The E3 ligase Ltn1 and the deubiquitylase Ubp3-Bre5 titrate the level of ribosomal subunit ubiquitylation and thereby set the rate of ribosomal protein degradation by ribophagy in response to nutrient supply and the level of protein translation.
Abstract
Autophagy, the process by which proteins or organelles are engulfed by autophagosomes and delivered for vacuolar/lysosomal degradation, is induced to ensure survival under starvation and other stresses. A selective autophagic pathway for 60S ribosomal subunits elicited by nitrogen starvation in yeast—ribophagy—was recently described and requires the Ubp3-Bre5 deubiquitylating enzyme. This discovery implied that an E3 ligases act upstream, whether inhibiting the process or providing an initial required signal. In this paper, we show that Ltn1/Rkr1, a 60S ribosome-associated E3 implicated in translational surveillance, acts as an inhibitor of 60S ribosomal subunit ribophagy and is antagonized by Ubp3. The ribosomal protein Rpl25 is a relevant target. Its ubiquitylation is Ltn1 dependent and Ubp3 reversed, and mutation of its ubiquitylation site rendered ribophagy less dependent on Ubp3. Consistently, the expression of Ltn1—but not Ubp3—rapidly decreased after starvation, presumably to allow ribophagy to proceed. Thus, Ltn1 and Ubp3-Bre5 likely contribute to adapt ribophagy activity to both nutrient supply and protein translation.
Introduction
Rapid degradation of mature ribosomes can occur under nutrient starvation conditions via a selective autophagic pathway, called ribophagy, presumably contributing to the maintenance of the intracellular amino acid pool (Kraft et al., 2008; Kristensen et al., 2008). Ribophagy promotes relocalization of mature ribosomes from the cytoplasm to the vacuole, resulting in their degradation. Importantly, 60S and 40S ribosomal subunits appear to be independently targeted for degradation; although the latter follows a pathway that remains to be elucidated, selective degradation of 60S ribosomal subunits specifically requires the Ubp3-Bre5 deubiquitinating enzyme complex (Kraft et al., 2008; Ossareh-Nazari et al., 2010; Dargemont and Ossareh-Nazari, 2012). Ubp3 and Bre5 form a heterotetrameric complex able to cleave off ubiquitin from diverse specific substrates (Cohen et al., 2003; Li et al., 2005, 2007). In addition, 60S ribophagy requires the AAA ATPase Cdc48/p97 and its cofactor, the ubiquitin binding protein Ufd3, which together play a key role in deciphering fates of ubiquitylated proteins (Ossareh-Nazari et al., 2010; Dargemont and Ossareh-Nazari, 2012). The relevant substrates of Ubp3-Bre5, the role of ubiquitylation/deubiquitylation in the process, and the E3 ligases acting upstream of Ubp3-Bre5 have remained to be elucidated.
Two simple models could describe the potential role of ubiquitin in ribophagy. Ubiquitylation would either prevent ribosomes from being degraded or be required for ribophagy by providing an initial degradation signal, with deubiquitylation being required to complete the process. Insights into these mechanisms should greatly benefit from the identification of E3 ligases involved. In a recent work, the Rsp5 E3 ligase has been proposed to cooperate with Ubp3-Bre5 in ribophagy by providing the “engulf me” signal per the second model (Kraft and Peter, 2008). However, Rsp5 mutation displays a synthetic ribophagy defect with loss of Ubp3, suggesting that the E3 most likely acts on a distinct molecular step rather than upstream of Ubp3-Bre5.
Among E3 ligases that have been characterized, Ltn1/Rkr1 has emerged as a likely 60S ribophagy regulator candidate by virtue of its association with the large ribosomal subunit (Bengtson and Joazeiro, 2010). Ltn1 has been previously shown to function in the context of mRNA surveillance and ribosome-associated quality control (Bengtson and Joazeiro, 2010; Brandman et al., 2012; Defenouillère et al., 2013; Shao et al., 2013). Cytoplasmic mRNA quality control mechanisms depend on translation and, in addition to mediating decay of aberrant messages, are associated with degradation of the encoded nascent protein and with recycling of stalled ribosomes. Ltn1 has been shown to function specifically in targeting the products of mRNA devoid of stop codons for degradation (Bengtson and Joazeiro, 2010). Consistent with Ltn1’s role in protein quality control, mutation of its mouse homologue, Listerin, causes neurodegeneration in mice (Chu et al., 2009).
Here, we show that Ltn1 protects ribosomes from degradation in an Ubp3-antagonistic manner. We identify the ribosomal protein Rpl25 as a substrate of both Ltn1 and Ubp3, along with an Rpl25 lysine residue involved in the ubiquitin-mediated inhibition of 60S ribophagy. Finally, consistent with a critical role for Ltn1 in inhibiting ribophagy, we show that expression of this E3 is severely reduced upon nutrient starvation.
Results and discussion
LTN1 deletion rescues the ribophagy defect of Ubp3-deficient yeast
The Ltn1 E3, which was previously implicated in cotranslational quality control, is predominantly associated with ribosomes, mostly with 60S and possibly with 80S particles (Bengtson and Joazeiro, 2010; Brandman et al., 2012; Defenouillère et al., 2013; Shao et al., 2013). Because ribophagy of the large ribosomal subunit involves ubiquitylation–deubiquitylation, it was reasonable to test whether Ltn1 was also implicated in this process.
To obtain evidence for a role of Ltn1 in ribophagy, ribosome degradation was monitored in vivo using GFP-tagged Rpl5 or Rpl25 ribosomal proteins as markers, as previously described (Kraft et al., 2008). These tagged proteins are competent to become efficiently incorporated into ribosomes and therefore were located throughout the cytoplasm in both wild-type and mutant cells before starvation (Fig. 1 B and not depicted). Nitrogen starvation indeed induces relocalization of the GFP signal from the cytoplasm to the vacuole, leading to degradation of the Rpl moiety and vacuolar accumulation of free GFP, which is relatively more resistant to vacuolar proteases (Kraft et al., 2008). Vacuolar targeting and cleavage of GFP from the reporters can then be easily followed both by direct GFP fluorescence and by Western blotting of total cell extracts. In agreement with a previous study, deletion of UBP3 led to delayed GFP cleavage and vacuolar accumulation upon starvation, indicating that this enzyme is required for optimal rates of ribophagy (Fig. 1; Kraft et al., 2008). Absence of the Ltn1 E3 ligase alone did not significantly affect either the ribophagy kinetics compared with wild-type cells (Fig. 1) or general autophagy, as measured by GFP-Atg8 degradation (Fig. S1 A). Strikingly, when combined, deletion of LTN1 and UBP3 led to rescue of the cleavage defect of both Rpl5- and Rpl25-GFP reporters compared with ubp3Δ cells (Fig. 1, A and C), consistent with an increased accumulation of the GFP fluorescence in the vacuole (Fig. 1, B and D, vacuole and cytoplasmic + vacuole). Ltn1 belongs to the RING (really interesting new gene) domain family of E3 ligases, and the interaction of this domain with ubiquitin-charged E2-conjugating enzymes underlies E3 activity (Deshaies and Joazeiro, 2009). The ribophagy process was thus examined in a strain whose chromosomal copy of LTN1 harbored a deletion of the C-terminal RING domain. Deletion of the RING domain was able to rescue the ribophagy defect of UBP3-null cells (Fig. S1 B).
Figure 1.
LTN1 deletion rescues the ribophagy defect of UBP3-null cells. (A–D) Cells expressing Rpl5-GFP (A and B) or Rpl25-GFP (C and D) were grown in rich medium (before starvation) and starved in SD-N for the indicated periods (starvation). (A and C, left) Degradation of GFP-tagged proteins was analyzed by anti-GFP blot of whole cell extracts. (right) The ratio between cleaved GFP and full-length protein was quantified for every time point in four independent experiments. a.u., arbitrary unit. (B and D) Cells before starvation or starved for 24 h (starvation) were examined both by fluorescence microscopy and differential interferential contrast (DIC). Note that 100% of wild-type (WT) and mutant cells expressed GFP-tagged proteins in the cytoplasm before starvation. C, cytoplasmic localization; V, vacuolar accumulation; C + V, localization in both cytoplasm and vacuole. Bars, 5 µm. The significance of the differences observed for the ribophagy efficiency was evaluated using Student’s t test. *, P = 0.01–0.05; **, P = 0.001–0.01. The errors bars correspond to standard deviations.
These results show that Ubp3-Bre5 deubiquitylation activity is less critical for ribophagy in the absence of Ltn1 or its catalytic activity, raising the possibility of an antagonistic action among these enzymes in the process and suggesting that Ltn1 may ubiquitylate a target whose Ubp3-Bre5–dependent deubiquitylation is required for ribophagy to occur. Two conserved proteins, Tae2 and Rqc1, have been recently reported to form a ribosome-bound quality control complex together with Ltn1 (Brandman et al., 2012; Defenouillère et al., 2013). However, deletion of TAE2 did not impact the ribophagy process in wild-type or in ubp3Δ cells, indicating that Tae2 does not represent a constitutive Ltn1 cofactor (Fig. S1 C).
The 60S ribosomal protein Rpl25 is a substrate of both Ltn1 and Ubp3
To investigate how Ltn1 and Ubp3 regulate ribophagy, we considered the possibility that this might be mediated by ubiquitylation of 60S ribosomal proteins. It had been previously shown that the ubiquitylation of ribosomal subunits and/or ribosome-associated proteins was specifically enriched in Ubp3-deficient cells after nitrogen starvation for 3 h, suggesting a direct mechanism (Kraft et al., 2008). We therefore set out to examine the ubiquitylation of ribosomal proteins more directly. In light of Ltn1’s known function in protein quality control, it was reasonable to hypothesize that its RING domain might become properly positioned to target nascent polypeptides via binding to the vicinity of the opening of the ribosomal exit tunnel. Therefore, if Ltn1 mediates ubiquitylation of ribosomal proteins, those that are surface exposed in the surroundings of the tunnel opening would represent good candidates for initial analysis, namely Rpl17, Rpl19, Rpl25, Rpl26, Rpl31, and Rpl35 (Jenner et al., 2012; Klinge et al., 2012).
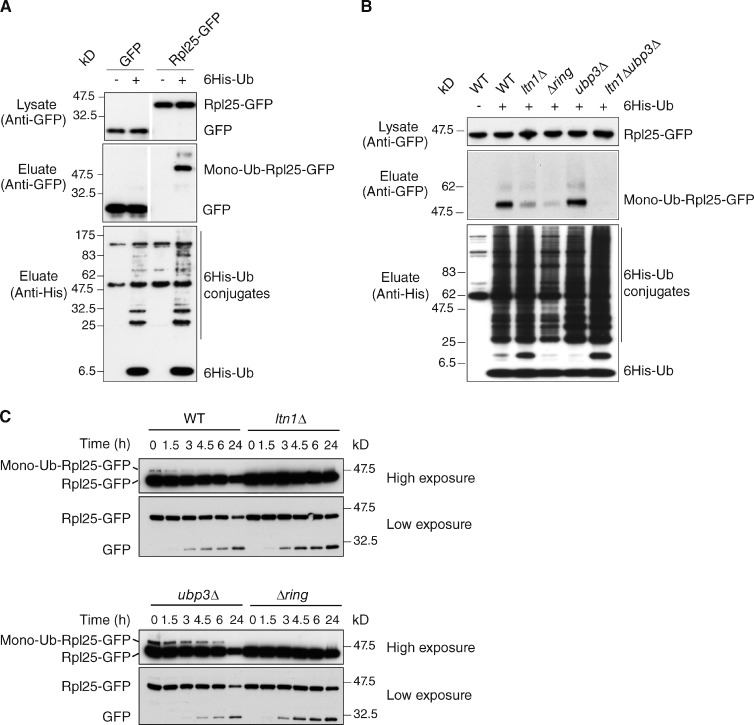
We decided to begin our analysis with Rpl25 because this protein is encoded by a single gene, is amenable to tagging, and is known to be modified with ubiquitin; the latter appeared to take place on a single lysine residue based on a mass spectrometry study, although the identification of additional ubiquitylation sites in Rpl25 could have been hampered by the detection limit of the analysis (Starita et al., 2012). A cupper-inducible His-tagged ubiquitin was coexpressed with GFP-tagged Rpl25 followed by purification of ubiquitylated proteins on a nickel column and anti-GFP blotting. This assay showed that Rpl25-GFP was monoubiquitylated, whereas the GFP alone was not (Fig. 2 A). Importantly, deletion of LTN1 or its RING domain severely impaired ubiquitylation of Rpl25-GFP consistent with the requirement for Ltn1 E3 enzymatic activity for this modification (Fig. 2 B). In agreement with these results, analysis of Rpl25-GFP modification by endogenous untagged ubiquitin in total cell extracts confirmed its Ltn1 RING domain dependency (Fig. 2 C, t = 0). The specificity of these results is underscored by the observation that ubiquitylation of a different 60S subunit protein, Rpl5, was not Ltn1 dependent (Fig. S2). Next, we examined whether Rpl25 ubiquitylation was also regulated by Ubp3 and found that the steady-state ubiquitylation of Rpl25-GFP, but not of Rpl5-GFP, was indeed increased in ubp3Δ cells (Fig. 2, B and C, t = 0; and Fig. S2). Therefore, Rpl25 appears to be a specific target for both the E3 ligase activity of Ltn1 and the deubiquitylating activity of Ubp3. Importantly, the low level of ubiquitylated Rpl25-GFP remaining upon LTN1 deletion was not increased by deletion of UBP3, suggesting that both enzymes catalyze conjugation and deconjugation of the same lysine residues of Rpl25 (Fig. 2 B). Together, these results raised the possibility that the ubiquitin modification of Rpl25 may provide a critical signal in the regulation of ribophagy.
Figure 2.
The 60S ribosomal protein Rpl25 is a substrate of both Ltn1 and Ubp3. (A) GFP or Rpl25-GFP were expressed in cells transformed (+) or not transformed (−) with a plasmid encoding 6His-ubiquitin. (top) Expression levels of GFP and Rpl25-GFP in whole cell lysates were verified with anti-GFP blotting. (middle) Purified 6His-ubiquitin conjugates were examined by anti-GFP blotting. (bottom) 6His-ubiquitin expression and efficiency of purification was controlled using an anti-6His antibody. White lines indicate that intervening lanes have been spliced out. (B) Analysis of ubiquitylated forms of Rpl25-GFP in wild-type and the indicated mutant cells as in A. (C) Expression of Rpl25-GFP and its modified form were analyzed by anti-GFP blot of extracts from wild-type and mutant cells starved for the indicated periods. Ub, ubiquitin; WT, wild type.
Interestingly, the steady-state levels of ubiquitylated Rpl25 were altered upon nutrient starvation, with a rapid decay of the modified compared with the unmodified form in wild-type cells. In contrast, in ubp3Δ cells, ubiquitylated Rpl25-GFP levels remained readily detectable even after 6 h of starvation (Fig. 2 C). These results thus suggest that starvation decreases the Ltn1-dependent ubiquitylation of Rpl25 and/or favors its deconjugation by Ubp3 before ribophagy.
Ubiquitylation of Rpl25 regulates 60S ribophagy
To further examine whether that Rpl25 ubiquitylation and deubiquitylation by Ltn1 and Ubp3-Bre5, respectively, are critical for ribophagy, we set out to generate a putative ubiquitylation-resistant Rpl25 mutant. Rpl25 lysine residue 74 (K74) had been identified as a ubiquitylation site in a recent large-scale proteomic study (Starita et al., 2012). Because Rpl25 K74’s neighboring residue is also a lysine (K75), both residues were therefore substituted for arginine in an attempt to generate a ubiquitylation-resistant mutant. Plasmids encoding wild-type or K74/75R mutant Rpl25-GFP (rpl25KR) were introduced into different strains followed by deletion of the endogenous RPL25 chromosomal copy. As shown in Fig. 3 A, mutation of K74 and 75 to arginine did not affect Rpl25 expression levels but did indeed reduce its ubiquitylation in both wild-type and ubp3Δ cells (Fig. 3 A, compare second with fourth lanes and sixth with eighth lanes), indicating that K74/75 thus constitutes the major ubiquitylation site of Rpl25. Residual monoubiquitylation of rpl25KR was detected that might result from modification on an alternative site or at a site artificially unmasked by the mutation (Hayakawa et al., 2012). Deleting LTN1 led to a severe decrease of Rpl25 ubiquitylation, including residual ubiquitylation observed for the rpl25KR mutant protein, thus indicating that Ltn1 represents the unique ubiquitin ligase for Rpl25 (Fig. 3 A, 10th and 12th lanes). Finally, combining deletion of UBP3 with deletion of LTN1 did not restore any ubiquitylation of wild-type or mutant Rpl25, consistent with the model that both enzymes catalyze conjugation and deconjugation of the same lysine residues of Rpl25 (Fig. 3 A, 14th and 16th lanes).
Figure 3.
Ubiquitylation of Rpl25 at K74 regulates 60S ribophagy. (A) Indicated strains were transformed with plasmids encoding either Rpl25-GFP (Rpl25) or rpl25 K74,75R-GFP (rpl25KR) before the deletion of a genomic copy of RPL25. (top) Comparable expression levels of Rpl25-GFP or rpl25 K74,75R-GFP in whole cell lysates were confirmed with anti-GFP blotting. Purified 6His-ubiquitin–conjugated forms of Rpl25-GFP or rpl25 K74,75R-GFP (middle) and 6His-ubiquitin expression (bottom) were analyzed as in Fig. 2. Ub, ubiquitin. (B) Wild-type and ubp3Δ cells expressing Rpl25-GFP or rpl25 K74,75R-GFP were starved for the indicated period of time. (C) Degradation of GFP-tagged proteins was analyzed by anti-GFP blot of whole cell extracts, and the ratio between cleaved GFP and full-length protein was quantified for all time points in four independent experiments. a.u., arbitrary units. (D) Localization of GFP-tagged proteins. C, cytoplasmic localization; V, vacuolar accumulation; C + V, localization in both cytoplasm and vacuole (E) Degradation of Rpl3 was analyzed at 24 h upon starvation by Western blotting of whole cell extracts using anti-Rpl3 (gift from V. Albanese, Institut Jacques Monod, Paris, France) or anti-Mex67 (as a control) antibodies. Rpl3 and Mex67 expression was quantified and normalized to the expression level at time 0. Significance of the differences observed for the ribophagy efficiency was evaluated using Student’s t test. *, P = 0.01–0.05; **, P = 0.001–0.01; ***, P < 0.001. White lanes indicate that intervening lanes have been spliced out. The errors bars correspond to standard deviations.
Our model predicted that if Rpl25 K74 is a critical target of Ltn1 in the inhibition of ribophagy, the rpl25KR mutant should phenocopy the LTN1 deletion in functional assays. We thus examined the ribophagy phenotype of strains expressing rpl25KR mutant proteins as their only source of Rpl25. The results in Fig. 3 (B–E) show that starvation-induced degradation of Rpl25-GFP as well as of the endogenous untagged Rpl3 60S component was similar in strains expressing wild-type or rpl25KR mutant protein. In contrast, expression of the ubiquitylation-resistant Rpl25 mutant (but not of wild-type Rpl25) partially but significantly restored the ribophagy defect observed in the ubp3Δ background (Fig. 3, B–D). This rescue could also be observed by monitoring the degradation of the Rpl3 60S component (Fig. 3 E). Thus, ubiquitylation of Rpl25 does not only protect Rpl25 from starvation-induced degradation but more generally protects the 60S subunit. Importantly, combining LTN1 deletion with the rpl25KR mutation did not lead to additional rescue of the ubp3Δ effect (Fig. S3), indicating that Rpl25 K74/75 represents the critical target for the role of Ltn1 in ribophagy. In conclusion, Ubp3-mediated deubiquitylation becomes less critical for efficient 60S ribophagy if Rpl25 ubiquitylation is impaired by either LTN1 deletion, K74/75 mutation, or both.
Ltn1 levels are markedly down-regulated on nitrogen starvation
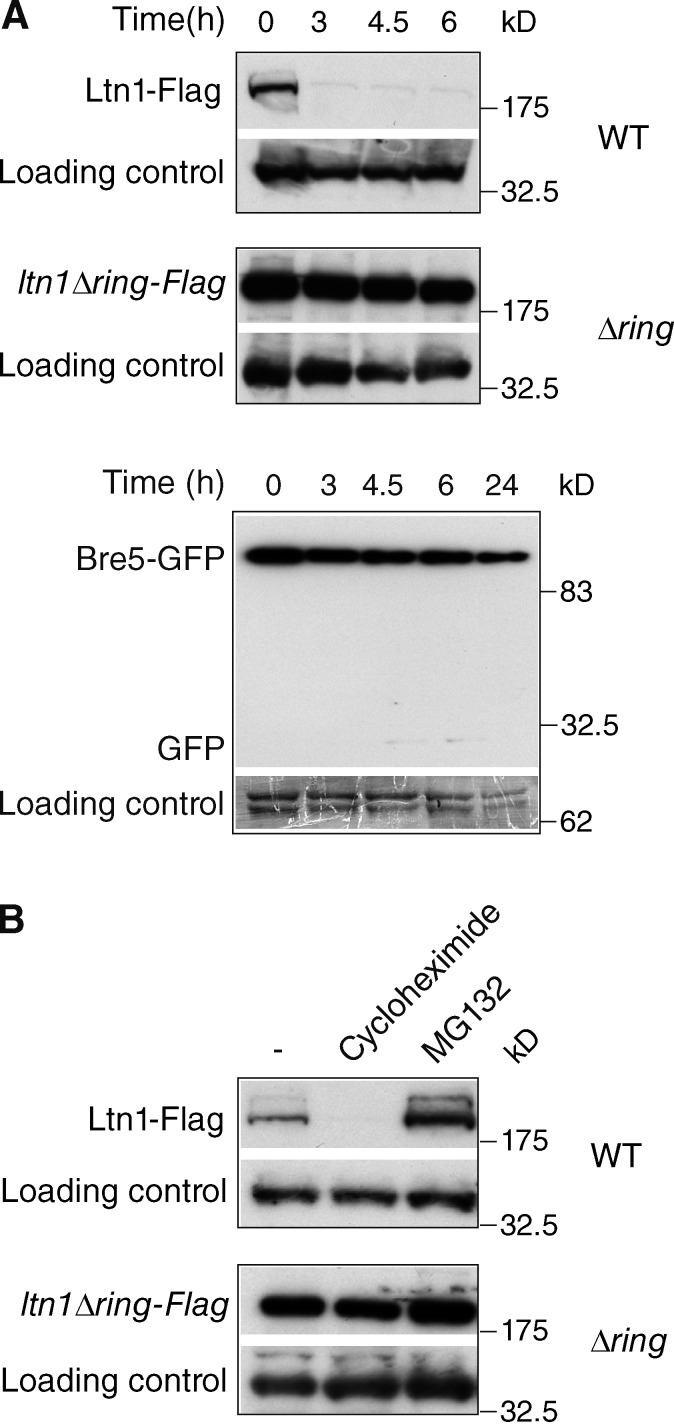
The aforementioned data led to the question of how the ubiquitylation/deubiquitylation balance is regulated under starvation conditions. In the course of examining whether Ltn1 and Ubp3-Bre5 would remain associated with ribosomes throughout the autophagic process, we discovered that nitrogen starvation induced a rapid and marked decay of Ltn1-Flag or Ltn1-GFP levels with a quasicomplete disappearance within 3 h; in contrast, Bre5-GFP degradation was only modest under the same conditions with a significant degradation only observed after 24-h starvation (Fig. 4 A and Fig. S3 B). The UBP3 deletion did not result in Ltn1 stabilization, thus arguing against a role of Ubp3-Bre5 upstream of Ltn1 (Fig. S3 B). In contrast, the RING domain was required for the starvation-induced degradation of Ltn1, indicating that the catalytic activity of Ltn1 is required for its decay (Fig. 4 A). This rapid turnover of Ltn1 not only occurred upon starvation but also in rich medium as shown by its disappearance upon 30-min treatment with cycloheximide. In rich medium, the efficient degradation of Ltn1 also required its RING domain and was inhibited by the proteasome inhibitor MG132 (Fig. 4 B).
Figure 4.
Ltn1 is rapidly degraded in a RING domain– and proteasome-dependent manner. (A) Wild-type cells expressing Bre5-GFP, Ltn1-Flag, or ltn1-ΔRing-Flag were starved for the indicated periods of time. (B) Cells were treated for 30 min with 100 µM cycloheximide or 100 µM MG132. Expression of tagged proteins was analyzed by anti-GFP or anti-Flag blotting of whole cell extracts. Five times less extracts were analyzed for ltn1-ΔRing-Flag cells. A nonspecific band served as a loading control. White lanes indicate that intervening lanes have been spliced out. WT, wild type.
Together, these results indicate that the efficient degradation of Ltn1 is mediated by autoubiquitylation (RING domain) and degradation by the proteasome and occurs both in rich medium and upon starvation. The general starvation-induced inhibition of protein synthesis likely results in a drastic change in the equilibrium between translation and degradation of Ltn1, leading to a severe reduction of Ltn1 expression and consequent Ltn1-dependent ubiquitylation of the ribosome.
The ubiquitin protease activity of Ubp3-Bre5 had been previously identified as a critical regulator of selective autophagy of 60S ribosomal subunits. The results presented here indicate that deficiency of the 60S ribosome-associated Ltn1 E3 ligase rescued the ribophagy defect of ubp3Δ cells, indicating that Ltn1 acts as a ribophagy inhibitor upstream of Ubp3. We found that this function of Ltn1 is mediated by the ubiquitylation of the lysine 74 in Rpl25, a protein in the large ribosomal subunit, identified as a presumably direct target for both Ltn1 and Ubp3. However, deficiency in Rpl25 ubiquitylation did not fully rescue the ribophagy defect of ubp3Δ cells, suggesting that additional Ubp3-dependent deubiquitylation events are required for ribophagy to proceed optimally. As the 60S ribosome is known to be extensively ubiquitylated (Starita et al., 2012), it will be interesting to identify what additional modification sites are implicated in regulating ribophagy efficiency.
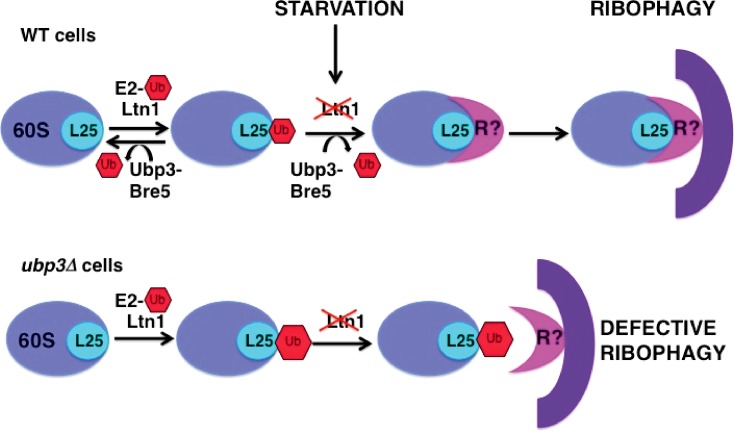
Our data are consistent with a model (Fig. 5) in which Ltn1 can leave ubiquitin marks on Rpl25 that prevent the selective degradation of 60S subunits and are constitutively cleaved off by Ubp3 in wild-type cells. Upon starvation, the rapid decay of Ltn1 combined with Ubp3-mediated removal of the ubiquitin from Rpl25 would accelerate the targeting of 60S ribosomes to autophagosomes. In the absence of Ubp3, ubiquitylated Rpl25 accumulates, thus resulting in a defective degradation of 60S ribosomes. Cargoes of selective autophagy pathways are recognized by specific receptors that simultaneously bind to autophagosome-associated proteins, such as the ubiquitin-like protein Atg8 (LC3 in mammalian cells; Shintani et al., 2002; Novak et al., 2010; Suzuki et al., 2010; Motley et al., 2012). Although no cargo adaptor has been yet identified for ribophagy, we speculate that Ltn1-mediated ubiquitylation of Rpl25 might prevent the interaction between such a receptor and 60S ribosomes (Fig. 5). Alternatively, the ubiquitin moiety could preclude engulfment or completion of the autophagosome through some other mechanism (Shaid et al., 2013).
Figure 5.
Model for the role of Ltn1 and Ubp3-Bre5 in starvation-induced autophagy of the 60S ribosome. R is a yet-unidentified specific autophagic receptor for ribosome. Ub, ubiquitin; WT, wild type.
The involvement of Ltn1 in ribophagy could potentially provide a cross talk mechanism between protein quality control and ribosomal disposal, whereby ribosomes that become stalled during translation of aberrant proteins would be targeted for ribophagic degradation. However, degradation of 60S ribosomes was not affected, at least to a measurable extent, in a strain harboring a mutation that increases nonstop protein production (unpublished data; Bradley et al., 2003; Bengtson and Joazeiro, 2010). Moreover Tae2, a partner of Ltn1 in the ribosome quality control complex (Brandman et al., 2012; Defenouillère et al., 2013) did not regulate ribophagy. Nonetheless, cotranslational quality control and ribosome degradation pathways clearly share at least one common factor, Cdc48, a ubiquitin-dependent AAA ATPase. Cdc48 has been indeed linked to ribophagy and nonstop protein degradation as well as to a distinct ribosomal degradation pathway that eliminates 60S subunits containing nonfunctional 25S ribosomal RNA (Ossareh-Nazari et al., 2010; Brandman et al., 2012; Fujii et al., 2012).
Materials and methods
Yeast strains, plasmids, and media
The Saccharomyces cerevisiae strains and plasmids used in this study are listed in Table 1 and Table 2. The plasmid encoding rpl25 K74,75R-GFP was constructed from the pRS315-RPL25-EGFP plasmid indicated in Table 2 (Gadal et al., 2001) using the site-directed mutagenesis kit (QuikChange; Agilent Technologies). Yeast cultures were grown at 30°C either in rich medium (YPD [yeast, peptone, dextrose]) or in synthetic medium (SD) containing 0.67% yeast nitrogen base with ammonium sulfate and 2% dextrose and supplemented with appropriate nutrients. Cells were starved in starvation medium (SD-N) containing 0.17% yeast nitrogen base without amino acids and 2% dextrose (Kraft et al., 2008).
Table 1.
Strains used in this study
| Strains | Genotype | Reference |
| WT (BY4741) | Mat a, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0 | EUROSCARF |
| ubp3Δ | Mat a, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0 UBP3::HIS | This study |
| ltn1Δ | Mat a, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, LTN1::KanMX6 | Bengtson and Joazeiro, 2010 |
| Δring | Mat a, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, ltn1Δ4500-3×FLAG::KanMX6 | Bengtson and Joazeiro, 2010 |
| ltn1Δ ubp3Δ | Mat a, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, LTN1::KanMX6, UBP3::HIS | This study |
| tae2Δ | Mat a, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, TAE2::KanMX6 | EUROSCARF |
| ubp3Δ tae2Δ | Mat a, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, TAE2::KanMX6 UBP3::HIS | This study |
| Ltn1-Flag | Mat a, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, LTN1-3×FLAG-KanMX6 | Bengtson and Joazeiro, 2010 |
| Ltn1-GFP | Mat a, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, LTN1-GFP-HIS | This study |
| Ubp3Δ Ltn1-GFP | Mat a, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, UBP3::KanMX6 LTN1-GFP-HIS | This study |
| Bre5-GFP | Mat a, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, BRE5-GFP-HIS | This study |
| rpl25Δ Rpl25 | Mat a, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, RLP25::HIS, pRS315-RPL25-EGFP | This study |
| rpl25Δ rpl25KR | Mat a, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, RPL25::HIS, pRS315-RPL25KR-EGFP | This study |
| ubp3Δ rpl25Δ Rpl25 | Mat a, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, UBP3::KanMX6, RPL25::HIS, pRS315-RPL25-EGFP | This study |
| ubp3Δ rpl25Δ rpl25KR | Mat a, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, UBP3::KanMX6, RPL25::HIS, pRS315-RPL25KR-EGFP | This study |
| ltn1Δ rpl25Δ Rpl25 | Mat a, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, LTN1::HPH RLP25::HIS, pRS315-RPL25-EGFP | This study |
| ltn1Δ rpl25Δ rpl25KR | Mat a, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, LTN1::HPH RPL25::HIS, pRS315-RPL25KR-EGFP | This study |
| ltn1Δ ubp3Δ rpl25Δ Rpl25 | Mat a, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, LTN1::HPH UBP3::KanMX6, RPL25::HIS, pRS315-RPL25-EGFP | This study |
| ltn1Δ ubp3Δ rpl25Δ rpl25KR | Mat a, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, LTN1::HPH UBP3::KanMX6, RPL25::HIS, pRS315-RPL25KR-EGFP | This study |
EUROSCARF, European Saccharomyces cerevisiae archive for functional analysis; WT, wild type.
Table 2.
Plasmids used in this study
| Plasmid | Description | Reference |
| YEp352-6HisUb | 2μ URA3 CUP-6His-Ub | Vitaliano-Prunier et al., 2008 |
| pRS315-RPL25-EGFP | CEN LEU2 | Gadal et al., 2001 |
| pRS315-RPL25KR-EGFP | CEN LEU2, RPL25K74,75R | This study |
| pRS315-RPL5-EGFP | CEN LEU2 | Gift from V. Pansea |
Plasmid pRS315-RPL25-EGFP was a gift from E. Hurt (Heidelberg University, Heidelberg, Germany).
Eidgenössische Technische Hochschule Zürich, Zürich, Switzerland.
Preparation of yeast total extracts
Cells grown in YPD or synthetic medium were collected during the exponential growth phase (OD600 of 1.5 or 0.8, respectively). Total protein extracts were prepared by the NaOH-TCA lysis method and analyzed by Western blotting using anti-GFP antibodies (Roche). Signals corresponding to GFP fusion proteins and GFP were quantified using the ImageJ software (National Institutes of Health), and the ratio of cleaved to noncleaved GFP protein was used to measure the ribophagy process (Kraft et al., 2008; Ossareh-Nazari et al., 2010).
Fluorescence microscopy
Wide-field fluorescence images of living cells were acquired using a microscope (DMR; Leica) with a 100× Plan Apochromat HCX oil immersion objective and a high-sensitive cooled interlined charge-coupled device camera (CoolSNAP HQ2; Photometrics). Rapid and precise z positioning was accomplished by a piezoelectric motor (Linear Variable Differential Transformer; Physik Instrumente) mounted underneath the objective lens. Maximum intensity projections were performed using MetaMorph software (Molecular Devices). The localization of GFP-tagged proteins was analyzed in ≥200 cells per condition and per experiment, and results were obtained from three independent experiments.
Purification of ubiquitylated proteins
Cells transformed with a plasmid encoding 6His-ubiquitin under the CUP1 promoter were grown on selective media and stimulated overnight with 0.1 mM CuSO4. Cells were lysed in 6 M guanidinium-HCl, 0.1 M Na2HPO4/NaH2PO4, 0.01 M Tris-HCl, pH 8.0, 0.1% Triton X-100 plus 5 mM imidazole, 10 mM β-mercaptoethanol, and protease inhibitors. Purifications of His6-ubiquitin–modified proteins were performed on Ni2+-nitrilotriacetic acid–agarose beads and eluted (QIAGEN). The beads were washed with 8 M urea, 0.1 M Na2HPO4/NaH2PO4, 0.01 M Tris-HCl, pH 6.3, 10 mM β-mercaptoethanol, and 0.2% Triton X-100 before elution and Western blot analysis using anti-GFP (Roche) and anti-6His (Covalab) antibodies (Vitaliano-Prunier et al., 2008; Hayakawa et al., 2012).
Statistical analyses
Significance of the differences observed for the ribophagy efficiency (Fig. 1, A–D; and Fig. 3, C–E) was evaluated using Student’s t test. No asterisk corresponds to P > 0.05, one asterisk corresponds to P = 0.01–0.05, and two asterisks correspond to P = 0.001–0.01.
Online supplemental material
Fig. S1 shows that Ltn1 does not affect general autophagy and that its RING domain, but not its cofactor Tae2, is required for its function in ribophagy. Fig. S2 shows that the ribosomal protein Rpl5 is ubiquitylated but in an Ltn1- and Ubp3-independent manner. Fig. S3 demonstrates that combining LTN1 deletion with K74/75R mutation in Rpl25 has no synergistic or additive effect. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201308139/DC1.
Supplementary Material
Acknowledgments
We are grateful to E. Hurt, V. Panse, and V. Albanese for the gift of reagents. We thank A. Babour and J. Weitzman for critically reading the manuscript.
This study was funded by grants from the Association for Research against Cancer to C. Dargemont, by the Fondation pour la Recherche Medicale (to C.A. Niño), by the National Institute of Neurological Disorders and Stroke of the National Institutes of Health (grant R01 NS075719), and by a Research Scholar Grant (RSG-08-298-01-TBE) from the American Cancer Society to C.A.P. Joazeiro.
The authors declare no competing financial interests.
References
- Bengtson M.H., Joazeiro C.A. 2010. Role of a ribosome-associated E3 ubiquitin ligase in protein quality control. Nature. 467:470–473 10.1038/nature09371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley M.E., Bagriantsev S., Vishveshwara N., Liebman S.W. 2003. Guanidine reduces stop codon read-through caused by missense mutations in SUP35 or SUP45. Yeast. 20:625–632 10.1002/yea.985 [DOI] [PubMed] [Google Scholar]
- Brandman O., Stewart-Ornstein J., Wong D., Larson A., Williams C.C., Li G.W., Zhou S., King D., Shen P.S., Weibezahn J., et al. 2012. A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell. 151:1042–1054 10.1016/j.cell.2012.10.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu J., Hong N.A., Masuda C.A., Jenkins B.V., Nelms K.A., Goodnow C.C., Glynne R.J., Wu H., Masliah E., Joazeiro C.A., Kay S.A. 2009. A mouse forward genetics screen identifies LISTERIN as an E3 ubiquitin ligase involved in neurodegeneration. Proc. Natl. Acad. Sci. USA. 106:2097–2103 10.1073/pnas.0812819106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen M., Stutz F., Belgareh N., Haguenauer-Tsapis R., Dargemont C. 2003. Ubp3 requires a cofactor, Bre5, to specifically de-ubiquitinate the COPII protein, Sec23. Nat. Cell Biol. 5:661–667 10.1038/ncb1003 [DOI] [PubMed] [Google Scholar]
- Dargemont C., Ossareh-Nazari B. 2012. Cdc48/p97, a key actor in the interplay between autophagy and ubiquitin/proteasome catabolic pathways. Biochim. Biophys. Acta. 1823:138–144 10.1016/j.bbamcr.2011.07.011 [DOI] [PubMed] [Google Scholar]
- Defenouillère Q., Yao Y., Mouaikel J., Namane A., Galopier A., Decourty L., Doyen A., Malabat C., Saveanu C., Jacquier A., Fromont-Racine M. 2013. Cdc48-associated complex bound to 60S particles is required for the clearance of aberrant translation products. Proc. Natl. Acad. Sci. USA. 110:5046–5051 10.1073/pnas.1221724110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies R.J., Joazeiro C.A. 2009. RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78:399–434 10.1146/annurev.biochem.78.101807.093809 [DOI] [PubMed] [Google Scholar]
- Fujii K., Kitabatake M., Sakata T., Ohno M. 2012. 40S subunit dissociation and proteasome-dependent RNA degradation in nonfunctional 25S rRNA decay. EMBO J. 31:2579–2589 10.1038/emboj.2012.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadal O., Strauss D., Braspenning J., Hoepfner D., Petfalski E., Philippsen P., Tollervey D., Hurt E. 2001. A nuclear AAA-type ATPase (Rix7p) is required for biogenesis and nuclear export of 60S ribosomal subunits. EMBO J. 20:3695–3704 10.1093/emboj/20.14.3695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayakawa A., Babour A., Sengmanivong L., Dargemont C. 2012. Ubiquitylation of the nuclear pore complex controls nuclear migration during mitosis in S. cerevisiae. J. Cell Biol. 196:19–27 10.1083/jcb.201108124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenner L., Melnikov S., Garreau de Loubresse N., Ben-Shem A., Iskakova M., Urzhumtsev A., Meskauskas A., Dinman J., Yusupova G., Yusupov M. 2012. Crystal structure of the 80S yeast ribosome. Curr. Opin. Struct. Biol. 22:759–767 10.1016/j.sbi.2012.07.013 [DOI] [PubMed] [Google Scholar]
- Klinge S., Voigts-Hoffmann F., Leibundgut M., Ban N. 2012. Atomic structures of the eukaryotic ribosome. Trends Biochem. Sci. 37:189–198 10.1016/j.tibs.2012.02.007 [DOI] [PubMed] [Google Scholar]
- Kraft C., Peter M. 2008. Is the Rsp5 ubiquitin ligase involved in the regulation of ribophagy? Autophagy. 4:838–840 [DOI] [PubMed] [Google Scholar]
- Kraft C., Deplazes A., Sohrmann M., Peter M. 2008. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat. Cell Biol. 10:602–610 10.1038/ncb1723 [DOI] [PubMed] [Google Scholar]
- Kristensen A.R., Schandorff S., Høyer-Hansen M., Nielsen M.O., Jäättelä M., Dengjel J., Andersen J.S. 2008. Ordered organelle degradation during starvation-induced autophagy. Mol. Cell. Proteomics. 7:2419–2428 10.1074/mcp.M800184-MCP200 [DOI] [PubMed] [Google Scholar]
- Li K., Zhao K., Ossareh-Nazari B., Da G., Dargemont C., Marmorstein R. 2005. Structural basis for interaction between the Ubp3 deubiquitinating enzyme and its Bre5 cofactor. J. Biol. Chem. 280:29176–29185 10.1074/jbc.M502975200 [DOI] [PubMed] [Google Scholar]
- Li K., Ossareh-Nazari B., Liu X., Dargemont C., Marmorstein R. 2007. Molecular basis for bre5 cofactor recognition by the ubp3 deubiquitylating enzyme. J. Mol. Biol. 372:194–204 10.1016/j.jmb.2007.06.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motley A.M., Nuttall J.M., Hettema E.H. 2012. Pex3-anchored Atg36 tags peroxisomes for degradation in Saccharomyces cerevisiae. EMBO J. 31:2852–2868 10.1038/emboj.2012.151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak I., Kirkin V., McEwan D.G., Zhang J., Wild P., Rozenknop A., Rogov V., Löhr F., Popovic D., Occhipinti A., et al. 2010. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 11:45–51 10.1038/embor.2009.256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossareh-Nazari B., Bonizec M., Cohen M., Dokudovskaya S., Delalande F., Schaeffer C., Van Dorsselaer A., Dargemont C. 2010. Cdc48 and Ufd3, new partners of the ubiquitin protease Ubp3, are required for ribophagy. EMBO Rep. 11:548–554 10.1038/embor.2010.74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaid S., Brandts C.H., Serve H., Dikic I. 2013. Ubiquitination and selective autophagy. Cell Death Differ. 20:21–30 10.1038/cdd.2012.72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao S., von der Malsburg K., Hegde R.S. 2013. Listerin-dependent nascent protein ubiquitination relies on ribosome subunit dissociation. Mol. Cell. 50:637–648 10.1016/j.molcel.2013.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T., Huang W.P., Stromhaug P.E., Klionsky D.J. 2002. Mechanism of cargo selection in the cytoplasm to vacuole targeting pathway. Dev. Cell. 3:825–837 10.1016/S1534-5807(02)00373-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starita L.M., Lo R.S., Eng J.K., von Haller P.D., Fields S. 2012. Sites of ubiquitin attachment in Saccharomyces cerevisiae. Proteomics. 12:236–240 10.1002/pmic.201100166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K., Kondo C., Morimoto M., Ohsumi Y. 2010. Selective transport of alpha-mannosidase by autophagic pathways: identification of a novel receptor, Atg34p. J. Biol. Chem. 285:30019–30025 10.1074/jbc.M110.143511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitaliano-Prunier A., Menant A., Hobeika M., Géli V., Gwizdek C., Dargemont C. 2008. Ubiquitylation of the COMPASS component Swd2 links H2B ubiquitylation to H3K4 trimethylation. Nat. Cell Biol. 10:1365–1371 10.1038/ncb1796 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.