Balu DT, Hodes GE, Anderson BT, Lucki I. Enhanced sensitivity of the MRL/MpJ mouse to the neuroplastic and behavioral effects of chronic antidepressant treatments. Neuropsychopharmacology. 2009;34:1764–1773. doi: 10.1038/npp.2008.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedelbaeva K, Snyder A, Gourevitch D, Clark L, Zhang XM, Leferovich J, et al. Lack of p21 expression links cell cycle control and appendage regeneration in mice. Proc Natl Acad Sci. 2010;107:5845–5850. doi: 10.1073/pnas.1000830107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenhorn E, Bryan G, Kossenkov A, Clark L, Zhang XM, Chang C, et al. Genetic loci that regulate healing and regeneration in LG/J and SM/J mice. Mamm Genome. 2009;20:720–733. doi: 10.1007/s00335-009-9216-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenhorn EP, Troutman S, Clark LD, Zhang XM, Chen P, Heber-Katz E. Sexually dimorphic genes regulate healing and regeneration in MRL mice. Mamm Genome. 2003;14:250–260. doi: 10.1007/s00335-002-2222-3. [DOI] [PubMed] [Google Scholar]
- Broman K, Sen S. A Guide to QTL Mapping with R/qtl. Springer: New York, NY, USA; 2009. [Google Scholar]
- Buckley G, Metcalfe AD, Ferguson MWJ. Peripheral nerve regeneration in the MRL/MpJ ear wound model. J Anat. 2011;218:163–172. doi: 10.1111/j.1469-7580.2010.01313.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhimschi CS, Zhao G, Sora N, Madri JA, Buhimschi IA. Myometrial wound healing postCesarean delivery in the MRL/MpJ mouse model of uterine scarring. Am J Pathol. 2010;177:197207. doi: 10.2353/ajpath.2010.091209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carre AL, James AW, MacLeod L, Kong W, Kawai K, Longaker MT, et al. Interaction of Wingless Protein (Wnt), Transforming Growth Factor-[beta]1, and hyaluronan production in fetal and postnatal fibroblasts. Plast Reconstr Surg. 2010;125:74–88. doi: 10.1097/PRS.0b013e3181c495d1. [DOI] [PubMed] [Google Scholar]
- Chadwick RB, Bu L, Yu H, Hu Y, Wergedal JE, Mohan S, et al. Digit tip regrowth and differential gene expression in MRL/MpJ, DBA/2, and C57BL/6 mice. Wound Repair Regen. 2007;15:275–284. doi: 10.1111/j.1524-475X.2007.00216.x. [DOI] [PubMed] [Google Scholar]
- Chai CK. Comparison of two inbred strains of mice and their F1 hybrid in response to androgen. Anat Rec. 1956;126:269–282. doi: 10.1002/ar.1091260302. [DOI] [PubMed] [Google Scholar]
- Chen L, Storey JD. Relaxed significance criteria for linkage analysis. Genetics. 2006;173:2371–l2381. doi: 10.1534/genetics.105.052506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheverud JM, Vaughn TT, Pletscher LS, King-Ellison K, Bailiff J, Adams E, et al. Epistasis and the evolution of additive genetic variance in populations that pass through a bottleneck. Evolution. 1999;53:1009–1018. doi: 10.1111/j.1558-5646.1999.tb04516.x. [DOI] [PubMed] [Google Scholar]
- Cheverud JM, Ehrich TH, Hrbek T, Kenney JP, Pletscher LS, Semenkovich CF. Quantitative trait loci for obesity- and diabetes-related traits and their dietary responses to high-fat feeding in LGXSM recombinant inbred mouse strains. Diabetes. 2004;53:3328–3336. doi: 10.2337/diabetes.53.12.3328. [DOI] [PubMed] [Google Scholar]
- Cheverud JM, Lawson HA, Funk R, Zhou J, Blankenhorn EP, Heber-Katz E. Healing quantitative trait loci in a combined cross analysis using related mouse strain crosses. Heredity. 2012;108:441–446. doi: 10.1038/hdy.2011.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun S, Fay JC. Identification of deleterious mutations within three human genomes. Genome Res. 2009;19:1553–1561. doi: 10.1101/gr.092619.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark LD, Clark RK, Heber-Katz E. A new murine model for mammalian wound repair and regeneration. Clin Immunol Immunopathol. 1998;88:35–45. doi: 10.1006/clin.1998.4519. [DOI] [PubMed] [Google Scholar]
- Darvasi A, Soller M. Advanced intercross lines, an experimental population for fine genetic mapping. Genetics. 1995;141:1199–1207. doi: 10.1093/genetics/141.3.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan H, Tsvetkov LM, Liu Y, et al. Promotion of S-phase entry and cell growth under serum starvation by SAG/ROC2/Rbx2/Hrt2, an E3 ubiquitin ligase component: association with inhibition of p27 accumulation. Mol Carcinog. 2001;30:37–46. doi: 10.1002/1098-2744(200101)30:1<37::aid-mc1011>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- Eppig JT, Blake JA, Bult CJ, Kadin JA, Richardson JE,, the Mouse Genome Database Group The Mouse Genome Database (MGD): comprehensive resource for genetics and genomics of the laboratory mouse. Nucleic Acids Res. 2012;40:D881–D886. doi: 10.1093/nar/gkr974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falconer DS, Mackay TFC.1996Introduction to Quantitative Genetics4th ednBenjamin Cummings: New York, NY, USA [Google Scholar]
- Fawcett GL, Roseman CC, Jarvis JP, Wang B, Wolf JB, Cheverud JM. Genetic architecture of adiposity and organ weight using combined generation QTL analysis. Obesity. 2008;16:1861–1868. doi: 10.1038/oby.2008.300. [DOI] [PubMed] [Google Scholar]
- Fitzgerald J, Rich C, Burkhardt D, Allen J, Herzka AS, Little CB. Evidence for articular cartilage regeneration in MRL/MpJ mice. Osteoarthritis Cartilage. 2008;16:1319–1326. doi: 10.1016/j.joca.2008.03.014. [DOI] [PubMed] [Google Scholar]
- Glotzer M. The molecular requirements for cytokinesis. Science. 2005;307:1735–1739. doi: 10.1126/science.1096896. [DOI] [PubMed] [Google Scholar]
- Goessling W, North TE, Loewer S, Lord AM, Lee S, Stoick-Cooper CL, et al. Genetic interaction of PGE2 and Wnt signaling regulates developmental specification of stem cells and regeneration. Cell. 2009;136:1136–1147. doi: 10.1016/j.cell.2009.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong X, Carmon KS, Lin Q, Thomas A, Yi J, Liu Q. LGR6 is a high affinity receptor of R-spondins and potentially functions as a tumor suppressor. PLoS One. 2012;7:e37137. doi: 10.1371/journal.pone.0037137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodale HD. A study of the inheritance of body weight in the albino mouse by selection. J Hered. 1938;29:101–112. [Google Scholar]
- Gourevitch DL, Clark L, Bedelbaeva K, Leferovich J, Heber-Katz E. Dynamic changes after murine digit amputation: The MRL mouse digit shows waves of tissue remodeling, growth, and apoptosis. Wound Repair and Regeneration. 2009;17:447–455. doi: 10.1111/j.1524-475X.2009.00492.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimes LN, Goss RJ. Regeneration of holes in rabbit ears. Am Zool. 1970;10:537. [Google Scholar]
- Haley CS, Knott SA. A simple regression method for mapping quantitative trait loci in line crosses using flanking markers. Heredity. 1992;69:315–324. doi: 10.1038/hdy.1992.131. [DOI] [PubMed] [Google Scholar]
- Hampton DW, Seitz A, Chen P, Heber-Katz E, Fawcett JW. Altered CNS response to injury in the MRL/MpJ mouse. Neuroscience. 2004;127:821–832. doi: 10.1016/j.neuroscience.2004.05.057. [DOI] [PubMed] [Google Scholar]
- Haris Naseem R, Meeson AP, Michael DiMaio J, White MD, Kallhoff J, Humphries C, et al. Reparative myocardial mechanisms in adult C57BL/6 and MRL mice following injury. Physiol Genomics. 2007;30:44–52. doi: 10.1152/physiolgenomics.00070.2006. [DOI] [PubMed] [Google Scholar]
- Heber-Katz E, Chen P, Dvm LC, Zhang X-M, Troutman S, Blankenhorn EP. Regeneration in MRL mice: further genetic loci controlling the ear hole closure trait using MRL and M. m. castaneus mice. Wound Repair Regen. 2004;12:384–392. doi: 10.1111/j.1067-1927.2004.012308.x. [DOI] [PubMed] [Google Scholar]
- Heber-Katz E, Zhang Y, Bedelbaeva K, Song F, Chen X, Stocum DL. Cell cycle regulation and regeneration. Curr Top Microbiol Immunol. 2013;367:253–276. doi: 10.1007/82_2012_294. [DOI] [PubMed] [Google Scholar]
- Hrbek T, de Brito RA, Wang B, Pletscher LS, Cheverud JM. Genetic characterization of a new set of recombinant inbred lines (LGXSM) formed from the intercross of SM/J and LG/J inbred mouse strains. Mamm Genome. 2006;17:417–429. doi: 10.1007/s00335-005-0038-7. [DOI] [PubMed] [Google Scholar]
- Joseph J, Dyson M. Tissue replacement in the rabbit's ear. Br J Surg. 1966;53:372–380. doi: 10.1002/bjs.1800530415. [DOI] [PubMed] [Google Scholar]
- Kawakami Y, Rodriguez EC, Raya M, Kawakami H, Mart M, Dubova I, Izpis BelmonteJC. Wnt/beta-catenin signaling regulates vertebrate limb regeneration. Genes Dev. 2006;20:3232–3237. doi: 10.1101/gad.1475106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kench JA, Russell DM, Fadok VA, Young SK, Worthen GS, Jones-Carson J, et al. Aberrant wound healing and TGF-beta production in the autoimmune-prone MRL/+ mouse. Clin Immunol. 1999;92:300–310. doi: 10.1006/clim.1999.4754. [DOI] [PubMed] [Google Scholar]
- Kramer MG, Vaughn TT, Pletscher LS, King-Ellison K, Adams E, Erikson C, et al. Genetic variation in body weight gain and composition in the intercross of Large (LG/J) and Small (SM/J) inbred strains of mice. Genet Mol Biol. 1998;21:211–218. [Google Scholar]
- Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- Kuriyama R, Gustus C, Terada Y, Uetake Y, Matuliene J. CHO1, a mammalian kinesin like protein, interacts with F-actin and is involved in the terminal phase of cytokinesis. J Cell Biol. 2002;156:783–790. doi: 10.1083/jcb.200109090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson HA, Cady JE, Partridge C, Wolf JB, Semenkovich CF, Cheverud JM. Genetic effects at pleiotropic loci are context-dependent with consequences for the maintenance of genetic variation in populations. PLOS Genet. 2011;7:e1002256. doi: 10.1371/journal.pgen.1002256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leferovich JM, Bedelbaeva K, Samulewicz S, Zhang XM, Zwas D, Lankford EB, Heber-Katz E. Heart regeneration in adult MRL mice. Proc Natl Acad Sci USA. 2001;98:9830–9835. doi: 10.1073/pnas.181329398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Ji L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity. 2005;95:221–227. doi: 10.1038/sj.hdy.6800717. [DOI] [PubMed] [Google Scholar]
- Li XM, Gu WK, Masinde G, Hamilton-Ulland M, Xu SZ, Mohan S, et al. Genetic control of the rate of wound healing in mice. Heredity. 2010;86:668–674. doi: 10.1046/j.1365-2540.2001.00879.x. [DOI] [PubMed] [Google Scholar]
- MacArthur J. Genetics of body size and related characters. Selection of small and large races of the laboratory mouse. Am Nat. 1944;78:142–157. [Google Scholar]
- Masinde GL, Li X, Gu W, Davidson H, Mohan S, Baylink DJ. Identification of wound healing/regeneration quantitative trait loci (QTL) at multiple time points that explain seventy percent of variance in (MRL/MpJ and SJL/J) mice F2 population. Genome Res. 2001;11:2027–2033. doi: 10.1101/gr.203701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBrearty BA, Clark LD, Zhang XM, Blankenhorn EP, Heber-Katz E. Genetic analysis of a mammalian wound-healing trait. Proc Natl Acad Sci USA. 1998;95:11792–11797. doi: 10.1073/pnas.95.20.11792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy ER, Roths JB. Autoimmunity and Lymphoproliferation: Induction by Mutant Gene Lpr, and Acceleration by a Male-Associated Factor in Strain BXSB Mice. Elsivier North Holland: New York, NY, USA; 1978. [Google Scholar]
- Naviaux RK, Le TP, Bedelbaeva K, Leferovich J, Gourevitch D, Sachadyn P, Zhang XM, Clark L, Heber-Katz E. Retained features of embryonic metabolism in the adult MRL mouse. Mol Genet Metab. 2009;96:133–144. doi: 10.1016/j.ymgme.2008.11.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neef R, Klein UR, Kopajtich R, Barr FA. Cooperation between mitotic kinesins controls the late stages of cytokinesis. Curr Biol. 2006;16:301–307. doi: 10.1016/j.cub.2005.12.030. [DOI] [PubMed] [Google Scholar]
- Nislow C, Lombillo VA, Kuriyama R, McIntosh JR. A plus-end-directed motor enzyme that moves antiparallel microtubules in vitro localizes to the interzone of mitotic spindles. Nature. 1992;359:543–547. doi: 10.1038/359543a0. [DOI] [PubMed] [Google Scholar]
- Norgard A, Lawson HA, Pletscher LS, Wang B, Brooks VR, Wolf JB, Cheverud JM. Genetic factors and diet affect long-bone length in the F34 LG,SM advanced intercross. Mamm Genome. 2011;22:178–196. doi: 10.1007/s00335-010-9311-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto A, Schmidt C, Luke G, Allen S, Valasek P, Muntoni F, Lawrence-Watt D, Patel K. Canonical Wnt signalling induces satellite-cell proliferation during adult skeletal muscle regeneration. J Cell Sci. 2008;121:2939–2950. doi: 10.1242/jcs.026534. [DOI] [PubMed] [Google Scholar]
- Portereiko MF, Saam J, Mango SE. ZEN-4/MKLP1 is required to polarize the foregut epithelium. Curr Biol. 2004;14:932–941. doi: 10.1016/j.cub.2004.05.052. [DOI] [PubMed] [Google Scholar]
- Poss KD, Nechiporuk A, Hillam AM, Johnson SL, Keating MT. Mps1 defines a proximal blastemal proliferative compartment essential for zebrafish fin regeneration. Development. 2002;800:5141–5149. doi: 10.1242/dev.129.22.5141. [DOI] [PubMed] [Google Scholar]
- Poss KD, Nechiporuk A, Stringer KF, Lee C, Keating MT. Germ cell aneuploidy in zebrafish with mutations in the mitotic checkpoint gene mps1. Genes Dev. 2004;18:1527–1532. doi: 10.1101/gad.1182604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin L, Chen Y, Niu Y, Chen W, Wang Q, Xiao S, et al. A deep investigation into the adipogenesis mechanism: profile of microRNAs regulating adipogenesis by modulating the canonical Wnt/beta-catenin signaling pathway. BMC Genomics. 2010;11:320. doi: 10.1186/1471-2164-11-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai MF, Hashimoto S, Eric E, Johnson EE, Janiszak KL, Fitzgerald J, et al. Heritability of articular cartilage regeneration and its association with ear-wound healing. Arthritis Rheum. 2012;64:2300–2310. doi: 10.1002/art.34396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raich WB, Moran AN, Rothman JH, Hardin J. Cytokinesis and midzone microtubule organization in Caenorhabditis elegans require the kinesin-like protein ZEN-4. Mol Biol Cell. 1998;9:2037–2049. doi: 10.1091/mbc.9.8.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao N, Jhamb D, Milner DJ, Li B, Song F, Wang M, et al. Proteomic analysis of blastema formation in regenerating axolotl limbs. BMC Biol. 2009;7:83. doi: 10.1186/1741-7007-7-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachadyn P, Zhang XM, Clark LD, Naviaux RK, Heber-Katz E. Naturally occurring mitochondrial DNA heteroplasmy in the MRL mouse. Mitochondrion. 2008;8:358–366. doi: 10.1016/j.mito.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt AJ. Forelimb regeneration of thyroidectomized adult newts. II. Histology. J Exp Zool. 1958;139:95–135. doi: 10.1002/jez.1401390108. [DOI] [PubMed] [Google Scholar]
- Semenza GL. Regulation of metabolism by hypoxia-inducible factor 1. Cold Spring Harb Symp Quant Biol. 2011;76:347–353. doi: 10.1101/sqb.2011.76.010678. [DOI] [PubMed] [Google Scholar]
- Shang Yc, Wang Sh, Xiong F, Zhao Cp, Fn Peng, et al. Wnt3a signaling promotes proliferation, myogenic differentiation, and migration of rat bone marrow mesenchymal stem cells. Acta Pharmacol Sin. 2007;28:1761–1774. doi: 10.1111/j.1745-7254.2007.00671.x. [DOI] [PubMed] [Google Scholar]
- Stocum DL, Cameron JA. Looking proximally and distally: 100 years of limb regeneration and beyond. Dev Dyn. 2011;240:943–968. doi: 10.1002/dvdy.22553. [DOI] [PubMed] [Google Scholar]
- Storey JohnD, Tibshirani Robert. Statistical significance for genome-wide studies. Proc Natl Acad Sci. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M, Gu Q, He H, Pamarthy D, Semenza GL, Sun Y. SAG/ROC2/RBX2 is a HIF-1 target gene that promotes HIF-1[alpha] ubiquitination and degradation. Oncogene. 2007;27:14041411. doi: 10.1038/sj.onc.1210780. [DOI] [PubMed] [Google Scholar]
- Thuret S, Toni N, Aigner S, Yeo GW, Gage FH. Hippocampus-dependent learning is associated with adult neurogenesis in MRL/MpJ mice. Hippocampus. 2009;19:658–669. doi: 10.1002/hipo.20550. [DOI] [PubMed] [Google Scholar]
- Thuret S, Thallmair M, Horky LL, Gage FH. Enhanced functional recovery in MRL/MpJ mice after spinal cord dorsal hemisection. PLoS One. 2012;7:e30904. doi: 10.1371/journal.pone.0030904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolba RH, Schildberg FA, Decker D, Abdullah Z, Büttner R, Minor T, Von Ruecker A. Mechanisms of improved wound healing in Murphy Roths Large (MRL) mice after skin transplantation. Wound Repair Regen. 2010;18:662–670. doi: 10.1111/j.1524-475X.2010.00631.x. [DOI] [PubMed] [Google Scholar]
- Ueno M, Lyons BL, Burzenski LM, Gott B, Shaffer DJ, Roopenian DC, Shultz LD. Accelerated wound healing of alkali-burned corneas in MRL mice Is associated with a reduced inflammatory signature. Invest Ophthalmol Vis Sci. 2005;46:40974106. doi: 10.1167/iovs.05-0548. [DOI] [PubMed] [Google Scholar]
- Vaughn TT, Pletscher LS, Peripato A, King-Ellison K, Adams E, Erikson C, Cheverud JM. Mapping quantitative trait loci for murine growth: a closer look at genetic architecture. Genet Res. 1999;74:313–322. doi: 10.1017/s0016672399004103. [DOI] [PubMed] [Google Scholar]
- Vorontsova MA, Liosner LD. Asexual Propagation and Regeneration. Permagon Press: New York, NY, USA; 1960. [Google Scholar]
- Ward BD, Furman BD, Huebner JL, Kraus VB, Guilak F, Olson SA. Absence of posttraumatic arthritis following intraarticular fracture in the MRL/MpJ mouse. Arthritis Rheum. 2008;58:744–753. doi: 10.1002/art.23288. [DOI] [PubMed] [Google Scholar]
- Wilson SP, Goodale HD, Kyle WH, Godfrey EF. Long term selection for body weight in mice. J Hered. 1971;62:228–235. doi: 10.1093/oxfordjournals.jhered.a108156. [DOI] [PubMed] [Google Scholar]
- Yokoyama H, Ogino H, Stoick-Cooper CL, Grainger RM, Moon RT. Wnt/[beta]-catenin signaling has an essential role in the initiation of limb regeneration. Dev Biol. 2007;306:170–178. doi: 10.1016/j.ydbio.2007.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Mohan S, Masinde G, Baylink D. Mapping the dominant wound healing and soft tissue regeneration QTL in MRL x CAST. Mamm Genome. 2005;16:918–924. doi: 10.1007/s00335-005-0077-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.