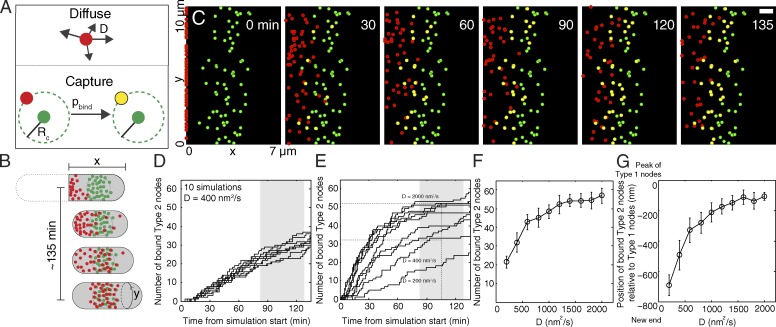
Two distinct classes of cortical nodes form separately during interphase in fission yeast cells and then merge at the cell equator by a diffuse-and-capture mechanism to prepare nodes to form the contractile ring for cytokinesis.
Abstract
We investigated the assembly of cortical nodes that generate the cytokinetic contractile ring in fission yeast. Observations of cells expressing fluorescent fusion proteins revealed two types of interphase nodes. Type 1 nodes containing kinase Cdr1p, kinase Cdr2p, and anillin Mid1p form in the cortex around the nucleus early in G2. Type 2 nodes with protein Blt1p, guanosine triphosphate exchange factor Gef2p, and kinesin Klp8p emerge from contractile ring remnants. Quantitative measurements and computer simulations showed that these two types of nodes come together by a diffuse-and-capture mechanism: type 2 nodes diffuse to the equator and are captured by stationary type 1 nodes. During mitosis, cytokinetic nodes with Mid1p and all of the type 2 node markers incorporate into the contractile ring, whereas type 1 nodes with Cdr1p and Cdr2p follow the separating nuclei before dispersing into the cytoplasm, dependent on septation initiation network signaling. The two types of interphase nodes follow parallel branches of the pathway to prepare nodes for cytokinesis.
Introduction
During cell division, fungi, amoebas, and animal cells assemble a contractile ring made of actin filaments and myosin to split the cell in two (Pollard and Wu, 2010). Understanding how the contractile ring is correctly placed and produces force during cytokinesis requires mechanistic information about contractile ring formation. The fission yeast Schizosaccharomyces pombe, a leading model organism to study cytokinesis, assembles a contractile ring around the cell equator during mitosis from ∼65 punctate structures called cytokinesis nodes containing anillin Mid1p (Moseley et al., 2009; Saha and Pollard, 2012a) and contractile ring proteins (Wu et al., 2006). Measurements of the timing of the appearance (Wu et al., 2003) and numbers of molecules of cytokinesis nodes (Wu and Pollard, 2005) led to a proposal that nodes self-assemble into the contractile ring by myosins pulling on actin filaments originating from other nodes (Wu et al., 2006). Successful simulations of a mathematical model of this hypothesis required that the connections between nodes break about every 20 s (Vavylonis et al., 2008), a process that depends on actin filament severing by cofilin (Chen and Pollard, 2011).
The septation initiation network (SIN) is a cascade of signaling proteins that terminates in the kinase Sid2p and promotes the formation and constriction of the contractile ring and the subsequent formation of the cell wall that divides daughter cells (Johnson et al., 2012). Components of SIN are associated with spindle pole bodies (SPBs), and the SPB that concentrates the SIN kinase Cdc7p is more active (Johnson et al., 2012).
Cytokinesis nodes originate during interphase (Paoletti and Chang, 2000; Moseley et al., 2009; Saha and Pollard, 2012a) as interphase nodes containing the cell cycle kinases Cdr1p, Cdr2p, and Wee1p as well as anillin-like Mid1p, kinesin Klp8p, node protein Blt1p, GTP exchange factor Gef2p (Paoletti and Chang, 2000; Morrell et al., 2004; Martin and Berthelot-Grosjean, 2009; Moseley et al., 2009; Ye et al., 2012), and Nod1p (Jourdain et al., 2013). Kinases Cdr1p and Cdr2p in interphase nodes negatively regulate Wee1p, which controls Cdk1p and the onset of mitosis (Martin and Berthelot-Grosjean, 2009; Moseley et al., 2009). Additional work established an extensive network of genetic and physical interactions among interphase node proteins (Breeding et al., 1998; Almonacid et al., 2009; Martin and Berthelot-Grosjean, 2009; Moseley et al., 2009; Ye et al., 2012; Guzman-Vendrell et al., 2013; Zhu et al., 2013). Nevertheless, the available information was fragmentary and left open many questions about the temporal and physical organization of interphase nodes and their relation to cytokinesis nodes. A basic understanding of the assembly pathway of interphase nodes was needed to interpret these observations.
To determine how nodes for cytokinesis assemble during interphase, we imaged fission yeast strains expressing six interphase node proteins tagged in the genome with fluorescent proteins. These proteins localized to two distinct types of nodes, and our analysis revealed that they represent two parallel branches of the pathway that prepares nodes for cytokinesis. Early in interphase, type 1 nodes containing Mid1p and kinases Cdr1p and Cdr2p form in a broad band around the nucleus as type 2 nodes containing Blt1p, Gef2p, and Klp8p emerge from the remnants of the constricted contractile ring. During interphase, type 2 nodes travel diffusively along the cell cortex for >1 h until they gradually associate with type 1 nodes to form cytokinesis nodes. Simulations of a simple diffuse and capture model reproduce the behavior of type 2 nodes. Early in mitosis, Mid1p transfers from type 1 nodes to cytokinesis nodes so that it incorporates into the contractile ring with all of the type 2 node markers. At the same time, type 1 nodes move from the equator and disperse their proteins into the cytoplasm during anaphase, a process that depends on active SIN signaling. We propose a node cycle that accounts for the organization of the constituent proteins across the entire cell cycle.
Results
Two types of interphase nodes
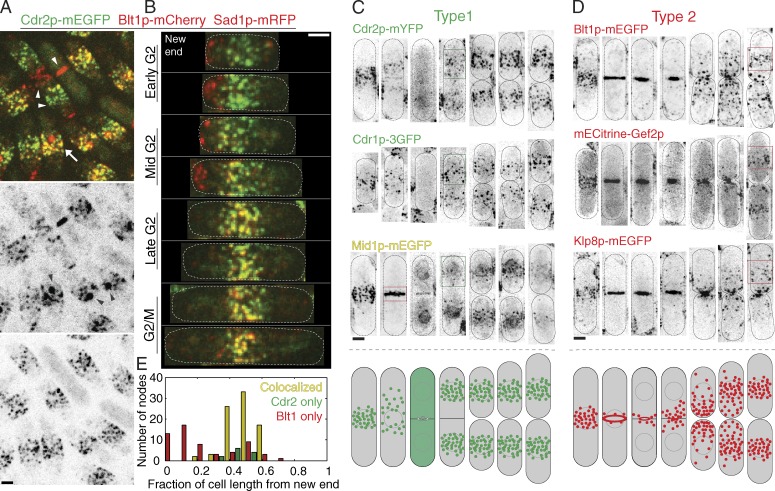
Images of cells expressing both kinase Cdr2p–monomeric EGFP (mEGFP) and node protein Blt1p-mCherry illustrate the presence of these proteins in spatially distinct structures for much of the cell cycle and their colocalization in nodes around the equator during mid- to late interphase and early mitosis (Figs. 1, A and B; and S1). We defined two proteins as being colocalized in a node if the centers of their fluorescent intensities were separated by less than the width of one point spread function (3 pixels). Images of cells expressing other proteins showed that the two types of interphase nodes have different compositions: Cdr1p, Cdr2p, and Mid1p concentrated in type 1 nodes, and Blt1p, Gef2p, and Klp8p concentrated in type 2 nodes (Figs. 1, C and D; and S1).
Figure 1.
Two types of interphase nodes observed by confocal fluorescence microscopy of asynchronous cells expressing fluorescent fusion proteins. (A and B) Localization of kinase Cdr2p-mEGFP, node protein Blt1p-mCherry, and SPB protein Sad1p-mRFP (black arrowheads in the reverse contrast image of the red channel). (A) Maximum intensity projections of five z sections in 400-nm steps including 2 µm (about half the thickness) of the cells. Cdr2p and Blt1p colocalize in nodes near the equator of some cells (arrow) but in separate nodes in other cells (paired white arrowheads). In cells with Blt1p concentrated in the contractile ring (single white arrowhead), Cdr2p is dispersed in the cytoplasm. (B) Cells such as in A were classified into interphase stages by length and other criteria (Materials and methods). (C and D) Localization of proteins in two types of nodes across the cell cycle. Reverse contrast, confocal fluorescence, and maximum intensity projections of 21 z sections taken at 260-nm steps and arranged in montages according to cell length. We adjusted the brightness and contrast of each image to account for differences in illumination intensity and exposure time. (bottom) Drawings of the distributions of the two types of nodes across the cell cycle. (C) Type 1 nodes located in broad bands around the equator (green boxes in one column) contain Cdr2p-mYFP and Cdr1p-3GFP. Mid1p-mEGFP is present in these nodes and transiently in the contractile ring (red box). (D) Type 2 nodes containing Blt1p-mYFP, mECitrine-Gef2p, and Klp8p-mYFP appear at the division site, redistribute to the equator during interphase (red boxes), and incorporate into the contractile ring. (E) Histogram of the distribution of nodes along the length of asynchronous interphase cells such as in A and B. Nodes were defined as colocalized if their intensity centers of mass in the red and green channels were less than one point spread function (3 pixels) apart. n = 188 nodes from 12 cells in two separate experiments. Dotted ovals outline cells. Bars, 2 µm.
The two types of nodes were separate in short cells during early interphase (Fig. 1 B). Before the time that daughter cells separated, type 1 nodes containing Cdr1p and Cdr2p appeared in cortical bands around the nuclei (Fig. 1 C), where they persisted throughout interphase (Morrell et al., 2004; Martin and Berthelot-Grosjean, 2009; Moseley et al., 2009). Markers for type 2 nodes (Blt1p, Gef2p, and Klp8p) were incorporated into the contractile ring during mitosis and reappeared as distinct nodes at the new cell tip as the contractile ring disassembled at the end of cytokinesis (Fig. 1 D). During interphase, type 2 nodes redistributed along the cell cortex (Saha and Pollard, 2012a) to the equator (Moseley et al., 2009; Ye et al., 2012), where up to 75% colocalized with type 1 nodes (Fig. 1 B). Thus, in a mixed population of interphase cells, centrally located nodes have type 1 markers or markers of both types, whereas nodes near the new end have only type 2 markers (Fig. 1 E). The results were similar when we reversed the fluorescent protein tags (Fig. S1 C). The following sections document the behaviors of the two types of nodes across the cell cycle.
Type 1 nodes separate from type 2 nodes during mitosis
Early in mitosis, ∼75% of cytokinesis nodes in the broad equatorial band had markers from both types of interphase nodes superimposed, but at time 0, when SPBs separated, these markers began to separate. This remarkable segregation of the molecules in the cytokinesis nodes took place in two phases.
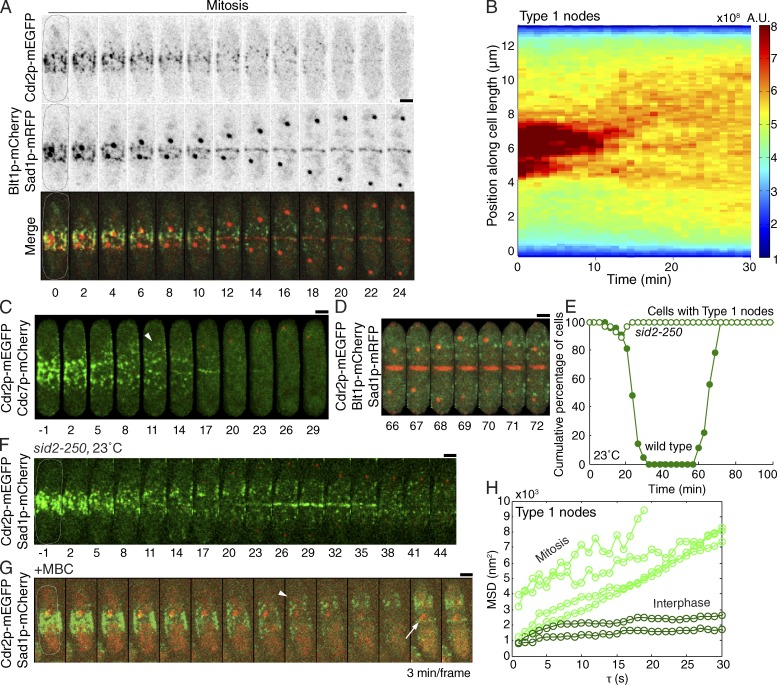
During the first 10–12 min after SPB separation, most type 1 nodes marked by Cdr2p-mEGFP separated from the cytokinesis nodes and started to move away from the division plane, whereas the cytokinesis nodes condensed into a contractile ring (Fig. 2 A and Video 1). These condensing cytokinesis nodes contained all type 2 node markers, Mid1p from type 1 nodes (Fig. 1 C), and the cytokinesis proteins, including myosin II, as observed previously (Moseley et al., 2009; Saha and Pollard, 2012a).
Figure 2.
Localization of type 1 nodes across the cell cycle. Images are time series of maximum intensity projections of confocal fluorescence micrographs with time in minutes from SPB separation except in G. (A) Reverse contrast and merged images early in mitosis of a cell expressing Cdr2p-mEGFP, Blt1p-mCherry, and Sad1p-mRFP. Blt1p incorporated into the contractile ring as type 1 nodes moved from the equator and dispersed Cdr2p into the cytoplasm. Projections of four z sections taken at 400-nm steps. (B) Demonstration of the asymmetric movements of the nodes from the midline by a false-color kymograph of the time course of Cdr2p-mEGFP fluorescence along the length of the cell in Fig. 2 A. At each time point, the fluorescence intensity across the cell width was summed to obtain a fluorescence profile and colored according to intensity per area. (C) Images of a cell expressing Cdr2p-mEGFP and Cdc7p-mCherry showing that Cdr2p nodes moved in greater numbers and to a larger extent (arrowhead) in the direction of the active SPB marked with Cdc7p-mCherry. The same correlation was present in every cell we observed (n > 13). (green images) 10 z sections taken at 360-nm steps comprising half the cell. (red images) 20 z sections taken at 360-nm steps comprising the entire cell. (D) Time series of confocal maximum intensity projection images of a cell expressing Cdr2p-mEGFP, Blt1p-mCherry, and Sad1p-mRFP showing Cdr2p nodes reappearing in a cortical band of type 1 nodes around the daughter nuclei ∼10 min before the cell separated at time 80 min. Same conditions as A. (E) Outcome plots of the time course of the localization of Cdr2p-mEGFP to nodes in wild-type (n = 21; closed circles) and temperature-sensitive sid2-250 cells (n = 28; open circles). Cells were imaged at a permissive temperature (23–25°C). In wild-type cells, Cdr2p disappeared from nodes into the cytoplasm for the 40-min eclipse period. Most sid2-250 cells never dispersed Cdr2p from their nodes; in 18% of cells, Cdr2p disappeared from nodes into the cytoplasm for an abbreviated 1–3-min eclipse period. (F) Time-lapse images of temperature-sensitive sid2-250 cells expressing Cdr2p-mEGFP and Sad1p-mCherry at a permissive temperature (23–25°C) showing incomplete dispersal of type 1 nodes marked with Cdr2p-mEGFP. Projections of 20 z sections taken at 360-nm steps. (G) Movement of type 1 nodes from the equator of cells treated with 131 µM MBC to depolymerize microtubules. Time series of images at 3-min intervals of cells expressing Cdr2p-mEGFP and Sad1p-mCherry. The arrowhead points to Cdr2 nodes after movement from the equator. The arrow points to divided SPBs. Images from a representative experiment repeated twice. (H) Plot of mean squared displacement (MSD) versus time step for a sample of type 1 nodes imaged at 1-s intervals and marked with Cdr2p-mEGFP during interphase (n = 23; dark green) and mitosis (n = 6; light green). Bars, 2 µm.
The type 1 nodes moved asymmetrically. Nodes near the active SPB containing Cdc7p moved first. They also moved in greater numbers and further from the equator than nodes on the other side of the division plane (Fig. 2 C).
Analysis of the trajectories of these type 1 nodes during mitosis showed that their motions were diffusive. For those nodes we could track, plots of mean squared displacement (MSD) versus time step were linear over short (30 s) time scales, consistent with a diffusion of D = 100 ± 50 nm2/s (Fig. 2 H and Video 2; Saxton, 1997). This diffusion coefficient represents a lower bound because about two thirds of type 1 nodes moved and/or disappeared so quickly that we were unable to track them reliably. Bayesian inference analysis (Monnier et al., 2012) of the MSD curves in Fig. 2 H confirmed that the node motions were diffusive with a probability of 0.99, rather than directed or a combination of directed plus diffusive.
During the second phase of segregation, after time 12 min, most type 1 nodes moved from the equator, and the Cdr1p and Cdr2p dispersed into the cytoplasm by time 30 min (Figs. 1 C and 2, A–C and E; Table S1; and Videos 1 and 2). This confirms previous observations made without timing (Morrell et al., 2004; Martin and Berthelot-Grosjean, 2009; Moseley et al., 2009; Saha and Pollard, 2012a). We call this phase without Cdr1p and Cdr2p organized in nodes the eclipse period. The total amount of Cdr2p per cell was constant across the cell cycle (Kanoh and Russell, 1998), so the fluorescence intensity of cytoplasmic Cdr2p-mEGFP increased as the type 1 nodes dispersed (Morrell et al., 2004). Rather than dispersing, ∼15% of the Cdr2p fluorescence from type 1 nodes incorporated into the contractile ring by time 1 min (Fig. 2, A and C) as observed previously (Morrell et al., 2004). Starting around time 17 min, Mid1p left the contractile ring in strands moving toward the cell tips (Fig. S2 C), which then dispersed into the cytoplasm and the nucleus (Fig. 1 C; Bähler et al., 1998a; Wu et al., 2003).
Dispersal of Cdr2p from type 1 nodes depended on active Sid2 kinase (Figs. 2, E and F; and S2 E; and Video 7). At the permissive temperature (23°C), nodes marked with Cdr2p moved normally from the equator in temperature-sensitive sid2-250 cells (Table S1), but 82% of these cells never completely dispersed Cdr2p from type 1 nodes into the cytoplasm (Fig. 2, E and F). Rather, nodes marked by Cdr2p-mEGFP persisted and moved intact away from the middle of the cell to reside in a wide band around the daughter nuclei (Fig. 2 F). In those sid2-250 cells that dispersed their Cdr2p nodes, the broad band of nodes returned quickly after a short eclipse period of only 4 ± 2 min (Fig. 2 E and Table S1). Similarly, at the semipermissive temperature of 31°C only 21% of sid2-250 cells completely dispersed their type 1 nodes, which reappeared just 3 ± 1 min later (Fig. S2, E and F).
Treatment of cells with methyl benzimidazol-2-yl-carbamate (MBC) to prevent polymerization of microtubules delayed the separation of SPBs, but type 1 nodes marked with Cdr2p-mEGFP moved normally from the equator and dispersed into the cytoplasm before SPB separation (Fig. 2 G). Thus, movement of Cdr2 nodes from the equator does not depend on microtubules, elongation of the spindle, or separation of the daughter nuclei.
Type 1 and 2 nodes are spatially distinct structures during early interphase
During the maturation and constriction of the contractile ring, no discrete nodes were visible, but both types of nodes reappeared between times 40 and 65 min. During the eclipse period, the type 1 node markers Cdr2p-mEGFP and Mid1p-mEGFP were dispersed in the cytoplasm and nucleus (Fig. 2 E and Table S1). Between anaphase B and cell separation, some nuclei transiently had a higher concentration of Cdr2p than the cytoplasm (Fig. S2 D), but Mid1p concentrated more highly and more consistently in the nucleus (Bähler et al., 1998a; Wu et al., 2003) than Cdr2p. Type 2 nodes reappeared first. They emerged as distinct structures from remnants of the contractile ring during the eclipse period at time 42 ± 6 min (Figs. 3 A and S3 A; Table S1; and Video 4). By the time the contractile ring completed constriction at time 50 ± 4 min, all of its other constituent proteins were dispersed into the cytoplasm (Wu et al., 2003).
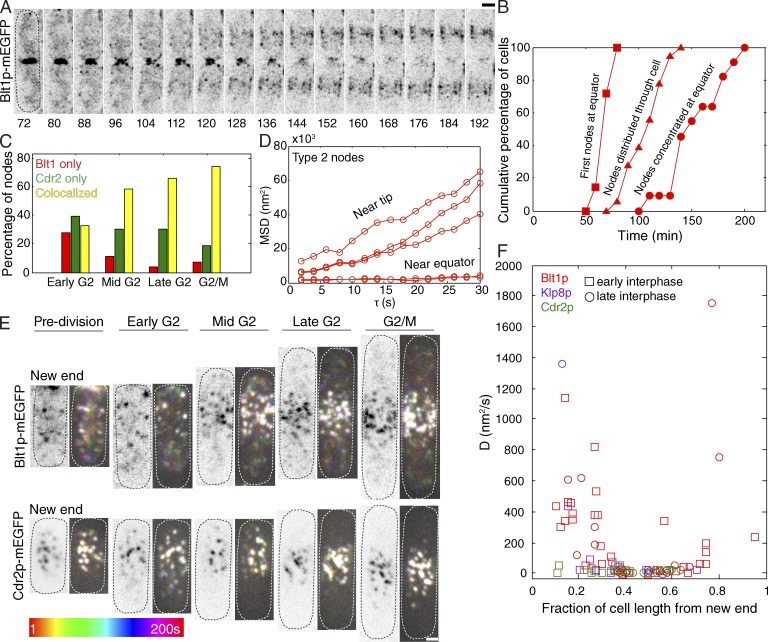
Figure 3.
Redistribution of type 2 nodes in cells during the cell cycle. (A) This time series of maximum intensity projections of reverse contrast fluorescence micrographs shows type 2 nodes marked with Blt1p-mEGFP emerging from the disassembling contractile ring and moving to the equator over 2 h. (B) Outcome plots of the time course of the localization of type 2 interphase nodes with time 0 at SPB separation, calculated as time from cell separation −80 min. Symbols: fraction of cells with ≥1 Blt1p node at new cell equator (closed squares); fraction of cells with nodes distributed along the cortex (defined as the first time point when the mean fluorescence at the old division site is less than or equal to the mean fluorescence at any other point along the length; closed triangles); and fraction of cells with Blt1p nodes maximally relocalized to the new cell equator (closed circles). (C) Histogram of the proportion of interphase nodes containing Blt1p only, Cdr2p only, or colocalized Blt1p and Cdr2p binned by stage of the cell cycle; same data as Fig. 1 E. (D) Plots of mean squared displacement (MSD) versus time step for five type 2 (Blt1p-mEGFP) nodes from a single interphase cell imaged at 2-s intervals. (E) Temporal color projections of node movements at 2-s intervals to compare the behavior of type 2 nodes marked with Blt1p-mEGFP (top) and type 1 nodes marked with Cdr2p-mEGFP at different stages of interphase (bottom). Each pair of micrographs has a reversed contrast sum fluorescence micrograph (three 300-nm z slices) on the left. Streaks of color show movements according to the time scale. White spots are stationary. The leftmost images are of one of two daughter cells still connected by a septum. Dotted ovals outline cells. (F) Plot of node diffusion coefficients in interphase cells as a function of the normalized distance from the new end of the cell showing that type 2 nodes diffuse faster near the cell tips than near the equator. Early interphase: cells still connected by septum or ≤8.5 µm in length; late interphase: cells >8.5 µm long. Type 2 Blt1p-mEGFP nodes (n = 49); type 2 Klp8p-mEGFP nodes (n = 23); type 1 Cdr2p-mEGFP nodes (n = 23). Bars, 2 µm.
Around time 65 min (∼15 min before cell separation), the eclipse period ended with the reappearance of type 1 nodes in cortical bands around the daughter nuclei (Figs. 1, A–C; and 2 D), first in one daughter cell and 1–4 min later in the other cell. The band of type 1 nodes marked with Cdr2p was centered around one third of the distance to the old pole in early interphase before new end takeoff (Figs. 1, B and C; and 3 E). These interphase type 1 nodes diffused slowly (Fig. 2 H), with a mean two-dimensional diffusion coefficient of 16 ± 12 nm2/s, similar to cytokinesis nodes before time 0 (Vavylonis et al., 2008) and <20% that of the old type 1 nodes that disappeared during anaphase (Fig. 2 H).
Reunion of type 1 and 2 nodes around the equator
Starting about time 55 min, interphase type 2 nodes containing Blt1p, Klp8p, and Gef2p gradually redistributed along the cell cortex from the new pole of the daughter cell to rejoin relatively static type 1 nodes located in a broad band around the equator (Figs. 1, B, D, and E; and 3 A; and Video 3). Treating cells with MBC to depolymerize microtubules or Latrunculin A to depolymerize actin filaments did not change the interphase diffusion of type 2 nodes (Fig. S3 C). Type 2 nodes diffused normally during interphase in cells arrested in G1 phase (cdc10-129 mutants) and arrested in G2 phase (cdc25-22 mutants) and sid2-250 cells at permissive temperatures (Figs. S2 G and S3 C and Video 9). The first type 2 nodes reached the new equator at time 67 ± 3 min (Fig. 3, A and B; and Table S1) and were scattered between the two poles around time 100 min (Fig. 3 B).
These redistributing type 2 nodes diffused quickly as discrete entities along the cell cortex with a wide range of diffusion coefficients (Fig. 3, D and F; and Video 3). Linear plots of MSD versus time step (Fig. 3 D) showed that interphase type 2 nodes diffused much faster (mean D = 250 ± 220 nm2/s) than type 1 nodes at this time in the cell cycle. Bayesian inference analysis (Monnier et al., 2012) of the MSD curves in Fig. 3 D confirmed that the node motions were diffusive (probability = 0.97) rather than directed or a combination of directed plus diffusive. Type 2 nodes diffused most rapidly in regions near the poles lacking type 1 nodes (Fig. 3 F and Video 3). Nodes with low levels of fluorescence tended to diffuse faster than brightly fluorescent nodes, which may be clusters of nodes (Fig. S3 I). Many rapidly diffusing nodes appeared and disappeared during 200 s of imaging, probably because of moving in and out of the imaging plane (Video 3). Type 2 nodes generally appeared to move in the cell cortex (Fig. S3 J), but we could not exclude rapid diffusion of some nodes in the cytoplasm.
Between cell separation at times 80 and 160 min, diffusing type 2 nodes gradually became immobilized at the new cell equator (Figs. 3, B–F; and S3 H; and Video 3) where their fluorescence overlapped with the fluorescence of type 1 nodes (Figs. 1 B and 3 C). Type 2 nodes accumulated at the new cell equator in cells arrested in G1 phase (cdc10-129 mutants), cells arrested in G2 phase (cdc25-22 mutants), and sid2-250 cells at permissive or restrictive temperatures (Figs. S2 G and S3 C and Video 9). Colocalization of the two types of nodes increased from 30% early in G2 to ∼75% by G2/M (Fig. 3 C). Once colocalized with type 1 nodes, type 2 nodes diffused slowly (D = 18 ± 12 nm2/s; Figs. 3, D–F; and S3 B).
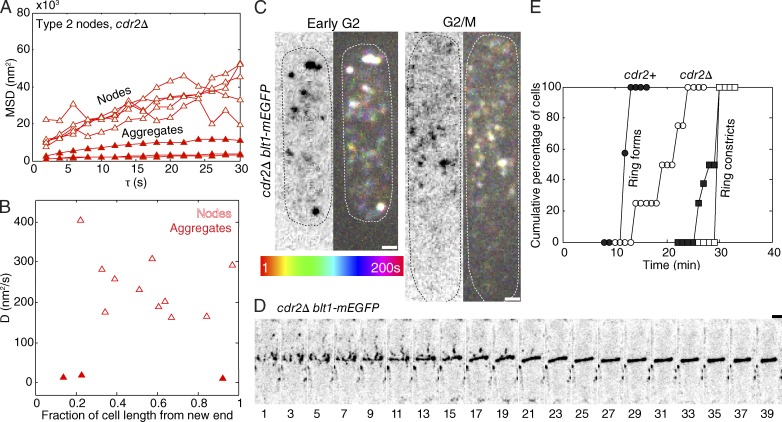
To test the hypothesis that equatorial type 1 nodes anchor and immobilize type 2 nodes, we imaged type 2 nodes in cdr2Δ cells, which lack type 1 nodes (Fig. 4 and Video 5; Almonacid et al., 2009; Martin and Berthelot-Grosjean, 2009; Moseley et al., 2009). Without Cdr2p, most type 2 nodes continued to diffuse rapidly (D = ∼200 ± 50 nm2/s; Fig. 4, A–C) well past the time that they normally became immobilized with the type 1 nodes in the middle of wild-type cells (Figs. 3 A and S4, A and B; and Video 3). Over time, in cdr2Δ cells, some type 2 nodes formed highly fluorescent, slowly diffusing (D = 6 ± 4 nm2/s) clusters (Fig. 4, A–C). By late interphase, about one third of type 2 nodes (judged by fluorescence) were aggregated in clusters around the equator of cdr2Δ cells. These clusters of type 2 nodes in cdr2Δ cells were less uniform in distribution and intensity (Figs. 4 C; and S4, A–C; Moseley et al., 2009) than cytokinesis nodes in wild-type cells (Figs. 1 B, 3 A, and S4 C and Video 5).
Figure 4.
Behavior of type 2 nodes marked with Blt1p-mEGFP in cdr2Δ cells lacking type 1 nodes. (A) MSDs versus time step of Blt1p-mEGFP nodes in cdr2Δ cells. Symbols: five type 2 nodes with intensity <12,000 A.U. (open triangles); three Blt1p-mEGFP particles with fluorescence intensity >12,000 A.U. (closed triangles), likely aggregates of nodes. (B) Plot of diffusion coefficients of Blt1p-mEGFP nodes in cdr2Δ cells during early interphase as a function of their fractional distance from the new end of the cell. Symbols: nodes with intensity <12,000 A.U. (open triangles); particles with intensity ≥12,000 A.U. (closed triangles). (C) Pairs of micrographs with a negative contrast sum intensity projection image of three 300-nm z slices (left) and a temporal color projection image at 2-s intervals for 200 s in early and late interphase cells expressing Blt1p-mEGFP in a cdr2Δ strain (right). Many more equatorial nodes were mobile in early G2 than G2/M (also see Fig. S4). Dotted ovals outline cells. (D) Time series of reverse contrast maximum intensity projections of seven z sections taken at 600-nm steps showing the assembly of a contractile ring in a cdr2Δ cell expressing Blt1p-mEGFP. See Fig. S4 for higher time resolution. (E) Outcome plots of the time course of contractile ring assembly and constriction in cells expressing type 2 node protein Blt1p-mEGFP ±Cdr2p. Symbols: wild-type cdr2+ cells (closed circles and squares); cdr2Δ cells (open circles and squares); fraction of cells with a compact contractile ring (circles); and fraction of cells with a constricting contractile ring (squares). Time is in minutes from SPB separation. Bars, 2 µm.
Cells lacking Cdr2p assembled a contractile ring in 20 ± 4 min, twice the time of wild-type cells (Fig. 4 E), from a combination of aggregated nodes and strands of nodes, which appeared by time 9 min (Figs. 4 D and S4 D and Video 6). These rings were often tilted with respect to the long axis of the cell (Figs. 4 D and S4 A), but most gradually straightened over 20 min (Fig. 4 D) and then constricted around time 30 min as in wild-type cells (Fig. 4, D and E).
Mathematical model of diffuse and capture
Numerical simulations of a mathematical model showed that a diffuse-and-capture mechanism is sufficient to explain the gradual redistribution of type 2 nodes from the pole to new cell equator (Fig. 5 and Video 8). In this computational model, we represented the cell cortex as a two-dimensional rectangular surface 7 µm long in x and 11 µm wide in y, corresponding to the length and circumference of the cell. We assumed that 65 type 2 nodes started at x = 0 (corresponding to the new cell tip), whereas type 1 nodes were spread in a Gaussian distribution around x = 3 µm (as observed, Fig. 1 B) with an SD of 800 nm (Fig. 5 B). In numerical, stochastic simulations, type 2 nodes diffused along the surface at each time step t according to their diffusion coefficient D. Stationary type 1 nodes “captured” diffusing type 2 nodes that came within a capture radius Rc at any time step with a probability of binding pbind and according to a defined stoichiometry (generally 1:1 in these simulations; Fig. 5 A).
Figure 5.
Simulations of a mathematical model of a diffuse-and-capture mechanism to redistribute type 2 nodes from the old to the new division site. (A) Model of diffusing type 2 nodes being captured irreversibly (yellow) by stationary type 1 nodes (green) with a probability of binding pbind if they reside within a defined capture radius Rc at any time point. (B) Drawing of the observations of type 2 nodes (red) redistributing from their origin at the new end of the cell (x = 0) until they colocalize with type 1 nodes in a broad band near the center of the cell. Y is the circumference of the cell. (C) Graphical output of a diffuse-and-capture simulation at the time points indicated. In this simulation, 65 type 2 nodes (red) started at x = 0 and diffused with D = 400 nm2/s until they were captured (yellow) by passing within 60 nm of one of the 65 type 1 nodes (green) located in a Gaussian distribution across a band centered 3 ± 0.8 µm from the new cell tip. The probability of capture was 50%. The simulation used steps of 2 s. Bar, 1 µm. Time expressed in minutes from the beginning of simulation, which corresponds to t = ∼40 min from SPB separation. Also see Fig. S5 J, in which some of the data are displayed for visual comparison with Fig. 1 B. Bar, 2 µm. (D) 10 simulations of the time course of binding of type 2 nodes to type 1 nodes with the parameters used in C. The gray region is the mean ± 1 SD of the time when type 2 nodes appeared maximally bound in live cells. (E) Dependence of simulated time courses of type 2 nodes binding to type 1 nodes on the diffusion coefficient of type 2 nodes. The horizontal dashed line indicates when 50% (bottom line) and 80% (top line) of the type 2 nodes were bound. Vertical dashed line is the mean time that type 2 nodes were maximally bound in live cells. The gray region is the mean ± 1 SD. (F) Number of captured type 2 nodes out of 65 at the end of the simulation as a function of diffusion coefficient D (n = 100). (G) Mean positions in simulations of captured type 2 nodes relative to the center of the band of type 1 nodes at 0, which is 3 µm from the new cell tip (n = 100). Error bars are ± 1 SD.
Stochastic simulations of this model reproduced the gradual equatorial accumulation and immobilization of type 2 nodes as they became associated with stationary type 1 nodes (Fig. 5 C). Measurements on cells provided estimates of some parameters, such as D and the numbers of the two types of nodes. We tested a range of physically reasonable values for Rc and pbind. For the simulation in Fig. 5 C, we used D = 400 nm2/s, Rc = 60 nm, and pbind = 0.5. With these parameter values, type 2 nodes diffused from their starting positions and slowly accumulated at the sites of type 1 nodes such that about half were immobilized by 135 min (Fig. 5, C–F), similar to experimental observations (Fig. 3 B). However, 135-min simulations with these parameter values differed in two ways from cells. First, many unbound type 2 nodes were still located between the new cell tip and the band of type 1 nodes (Fig. 5 C). Second, the simulated distributions of bound type 2 nodes were more biased toward the type 1 nodes closest to the new cell tip (Fig. 5, C and G) than in cells (Fig. 1 B).
We varied D, Rc, and pbind to determine how the value of each parameter affects the simulations. We tested a range of diffusion coefficients for type 2 nodes (Fig. 5 E) beyond the highest value we measured in cells (1,800 nm2/s in cells; Fig. 3 E). The rate of type 2 node binding to type 1 nodes increased with D, such that 135 min of simulation immobilized 50% of type 2 nodes with D = 400 nm2/s and >75% of nodes with D ≥ 1,000 nm2/s (Fig. 5, E and F). This broadly agreed with the proportion of type 1 nodes coincident with a type 2 node in mid-G2 cells (∼65%) and G2/M cells (∼75%). With higher D, type 2 nodes were distributed more uniformly across the band of type 1 nodes at the end of the simulation (Fig. 5 G). Assuming that type 2 nodes can bind more than one type 1 node reduced the number of type 1 nodes with a bound type 2 node and shifted the distribution of bound type 2 nodes toward their starting position at the new cell tip (Fig. S5, D and E). Assuming >65 type 2 nodes resulted in more type 1 nodes with a bound type 2 node, with a more uniform distribution along the x axis (Fig. S5, H and I). No nodes were captured with Rc = 0, and the total number of bound nodes was very sensitive to Rc up to ∼30 nm (Fig. S5 F), but the whole range of Rc values tested influenced the final positions of the captured type 2 nodes (Fig. S5 G). Varying the probability of binding from 0.1 to 1.0 had little effect on the outcome of the simulations (Fig. S5, A–C). Thus, the diffuse-and-capture mechanism is relatively robust, and the parameter values have more influence on the final positions of the captured type 2 nodes than other features of the process.
Discussion
Our new observations allow us to trace the history of the interphase node proteins around the full cell cycle and to provide a unifying model for the node cycle (Fig. 6). Previous investigators (Paoletti and Chang, 2000; Morrell et al., 2004; Almonacid et al., 2009; Moseley et al., 2009; Laporte et al., 2011; Saha and Pollard, 2012a; Ye et al., 2012) observed many aspects of interphase node behavior in isolation, but no one recognized the existence of two types of interphase nodes or assembled the information into a complete node cycle. Paoletti and Chang (2000) discovered interphase nodes in confocal micrographs of cells expressing Mid1p-GFP. They correctly described the node cycle: “1) a central broad band through much of G2 phase and during very early mitosis; 2) a tight ring later in mitosis; and 3) no specific cell surface localization in G1 and S phases, between septum formation and cell separation.” Morrell et al. (2004) also described three different distributions of kinase Cdr2p, which they found weakly associated with the contractile ring early in mitosis, diffuse in the cytoplasm later in mitosis, and concentrated in nodes around the equator during interphase. Moseley et al. (2009) expanded the inventory of interphase node proteins to include Cdr1p, Blt1p, Gef2p, and Klp8p. Their Fig. S3 B showed how nodes marked by Blt1p redistribute from the new cell tip to the equatorial band of interphase nodes to the contractile ring. They also discovered that targeting interphase nodes marked by Blt1p to the equator depends on the presence of Cdr2p. Other work (Almonacid et al., 2009; Martin and Berthelot-Grosjean, 2009; Moseley et al., 2009; Ye et al., 2012; Guzman-Vendrell et al., 2013; Jourdain et al., 2013; Zhu et al., 2013) established genetic interactions among the interphase node proteins and mapped pairwise direct or indirect interactions between these proteins in cell extracts. We confirmed the findings of these pioneering studies, and our new observations provide the physical and temporal context to interpret that work and propose specific physical pathways.
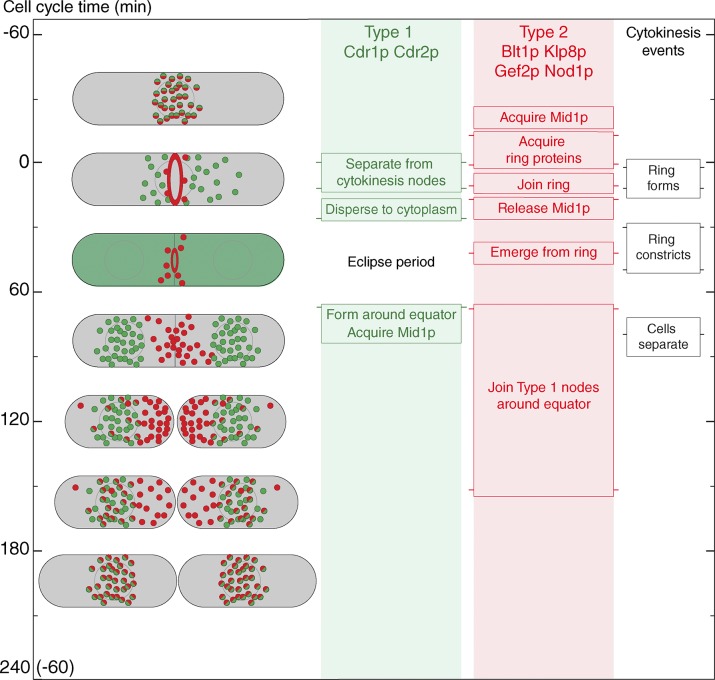
Figure 6.
Summary of the node cycle with cell cycle time on the y axis. Column 1 shows cartoons of cells with type 1 nodes in green, type 2 nodes in red, and colocalized type 1 and 2 nodes in red/green. The green cell shows Cdr1p and Cdr2p dispersed in the cytoplasm during the ∼40-min eclipse period (Fig. 2 E and Table S1). Columns 2, 3, and 4 indicate the times of events. The light green and red background shading shows when node markers are organized in structures. Horizontal lines on the side of each box mark the mean time of each event. Pairs of horizontal lines represent the beginning and end of events spread over time. The time when Mid1p is handed off from type 1 to type 2 nodes is not known.
We show that cytokinesis nodes, the structural precursors of the cytokinetic contractile ring, assemble from two distinct types of nodes that arise separately and merge during interphase. Type 1 nodes originate from cytoplasmic pools of Cdr1p and Cdr2p at the future division site where they serve as receptors for the second type of node that is released from the contractile ring and diffuses in the cortex, consistent with simulations of a mathematical model. These two types of nodes represent parallel branches of the pathway that prepares nodes for cytokinesis. The following sections explain what we know about the main transitions in this cycle of nodes. We also point out the key missing information and suggest the experiments that will be most revealing about how the node cycle works.
Type 1 nodes form in a broad cortical band around the nucleus early in interphase
Even before daughter cells separate, type 1 nodes assemble in the cortex around the nucleus as Cdr2p and Cdr1p transition from a diffuse cytoplasmic pool into discrete nodes. This ends the “eclipse period” of ∼40 min, the only time in the cycle when Cdr1p and Cdr2p are not organized into nodes.
Kinase Cdr2p is required to form type 1 nodes (Almonacid et al., 2009; Martin and Berthelot-Grosjean, 2009; Moseley et al., 2009) and appears to organize the other proteins through direct interactions with Cdr1p and Mid1p (Martin and Berthelot-Grosjean, 2009; Moseley et al., 2009; Guzman-Vendrell et al., 2013). Shortly after appearing, type 1 nodes begin to recruit anillin Mid1p from the cytoplasm and nucleus. At this stage in the cell cycle, Mid1p presumably interacts directly with Cdr2p (Moseley et al., 2009). The localization of kinase Wee1p (with GFP tags) to interphase nodes depends on the presence of Cdr2p but not Cdr1p or Blt1p (Moseley et al., 2009), but the timing of its association with nodes is less clear. Signals from the poles provided by Pom1p confine type 1 nodes to the middle of the cell, although for an unknown reason, type 1 nodes are still restricted from one tip of the cell in the absence of Pom1p (Martin and Berthelot-Grosjean, 2009; Moseley et al., 2009). Type 1 nodes reappear around the divided daughter nuclei at time 65 min, approximately when the SIN declines as indicated by the loss of Cdc7p-GFP fluorescence from the SPB (García-Cortés and McCollum, 2009). The precise mechanisms controlling assembly and disassembly of type 1 nodes remain to be discovered along with understanding of how the full complement of type 1 node proteins is organized and how their assembly in nodes relates to their regulation of Cdk1p.
During mitosis, type 1 nodes move in the cortex with a diffusion coefficient of ∼100 nm2/s, so something must restrict their lateral diffusion during interphase when D < 20 nm2/s. No known type 1 node protein has characteristics that make it a candidate to restrict diffusion, so it is likely that the nodes interact with one or more proteins associated with the plasma membrane. Previous work (Vavylonis et al., 2008) showed that the low diffusion coefficient of cytokinesis nodes does not depend on actin filaments. Characterizing this anchoring mechanism should be a future goal.
Type 2 nodes emerge from disassembling contractile ring at the end of cytokinesis
Type 2 nodes containing Blt1p, Klp8p, and Gef2p as well as Nod1p (Jourdain et al., 2013) are the only structural remnant known to survive disassembly of the contractile ring. These nodes remain as discrete structures (Figs. 3 and S3; and Fig. S3 A in Moseley et al., 2009) as all other contractile ring proteins disperse into the cytoplasm (Wu and Pollard, 2005). Their reemergence from rings suggests that type 2 nodes are present continuously in contractile rings where they are packed too tightly to be resolved by confocal microscopy. Observations of cofilin mutants support this concept of the continuous presence of type 2 nodes through cytokinesis; mature contractile rings fragment into nodelike structures containing Myo2 when adf1-1 cells are shifted to the restrictive temperature (Chen and Pollard, 2011). Loss of type 2 nodes in blt1Δ cells did not affect the growth of the new end as judged by cells stained with calcofluor (Fig. S4 E). Type 2 nodes did not colocalize with polarity “nodes” containing Tea1p, Tea3p, Pom1p, or Bud6p (Fig. S1, F–I).
Several lines of evidence suggest that Blt1p is a scaffold for type 2 nodes. Blt1p is a tetramer (unpublished data; Goss, J., personal communication) that interacts with Gef2p, Klp8p, Cdc15p (Moseley et al., 2009), and Nod1p (Jourdain et al., 2013; Zhu et al., 2013) in immunoprecipitation assays. Furthermore, neither Gef2p nor Nod1p concentrates in interphase nodes in cells lacking Blt1p (Moseley et al., 2009; Jourdain et al., 2013). We expect that type 2 nodes contain other proteins that have yet to be characterized.
Type 2 nodes diffuse in the cortex, presumably while tethered to the plasma membrane. Interactions of the C-terminal half of Blt1p with phospholipids may help tether type 2 nodes to the membrane (Guzman-Vendrell et al., 2013). Our Bayesian analysis of node trajectories is consistent with free diffusion and argues against confined diffusion or directed motion, so this tether is presumably moving in the lipid bilayer. We note, however, that simple transmembrane proteins diffuse faster by orders of magnitude (Kusumi et al., 2005), so the large size of type 2 nodes or other factors must limit their diffusion. Interactions with Mid1p may complement this interaction with the membrane after cytokinesis nodes form (Guzman-Vendrell et al., 2013), but the tethering mechanism deserves better characterization.
Cytokinesis nodes form by a diffuse-and-capture mechanism
Type 2 nodes diffuse along the inside of the plasma membrane until they associate with static type 1 nodes located around the equator of the cell. The fact that mobile type 2 nodes assume the low diffusion coefficient of type 1 nodes once their fluorescence signals are coincident is evidence that the two types of nodes are physically associated.
Previous work identified two candidates to mediate interactions between the nodes: (1) Blt1p from type 2 nodes coimmunoprecipitates with Cdr2p from type 1 nodes (Moseley et al., 2009). (2) Mid1p from type 1 nodes and Gef2p from type 2 nodes coimmunoprecipitate, and Gef2p couples Mid1p indirectly to Blt1p (Guzman-Vendrell et al., 2013). However, cytokinesis nodes form in cells without Blt1p (Guzman-Vendrell et al., 2013), so we expect that the actual interactions between nodes are more complicated than a simple link between single copies of proteins from the two nodes. More information about the structures of the nodes will help us understand how they interact and mature into cytokinesis nodes capable of adding contractile ring proteins.
Type 2 nodes travel from the new end of the daughter cell to the equator with a wide range of diffusion coefficients (D = 50–1,800 nm2/s). This variation is related in part to the size of the diffusing particle. Slowly moving fluorescent particles with many copies of a node marker protein are probably aggregates of smaller units that diffuse faster on their own (Fig. S3 I). Local differences in the lipid composition of the plasma membrane (Wachtler et al., 2003) might also contribute to the wide range of diffusion coefficients of these nodes.
Our observations and simulations show that a simple diffuse-and-capture mechanism can explain the redistribution of type 2 nodes to the equator in the time observed (Fig. 5 E). Because type 1 nodes are required to capture the type 2 nodes (Fig. 4), the model simply posits that type 2 nodes diffuse randomly along the plasma membrane from their origins until they bind to a static type 1 node. The range of diffusion coefficients measured in cells quantitatively recapitulates the fraction of type 2 binding to type 1 nodes. The stochastic binding underlying this model accounts for a wide range of times required for type 2 nodes to concentrate at the equator (152 ± 29 or ∼110 min after type 2 nodes appeared; Table S1). The model is also consistent with the observation that ∼30% of mECitrine-Gef2p fluorescence in interphase nodes recovers slowly from photobleaching (t1/2 of ∼2 min; Ye et al., 2012), which we interpret as a mixed population of immobilized and diffusing nodes, although some Gef2p may exchange between nodes and the cytoplasm.
Our simulations reproduced a subtle feature of the distribution of nodes with type 1 and type 2 proteins in cells (Fig. 1 B; Fig. 1 in Moseley et al., 2009). Nodes closer to the new tip are more likely to contain both Blt1p and Cdr2p than nodes further from the new tip. Our simulations show that this gradient results from type 1 nodes near the new pole capturing diffusing type 2 nodes before they diffuse across the midline. The fact that cells without Cdr2p or type 1 nodes form contractile rings from type 2 nodes positioned imperfectly near the middle of the cell (Figs. 4 D and S4 D; Almonacid et al., 2009; Moseley et al., 2009; Ye et al., 2012; Jourdain et al., 2013) points to the existence of secondary mechanisms to concentrate type 2 nodes around the equator at the G2/M transition and for these type 2 nodes to acquire Mid1p from a source other than type 1 nodes.
Cytokinesis nodes acquire additional proteins and condense into the contractile ring
Previous work showed that cytokinesis nodes with Mid1p and markers from type 2 nodes accumulate contractile ring precursors including Myo2, IQGAP Rng2p, F-BAR Cdc15p, and formin Cdc12p (Wu et al., 2003; Moseley et al., 2009; Saha and Pollard, 2012a) and then condense into a contractile ring (Vavylonis et al., 2008). Mid1p comes to cytokinesis nodes with type 1 nodes, but the time that it transfers from type 1 nodes to cytokinesis nodes is not known. At the latest, Mid1p must dissociate from Cdr2p and establish new connections to other node proteins before the Cdr2p nodes separate from cytokinesis nodes during mitosis. Gef2p, Rng2p, and Cdc15p are all candidates to anchor Mid1p in cytokinesis nodes (Laporte et al., 2011; Saha and Pollard, 2012a). Phosphorylation of Mid1p by Plo1p (Almonacid et al., 2011) may promote this transfer of Mid1p from type 1 nodes to type 2 nodes.
The transfer of Mid1p might be one factor that activates cytokinesis nodes to bind Myo2, Rng2p, and Cdc15p. Perhaps the stepwise increase in the numbers of Mid1p molecules per cytokinesis node (from ∼17 to ∼22 to ∼28; Laporte et al., 2011) corresponds to the transfer of Mid1p octamers (Saha and Pollard, 2012b) to type 2 nodes, a process that occurs before myosin II accumulates and depends on Rng2p and Cdc4p.
Disassembly of type 1 nodes during mitosis depends on SIN activity
The most complicated transition in the node cycle happens as cells enter anaphase: nodes containing most of the Cdr1p and Cdr2p are released from cytokinesis nodes and from the membrane anchors that immobilized them up to this point in the cell cycle. Once released, type 1 nodes diffuse more rapidly in the cortex as they shed their proteins into the cytoplasm during the 40-min eclipse period in early interphase. Morrell et al. (2004) discovered the eclipse period but did not report the diffusive movements of type 1 nodes away from the forming contractile ring.
Three observations suggest that type 1 nodes disassemble when SIN activity is high. First, type 1 nodes are stable in SIN mutant strains even though they separate from cytokinesis nodes. Second, type 1 nodes disperse when the SIN turns on early in mitosis and reappear around the divided daughter nuclei when SIN activity drops (García-Cortés and McCollum, 2009). Third, diffusing type 1 nodes separate asymmetrically from the division plane toward the active SPB (Fig. 2 C). Earlier in the cell cycle, Sid2p promotes entry into mitosis by phosphorylating Fin1p (Grallert et al., 2012), a reaction that might subsequently influence the behavior of type 1 nodes during anaphase. The genetic interactions of gef2Δ and nod1Δ with the SIN pathway genes suggest another potential mechanistic connection (Ye et al., 2012; Zhu et al., 2013).
A third type of node
Both type 1 and 2 nodes differ from Skb1p nodes, which are proposed to regulate signaling by type 1 nodes (Deng and Moseley, 2013). Each of the three types of interphase nodes has a distinct protein composition and localization pattern. The existence of multiple types of interphase nodes suggests that they might have additional functions during interphase, outside of their roles in controlling the timing of mitosis and the assembly of the contractile ring.
Prospects
Now that we understand the general features of the node cycle (Fig. 6), many new questions arise about how cells regulate the transitions along the two pathways. We note that the node cycle shares some architectural features with the stepwise assembly of bacteriophages from subassemblies on parallel pathways (Wood et al., 1968). Like bacteriophages, new properties, such as binding sites, appear to emerge when subassemblies (nodes in this case) come together. If this analogy holds, we expect that more information about the structures of nodes will be essential to understand their functions.
Materials and methods
Strain construction
We constructed strains of S. pombe with genetically encoded fluorescent fusion proteins using strains JW728 and JW729. We used plasmids PFA6a-mEGFP-kanMX6, PFA6a-3GFP, PFA6a-mYFP-kanMX6, and PFA6a-mCherry-natMX6 to insert genes for fluorescent proteins mEGFP, mYFP, mCherry, or 3GFP directly upstream or downstream of the open reading frame in the endogenous chromosomal locus using a PCR-based gene-targeting method (Bähler et al., 1998b). We constructed all other strains by genetic crosses to laboratory stock strains or strains provided by the following labs: F. Chang (Columbia University, New York, NY), J. Moseley (Dartmouth College, Hanover, NH), and J.-Q. Wu (The Ohio State University, Columbus, OH; Table S2). We tested strains depending on fluorescent fusion proteins for functionality by measuring the timing of cell cycle events: SPB separation, contractile ring constriction, septation, and cell separation. We also used cell length as a metric for functionality, as deleting or overproducing Cdr1p, Cdr2p, or Blt1p alters cell length (Breeding et al., 1998; Martin and Berthelot-Grosjean, 2009; Moseley et al., 2009). Compared with wild-type cells, cdr1-3GFP cells were shorter at the time of division (Martin and Berthelot-Grosjean, 2009; Moseley et al., 2009), indicating that this tag has mild effects on the functions of this protein.
Microscopy
We observed cells on gelatin pads in growth chambers of EMM5S media at 25°C on an inverted microscope (IX71; Olympus) with a 100×, 1.4 NA Plan Apochromat objective (Olympus), argon ion lasers (Melles Griot), a spinning-disk confocal head (CSU-X1; Yokogawa Corporation of America), and an electron-multiplying charge-coupled device camera (iXon 897; Andor Technology) using iQ2 acquisition software (Andor Technology). We imaged cells at 31 or 36°C on agar rather than gelatin pads. For time-lapse images, we took z series of 13–20 confocal slices at 300–400-nm intervals encompassing the entire cell or three to seven slices closest to the coverslip.
Data analysis
Image correction.
We corrected for the camera offset and uneven illumination with an ImageJ macro (National Institutes of Health; McCormick et al., 2013) that automates the image correction process described by Wu et al. (2008).
Criteria to identify new ends.
We identified the new end originating from the previous cell division based on three criteria: (1) the end with more Blt1p nodes (originating from the previous cell division; Fig. 1 D); (2) the end nearer the division scar, which marks previous sites of septation (Mitchison and Nurse, 1985); and (3) the end nearer the broad band of Cdr2 nodes that grows slower before “new end takeoff.” These criteria always agreed on the designation of the new end.
Criterion for colocalization in nodes.
We manually identified fluorescent spots in each fluorescence channel with a region of interest and used a custom ImageJ plugin (McCormick et al., 2013) to automatically recenter each region of interest based on the spot’s intensity-based center of mass. We defined colocalization between two node proteins when their centers of mass were separated by less than the width of one point spread function (3 pixels).
Analysis of time-lapse images.
An ImageJ plugin created montages of cells from defined regions and measured the fluorescence intensity profiles over time. The profiles are sums of the fluorescence intensity across the width of the cell for each pixel along the cell length. We defined SPB separation as time 0 of the cellular clock (Wu et al., 2003). When information on SPB separation was not available, we used other cellular events for timing: the onset of anaphase B at t = 12 min, maximum SPB separation at t = 26 min, or cell separation at t = 80 min (Table S1). We used cell length as a proxy for cell cycle time (Baumgärtner and Tolić-Nørrelykke, 2009) when we lacked time-lapse data. For presentation purposes, we subdivided the 4–5 h of interphase (Baumgärtner and Tolić-Nørrelykke, 2009) into five stages based on cell morphology or length: predivision cells were still connected by a septum to the sister cell, early G2 cells were <8.5 µm long, mid-G2 cells were 8.5–10.5 µm long, late G2 cells were 10.6–12.5 µm long, and G2/M cells were >12.5 µm long. Cells lacking Cdr2p are longer than wild-type cells (Martin and Berthelot-Grosjean, 2009; Moseley et al., 2009), so we define the latter four stages of interphase as >3 µm. Cells expressing Cdr1p-3GFP are shorter than wild-type cells (Martin and Berthelot-Grosjean, 2009; Moseley et al., 2009), so we define the latter four stages of interphase as for wild type minus 1.5 µm.
Temporal color coding of images.
We made color-coded images to depict the motion of nodes in cells with an ImageJ plugin developed by K. Miura (Centre for Molecular and Cellular Imaging, European Molecular Biology Laboratory, Heidelberg, Germany). The plugin assigns a different color to an object at each time point of a time-lapse series and generates a maximum intensity projection of the resultant stack of images. White spots represent nodes that did not move during the time series, whereas colored spots or tracks represent nodes that disappeared or moved.
Particle tracking.
We tracked nodes at intervals of 1–5 s using two algorithms: custom ImageJ plugins that identify and track the intensity-based centers of mass of user-defined circular regions 7 pixels in diameter (Supplemental material) or the plugin TrackMate, based on methods to minimize the linear assignment problem (Jaqaman et al., 2008). From the corresponding tracks of node position over time, we measured the MSD as a function of time step τ between measurements. The two tracking algorithms produced similar results. We measured the slope of the first half of the data points and calculated the two-dimensional diffusion coefficient D according to the equation MSD = 4Dτ, which agreed with least-squares fitting with an error covariance matrix incorporated (Monnier et al., 2012). We measured the intensity of each node by summing the fluorescence intensity of the 7-pixel-diameter region and subtracting the fluorescence intensity of a concentric 8-pixel-diameter region, while correcting for photobleaching. To compare datasets with different imaging conditions, we report the intensity of each node relative to the mean node intensity in the field of asynchronous cells. For mitotic nodes, we did not track the nodes nearest the equator that were pulled into the contractile ring, as the heterogeneous stop–start motion cannot be used in MSD analyses, which average over all time points. The motion of mitotic nodes being pulled into the contractile ring has been carefully measured elsewhere (Vavylonis et al., 2008; Chen and Pollard, 2011).
Determination of diffusive motion.
We used the Bayesian inference method (Monnier et al., 2012) to assess whether the nodes were moving diffusively, with directed motion, or by a combination of the two. Bayesian inference analysis uses fits of plots of MSD versus time step to assign a relative probability of known models for the motion of particles. The original paper (Monnier et al., 2012) and our Fig. S3 validated the method for known and simulated data. Generally, we set the algorithm to test between three possibilities—diffusive only, D; diffusive plus directed (velocity), DV; and directed (velocity) only, V—and report the resultant probability of one type of motion relative to the other two. For a subset of curves, we added a term for error (a constant term at the y intercept of MSD curves), which affected the probability by <1%, so in general, we did not include error terms in the analysis. The probability of anomalous or confined diffusion for a subset of type 1 and type 2 nodes was ∼0%, so generally, we did not include these terms in the analysis. The Bayesian inference method accurately distinguished whether simulated tracks came from diffusive or ballistic particles (or a combination of the two), with a probability of 0.98–0.99 (Fig. S3 F) as demonstrated previously (Monnier et al., 2012). As a positive control for directed motion, we tracked SPBs during anaphase B (Fig. 2 A), which the algorithm identified as either ballistic (P = ∼0.6) or a combination of diffusive and ballistic (P = ∼0.4) but not diffusive (P = ∼0; Fig. S3, F and G).
Mathematical model of diffuse-and-capture mechanism
Setup of model.
We used MATLAB (MathWorks, Inc.) to make a computational model of objects diffusing at each time step t with diffusion coefficient D (Supplemental materials). These objects (nodes) were placed in a 7 × 3.5π–µm box corresponding to the length and circumference of the cell as follows. One set of objects (type 2 nodes) were placed in a random, uniform distribution along the y axis at x = 0, whereas the other set of objects (stationary type 1 nodes) were placed in a random, uniform distribution along the y axis and in a Gaussian distribution around x = 3,000 nm (measured from Fig. 1, A and B) with an SD of 800 nm, the same width as cytokinesis nodes (Vavylonis et al., 2008).
Simulations.
At each time step, each type 2 node diffused in the space according to its diffusion coefficient and bound to stationary type 1 nodes with a probability of binding pbind, when the centers of the two nodes were separated within an effective capture radius (Rc). Note that pbind and Rc are “effective” rather than physical parameters, as they take into account the discrete time step (here, 2 s) for which nodes are able to bind in the simulation. Nodes bound each other with a defined stoichiometry, usually set to 1:1. The limits of the surface in x (cell tips) were subjected to reflecting boundary conditions, and the limits of the surface in y (continuous circumference of the cell) were subjected to periodic boundary conditions. We calculated the mean position of captured type 2 nodes relative to the mean position of type 1 nodes to measure how evenly the captured type 2 nodes were distributed across the band of type 1 nodes.
Online supplemental material
Fig. S1 shows colocalization of proteins in nodes of cells expressing pairs of tagged proteins. Fig. S2 shows detailed behavior of type 1 node markers during mitosis and early G2. Fig. S3 shows observations and simulations of type 2 node movements during G2. Fig. S4 shows detailed behavior of type 2 nodes and assembly of the contractile ring in the absence of type 1 nodes in cdr2Δ cells. Fig. S5 shows dependence of the outcomes of simulations of the diffuse-and-capture model on the values of four parameters. Table S1 lists the timing of the movements of interphase node proteins during the cell cycle ± SD. Table S2 lists strains used in this study. Video 1 shows type 1 and 2 nodes separating in a cell during mitosis. Video 2 shows type 1 nodes diffusing away and dispersing into the cytoplasm of a cell during mitosis. Video 3 shows diffusing type 2 nodes in five cells at progressive stages of interphase. Video 4 shows type 2 nodes emerging from a cell’s disassembling contractile ring during cytokinesis. Video 5 shows type 2 nodes diffusing along the length of two interphase cdr2Δ cells lacking type 1 nodes. Video 6 shows slow contractile ring assembly in a cdr2Δ cell lacking type 1 nodes. Video 7 shows type 1 nodes in sid2-250 and wild-type mitotic cells at 36°C. Video 8 is a graphical output of a diffuse-and-capture simulation showing simulated type 2 nodes diffusing and being captured by stationary type 1 nodes. Video 9 shows type 2 nodes diffusing in wild-type, sid2-250, cdc25-22–arrested, and cdc10-129–arrested cells. A suite of MATLAB programs (diffuse-and-capture model) that stochastically simulate diffusing nodes according to the diffuse-and-capture model described in this paper is provided online as a ZIP file. A suite of ImageJ plugins (patch tracking tools) that correct for image offset and uneven illumination and find and track the centers of fluorescent spots in an image is provided online as a ZIP file. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201307174/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201307174.dv.
Supplementary Material
Acknowledgments
The authors thank Jian-Qiu Wu, Fred Chang, and James Moseley for providing strains, Sofia Espinoza and John Goss for help with experiments, Logan Akamatsu for help optimizing the MATLAB code, and Chad McCormick and Kota Miura for ImageJ macros.
This work was supported by National Institutes of Health research grants GM-026132 and GM-026338.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- A.U.
- arbitrary unit
- MBC
- methyl benzimidazol-2-yl-carbamate
- mEGFP
- monomeric EGFP
- MSD
- mean squared displacement
- SIN
- septation initiation network
- SPB
- spindle pole body
References
- Almonacid M., Moseley J.B., Janvore J., Mayeux A., Fraisier V., Nurse P., Paoletti A. 2009. Spatial control of cytokinesis by Cdr2 kinase and Mid1/anillin nuclear export. Curr. Biol. 19:961–966 10.1016/j.cub.2009.04.024 [DOI] [PubMed] [Google Scholar]
- Almonacid M., Celton-Morizur S., Jakubowski J.L., Dingli F., Loew D., Mayeux A., Chen J.-S., Gould K.L., Clifford D.M., Paoletti A. 2011. Temporal control of contractile ring assembly by Plo1 regulation of myosin II recruitment by Mid1/anillin. Curr. Biol. 21:473–479 10.1016/j.cub.2011.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bähler J., Steever A.B., Wheatley S., Wang Y., Pringle J.R., Gould K.L., McCollum D. 1998a. Role of polo kinase and Mid1p in determining the site of cell division in fission yeast. J. Cell Biol. 143:1603–1616 10.1083/jcb.143.6.1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bähler J., Wu J.Q., Longtine M.S., Shah N.G., McKenzie A., III, Steever A.B., Wach A., Philippsen P., Pringle J.R. 1998b. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 14:943–951 [DOI] [PubMed] [Google Scholar]
- Baumgärtner S., Tolić-Nørrelykke I.M. 2009. Growth pattern of single fission yeast cells is bilinear and depends on temperature and DNA synthesis. Biophys. J. 96:4336–4347 10.1016/j.bpj.2009.02.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breeding C.S., Hudson J., Balasubramanian M.K., Hemmingsen S.M., Young P.G., Gould K.L. 1998. The cdr2(+) gene encodes a regulator of G2/M progression and cytokinesis in Schizosaccharomyces pombe. Mol. Biol. Cell. 9:3399–3415 10.1091/mbc.9.12.3399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Pollard T.D. 2011. Actin filament severing by cofilin is more important for assembly than constriction of the cytokinetic contractile ring. J. Cell Biol. 195:485–498 10.1083/jcb.201103067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L., Moseley J.B. 2013. Compartmentalized nodes control mitotic entry signaling in fission yeast. Mol. Biol. Cell. 24:1872–1881 10.1091/mbc.E13-02-0104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Cortés J.C., McCollum D. 2009. Proper timing of cytokinesis is regulated by Schizosaccharomyces pombe Etd1. J. Cell Biol. 186:739–753 10.1083/jcb.200902116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grallert A., Connolly Y., Smith D.L., Simanis V., Hagan I.M. 2012. The S. pombe cytokinesis NDR kinase Sid2 activates Fin1 NIMA kinase to control mitotic commitment through Pom1/Wee1. Nat. Cell Biol. 14:738–745 10.1038/ncb2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman-Vendrell M., Baldissard S., Almonacid M., Mayeux A., Paoletti A., Moseley J.B. 2013. Blt1 and Mid1 provide overlapping membrane anchors to position the division plane in fission yeast. Mol. Cell. Biol. 33:418–428 10.1128/MCB.01286-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaqaman K., Loerke D., Mettlen M., Kuwata H., Grinstein S., Schmid S.L., Danuser G. 2008. Robust single-particle tracking in live-cell time-lapse sequences. Nat. Methods. 5:695–702 10.1038/nmeth.1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A.E., McCollum D., Gould K.L. 2012. Polar opposites: Fine-tuning cytokinesis through SIN asymmetry. Cytoskeleton (Hoboken). 69:686–699 10.1002/cm.21044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jourdain I., Brzezińska E.A., Toda T. 2013. Fission yeast Nod1 is a component of cortical nodes involved in cell size control and division site placement. PLoS ONE. 8:e54142 10.1371/journal.pone.0054142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanoh J., Russell P. 1998. The protein kinase Cdr2, related to Nim1/Cdr1 mitotic inducer, regulates the onset of mitosis in fission yeast. Mol. Biol. Cell. 9:3321–3334 10.1091/mbc.9.12.3321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusumi A., Nakada C., Ritchie K., Murase K., Suzuki K., Murakoshi H., Kasai R.S., Kondo J., Fujiwara T. 2005. Paradigm shift of the plasma membrane concept from the two-dimensional continuum fluid to the partitioned fluid: high-speed single-molecule tracking of membrane molecules. Annu. Rev. Biophys. Biomol. Struct. 34:351–378 10.1146/annurev.biophys.34.040204.144637 [DOI] [PubMed] [Google Scholar]
- Laporte D., Coffman V.C., Lee I.-J., Wu J.-Q. 2011. Assembly and architecture of precursor nodes during fission yeast cytokinesis. J. Cell Biol. 192:1005–1021 10.1083/jcb.201008171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S.G., Berthelot-Grosjean M. 2009. Polar gradients of the DYRK-family kinase Pom1 couple cell length with the cell cycle. Nature. 459:852–856 10.1038/nature08054 [DOI] [PubMed] [Google Scholar]
- McCormick C.D., Akamatsu M.S., Ti S.-C., Pollard T.D. 2013. Measuring affinities of fission yeast spindle pole body proteins in live cells across the cell cycle. Biophys. J. 105:1324–1335 10.1016/j.bpj.2013.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison J.M., Nurse P. 1985. Growth in cell length in the fission yeast Schizosaccharomyces pombe. J. Cell Sci. 75:357–376 [DOI] [PubMed] [Google Scholar]
- Monnier N., Guo S.-M., Mori M., He J., Lénárt P., Bathe M. 2012. Bayesian approach to MSD-based analysis of particle motion in live cells. Biophys. J. 103:616–626 10.1016/j.bpj.2012.06.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrell J.L., Nichols C.B., Gould K.L. 2004. The GIN4 family kinase, Cdr2p, acts independently of septins in fission yeast. J. Cell Sci. 117:5293–5302 10.1242/jcs.01409 [DOI] [PubMed] [Google Scholar]
- Moseley J.B., Mayeux A., Paoletti A., Nurse P. 2009. A spatial gradient coordinates cell size and mitotic entry in fission yeast. Nature. 459:857–860 10.1038/nature08074 [DOI] [PubMed] [Google Scholar]
- Paoletti A., Chang F. 2000. Analysis of mid1p, a protein required for placement of the cell division site, reveals a link between the nucleus and the cell surface in fission yeast. Mol. Biol. Cell. 11:2757–2773 10.1091/mbc.11.8.2757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard T.D., Wu J.-Q. 2010. Understanding cytokinesis: lessons from fission yeast. Nat. Rev. Mol. Cell Biol. 11:149–155 10.1038/nrm2834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S., Pollard T.D. 2012a. Anillin-related protein Mid1p coordinates the assembly of the cytokinetic contractile ring in fission yeast. Mol. Biol. Cell. 23:3982–3992 10.1091/mbc.E12-07-0535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S., Pollard T.D. 2012b. Characterization of structural and functional domains of the anillin-related protein Mid1p that contribute to cytokinesis in fission yeast. Mol. Biol. Cell. 23:3993–4007 10.1091/mbc.E12-07-0536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton M.J. 1997. Single-particle tracking: the distribution of diffusion coefficients. Biophys. J. 72:1744–1753 10.1016/S0006-3495(97)78820-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vavylonis D., Wu J.Q., Hao S., O’Shaughnessy B., Pollard T.D. 2008. Assembly mechanism of the contractile ring for cytokinesis by fission yeast. Science. 319:97–100 10.1126/science.1151086 [DOI] [PubMed] [Google Scholar]
- Wachtler V., Rajagopalan S., Balasubramanian M.K. 2003. Sterol-rich plasma membrane domains in the fission yeast Schizosaccharomyces pombe. J. Cell Sci. 116:867–874 10.1242/jcs.00299 [DOI] [PubMed] [Google Scholar]
- Wood W.B., Edgar R.S., King J., Lielausis I., Henninger M. 1968. Bacteriophage assembly. Fed. Proc. 27:1160–1166 [PubMed] [Google Scholar]
- Wu J.-Q., Pollard T.D. 2005. Counting cytokinesis proteins globally and locally in fission yeast. Science. 310:310–314 10.1126/science.1113230 [DOI] [PubMed] [Google Scholar]
- Wu J.-Q., Kuhn J.R., Kovar D.R., Pollard T.D. 2003. Spatial and temporal pathway for assembly and constriction of the contractile ring in fission yeast cytokinesis. Dev. Cell. 5:723–734 10.1016/S1534-5807(03)00324-1 [DOI] [PubMed] [Google Scholar]
- Wu J.-Q., Sirotkin V., Kovar D.R., Lord M., Beltzner C.C., Kuhn J.R., Pollard T.D. 2006. Assembly of the cytokinetic contractile ring from a broad band of nodes in fission yeast. J. Cell Biol. 174:391–402 10.1083/jcb.200602032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J.-Q., McCormick C.D., Pollard T.D. 2008. Chapter 9: Counting proteins in living cells by quantitative fluorescence microscopy with internal standards. Methods Cell Biol. 89:253–273 10.1016/S0091-679X(08)00609-2 [DOI] [PubMed] [Google Scholar]
- Ye Y., Lee I.-J., Runge K.W., Wu J.-Q. 2012. Roles of putative Rho-GEF Gef2 in division-site positioning and contractile-ring function in fission yeast cytokinesis. Mol. Biol. Cell. 23:1181–1195 10.1091/mbc.E11-09-0800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y.H., Ye Y., Wu Z., Wu J.Q. 2013. Cooperation between Rho-GEF Gef2 and its binding partner Nod1 in the regulation of fission yeast cytokinesis. Mol. Biol. Cell. 24:3187–3204 10.1091/mbc.E13-06-0301 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.