OPA1 processing by YEM1L and OMA1 is dispensable for mitochondrial fusion and instead drives mitochondrial fragmentation, which is crucial for mitochondrial integrity and quality control.
Abstract
Mitochondrial fusion and structure depend on the dynamin-like GTPase OPA1, whose activity is regulated by proteolytic processing. Constitutive OPA1 cleavage by YME1L and OMA1 at two distinct sites leads to the accumulation of both long and short forms of OPA1 and maintains mitochondrial fusion. Stress-induced OPA1 processing by OMA1 converts OPA1 completely into short isoforms, inhibits fusion, and triggers mitochondrial fragmentation. Here, we have analyzed the function of different OPA1 forms in cells lacking YME1L, OMA1, or both. Unexpectedly, deletion of Oma1 restored mitochondrial tubulation, cristae morphogenesis, and apoptotic resistance in cells lacking YME1L. Long OPA1 forms were sufficient to mediate mitochondrial fusion in these cells. Expression of short OPA1 forms promoted mitochondrial fragmentation, which indicates that they are associated with fission. Consistently, GTPase-inactive, short OPA1 forms partially colocalize with ER–mitochondria contact sites and the mitochondrial fission machinery. Thus, OPA1 processing is dispensable for fusion but coordinates the dynamic behavior of mitochondria and is crucial for mitochondrial integrity and quality control.
Introduction
Mitochondria undergo continuous fusion and fission to maintain their morphology and function (Westermann, 2010; Tamura et al., 2011; Chan, 2012; Liesa and Shirihai, 2013; van der Bliek et al., 2013). Mitochondrial dynamics are implicated in various cellular processes such as apoptosis, cell differentiation, cell division, and development (Nunnari and Suomalainen, 2012; Escobar-Henriques and Anton, 2013; Otera et al., 2013). It acts as an important quality control mechanism, where fusion contributes to mitochondrial maintenance and fission allows for the segregation of dysfunctional mitochondria (Twig et al., 2008; Youle and van der Bliek, 2012).
Fusion and fission events occur in a regulated, cyclic manner, determining the shape, size, and distribution of mitochondria (Twig et al., 2008; Liu et al., 2009; Cagalinec et al., 2013). Conserved GTPases of the dynamin family mediate mitochondrial fission and fusion: mitofusins (MFN1 and MFN2) and optic atrophy 1 (OPA1) are required for the fusion of mitochondrial outer (OM) and inner membranes (IM), respectively; dynamin-related protein 1 (DRP1) mediates mitochondrial fission. Fission sites are marked by the ER, which closely associates with the OM, generating defined membrane domains to which DRP1 are recruited (Friedman et al., 2011; Murley et al., 2013).
Disturbances in the dynamic behavior of mitochondria cause various neurodegenerative diseases (Knott and Bossy-Wetzel, 2008; Itoh et al., 2013). Mutations in OPA1 cause dominant optic atrophy (Alexander et al., 2000; Delettre et al., 2000). The loss of OPA1 impairs mitochondrial fusion, perturbs cristae structure, and increases the susceptibility of cells toward apoptosis (Olichon et al., 2003; Cipolat et al., 2004, 2006; Lee et al., 2004; Meeusen et al., 2006). Overexpression of OPA1, however, protects against various apoptotic stimuli (Cipolat et al., 2006). The biogenesis of OPA1 is regulated both at the transcriptional and posttranscriptional level (Müller-Rischart et al., 2013). The alternative splicing of Opa1 pre-mRNA at exons 4, 4b, and 5b yields a total of eight isoforms expressed in a tissue-dependent manner (Delettre et al., 2001). These isoforms can modulate different functions of OPA1, as indicated by isoform-specific silencing of OPA1 variants (Olichon et al., 2007). The presence of proteolytic cleavage sites S1 and S2, encoded by exons 5 and 5b, respectively, introduces additional complexity (Ishihara et al., 2006). Proteolysis at these sites results in the loss of the transmembrane domain of OPA1 and leads to the formation of short OPA1 forms (S-OPA1). At steady state, mature OPA1 undergoes constitutive processing at S1 and S2, leading to the accumulation of noncleaved, long OPA1 (L-OPA1) and short OPA1 (S-OPA1) forms. Mitochondrial fusion is thought to depend on the presence of L- and S-OPA1 (Song et al., 2007), which assemble into oligomeric complexes maintaining cristae structure (Frezza et al., 2006; Yamaguchi et al., 2008). Various stress conditions including apoptotic stimulation disrupt these complexes and trigger the complete conversion of L-OPA1 into S-OPA1, inhibiting mitochondrial fusion (Duvezin-Caubet et al., 2006; Ishihara et al., 2006; Baricault et al., 2007; Song et al., 2007; Guillery et al., 2008). Ongoing fission events fragment the mitochondrial network, allowing the selective removal of damaged mitochondria by mitophagy or the progression of apoptosis (Youle and van der Bliek, 2012). Proteolysis of OPA1 is therefore crucial for mitochondrial integrity and quality control.
Recent evidence revealed that the IM peptidase OMA1 and the i-AAA protease YME1L cleave OPA1 at S1 and S2, respectively. Loss of OMA1 impairs OPA1 processing at S1 but does not grossly affect mitochondrial morphology (Ehses et al., 2009; Head et al., 2009). However, OMA1 is required for the stress-induced cleavage of OPA1 and mitochondrial fragmentation. OMA1-deficient mice are viable but suffer from diet-induced obesity and show impaired thermogenesis, which suggests important functions of OMA1 and stress-induced OPA1 processing for metabolic homeostasis (Quirós et al., 2012). Depletion of YME1L, however, impairs constitutive processing of OPA1 at S2 (Griparic et al., 2007; Song et al., 2007) and causes the fragmentation of the mitochondrial network, perturbs cristae morphogenesis, and renders cells susceptible to apoptosis (Stiburek et al., 2012).
To further define the role of YME1L and OMA1 in OPA1 processing and mitochondrial morphology, we generated cell lines lacking either or both proteases and examined the dynamic behavior of mitochondria. Our experiments demonstrate that OPA1 processing is dispensable for mitochondrial fusion and suggest a stimulatory role of S-OPA1 for mitochondrial fission.
Results and discussion
Long OPA1 forms are sufficient to mediate mitochondrial fusion
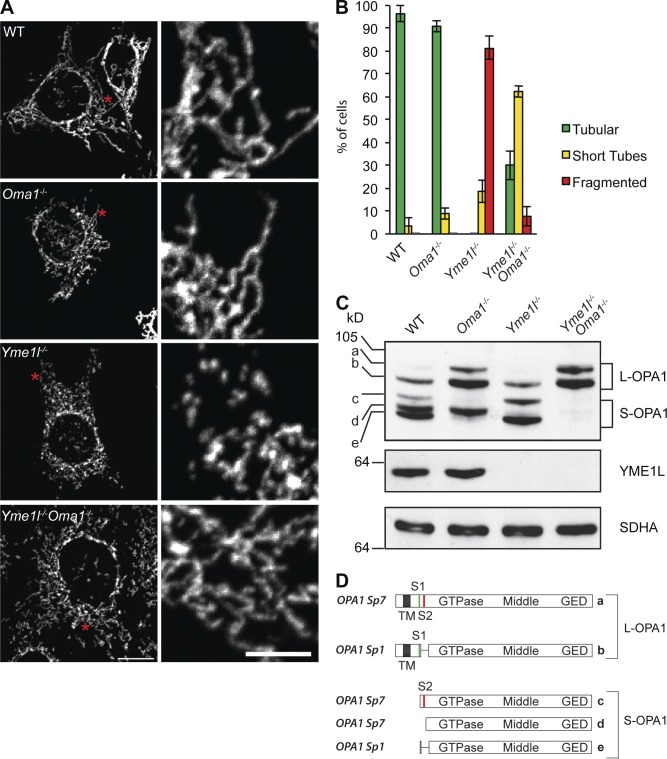
We generated mouse embryonic fibroblasts (MEFs) that were deficient for the proteases required for OPA1 processing, Yme1l−/−, Oma1−/−, and Yme1l−/−Oma1−/− (double knockout [DKO]). These cells propagated normally, which indicates that YME1L and OMA1 are dispensable for cell growth. As expected, Yme1l−/− cells showed fragmented mitochondria, whereas deletion of Oma1 did not grossly impair the mitochondrial network (Fig. 1, A and B). Surprisingly, we observed tubular mitochondria in DKO cells lacking both YME1L and OMA1 (Fig. 1, A and B). Mitochondria formed short tubules in DKO cells, which were different from the fragmented mitochondria of Yme1l−/− cells (Fig. 1, A and B). Mitochondrial tubulation in DKO cells depended on OPA1, as siRNA-mediated down-regulation of OPA1 led to mitochondrial fragmentation (Fig. S1, A and B). Although total OPA1 levels were moderately increased, steady-state levels of proteins involved in mitochondrial morphogenesis were unchanged in cells lacking both proteases (Fig. S1 C).
Figure 1.
Loss of OMA1 restores tubular mitochondria in Yme1l−/− cells. (A) Representative images of mitochondrial morphology in MEFs. The asterisks denote the area of magnification enlarged on the right. Bars: (left) 15 µm; (right) 5 µm. (B) Quantification of three independent experiments (error bars indicate mean ± SD), n ≥ 100. (C) Accumulation of OPA1 forms in MEFs lacking YME1L, OMA1, or both. Cells lysates were analyzed by SDS-PAGE analysis and immunoblotting using the indicated antibodies. a–e, OPA1 forms. (D) Schematic representation of mature L-OPA1 forms derived from splice variants 1 and 7 and S-OPA1 forms produced by cleavage at proteolytic sites S1 or S2 by OMA1 or YME1L, respectively.
We monitored OPA1 processing in cells lacking YME1L and OMA1 by immunoblotting. At least five distinct forms (a–e) of OPA1 generated from splice variants 1 and 7 accumulate in mitochondria in MEFs (Fig. 1, C and D). OMA1 cleaves L-OPA1 at S1 and generates S-OPA1 forms c and e (Ehses et al., 2009; Head et al., 2009; Quirós et al., 2012), which were absent in Oma1−/− cells (Fig. 1 C). However, the S-OPA1 form d produced by S2 cleavage of L-OPA1 was not present in Yme1l−/− cells (Fig. 1 C), as previously observed in YME1L-depleted cells (Song et al., 2007; Stiburek et al., 2012). We observed increased amounts of S-OPA1 forms c and e in Yme1l−/− cells, which indicates accelerated OPA1 processing at S1 by OMA1 (Fig. 1 C). Notably, DKO cells contained L-OPA1 forms but were completely devoid of S-OPA1 (Fig. 1 C). We therefore conclude that YME1L and OMA1 are the only proteases that process L-OPA1 and generate S-OPA1 forms under these conditions.
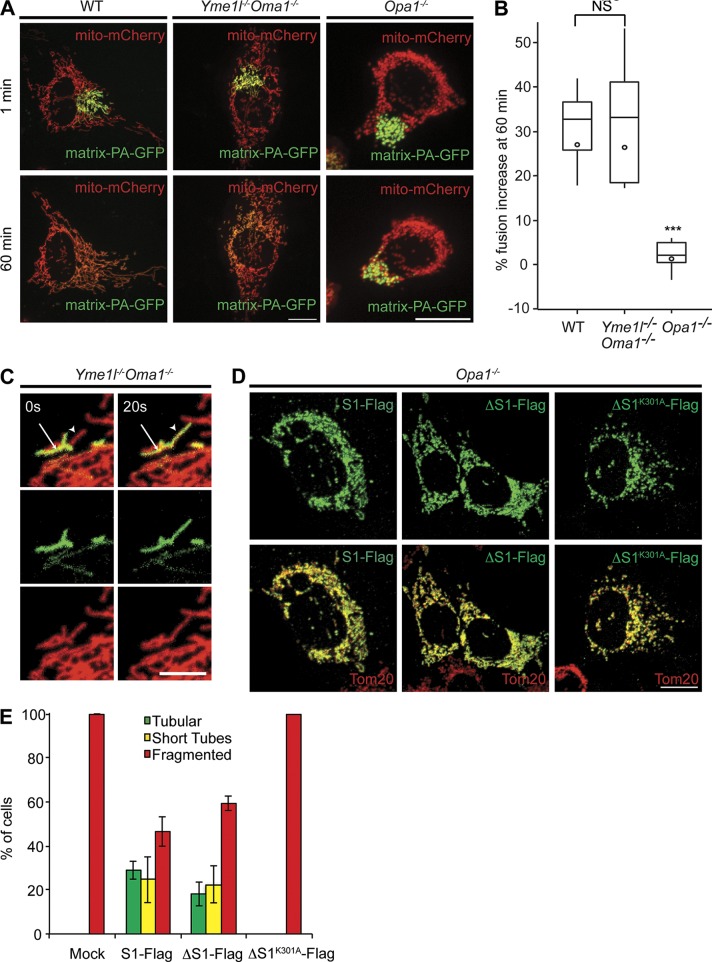
The presence of tubular mitochondria in DKO cells containing only L-OPA1 suggests that mitochondrial membranes can fuse. To directly assess mitochondrial fusion, we transiently expressed a photoactivatable GFP variant targeted to the mitochondrial matrix (matrix-PA-GFP) in wild type (WT) and DKO cells. Upon photoactivation of a small fraction of the mitochondrial network, we examined fusion of mitochondria by tracking the redistribution of GFP fluorescence over time (Fig. 2 A). We observed efficient mitochondrial fusion in DKO but not in Opa1−/− cells (Fig. 2, A and B; and Fig. S2 A). Mitochondrial fusion was also observed in DKO cells grown under respiring conditions (Fig. S2 B). We further substantiated the fusion competence of DKO cells by monitoring the fusion of individual mitochondria after photoactivation (Fig. 2 C). Moreover, assessment of mitochondrial fusion upon expression of PA-GFP targeted to the IM revealed that matrix as well as IM contents are efficiently exchanged in DKO cells (Fig. S2 C), which demonstrates that L-OPA1 is sufficient to mediate complete fusion of mitochondria.
Figure 2.
OPA1 processing is dispensable for mitochondrial fusion. (A and B) Photoactivatable GFP (matrix-PA-GFP) and mito-mCherry both targeted to the mitochondrial matrix were expressed in WT, Yme1l−/−Oma1−/−, and Opa1−/− MEFs (A). Fusion was monitored by the time-dependent dilution and redistribution of PA-GFP fluorescence (top, 1 min; bottom, 60 min). Bars, 15 µm. (B) Quantification of mitochondrial fusion. Results are represented in the form of a box plot: boxes represent data between the 25th and 75th percentiles, lines extend between the 10th and 90th percentiles, the horizontal line indicates the median, and the circle indicates the mean value (n ≥ 14; Yme1l−/−Oma1−/− vs. WT, P = 0.8; Yme1l−/−Oma1−/− vs. Opa1−/−, P = 1.6 × 10−12; WT vs. Opa1−/−, P = 4.6 × 10−8; ***, P ≤ 0.001). (C) Visualization of individual fusion events in Yme1l−/−Oma1−/− MEFs using matrix-PA-GFP and mito-mCherry. The arrowheads indicate mitochondria that have acquired photoactivated GFP due to fusion with photoactivated mitochondria. Bar, 5 µm. (D) Mitochondrial morphology in Opa1−/− MEFs expressing cleavable or noncleavable OPA1 forms. Flag-tagged variants of human OPA1 isoform 1 (S1-Flag), isoform 1 lacking the S1 site (ΔS1-Flag), or ΔS1-FlagK301A were transiently expressed in Opa1−/− cells. The mitochondrial network was visualized using a TOM20-specific antibody. Bar, 15 µm. (E) Quantification of three independent experiments (error bars indicate mean ± SD), n ≥ 100.
We expressed the human OPA1 isoform 1 and a variant thereof lacking S1 in Opa1−/− cells and assessed mitochondrial morphology. The loss of OPA1 results in the fragmentation of the mitochondrial network, which was at least partially restored upon expression of human isoform 1 of OPA1 (Fig. 2, D and E). Unlike previous observations (Song et al., 2007), deletion of S1 did not significantly affect the ability of OPA1 to maintain tubular mitochondria, which indicates that OPA1 processing is dispensable (Fig. 2, D and E). However, tubulation was not observed upon expression of noncleavable OPA1 harboring a GTPase mutation in Opa1−/− cells (Fig. 2, D and E).
These experiments indicate that L-OPA1 is sufficient to mediate mitochondrial fusion. L-OPA1 appears to be matured completely by the matrix-localized mitochondrial processing peptidase in DKO cells. Alkaline extraction of mitochondrial membranes at different pH indicated that membrane insertion of L-OPA1 was not significantly altered in DKO cells when compared with WT cells (Fig. S1 D). However, these experiments do not exclude the possibility that a small fraction of L-OPA1 fails to insert properly in the IM of DKO cells, thereby mimicking the function of S-OPA1. Nevertheless, our experiments in DKO cells indicate unambiguously that mitochondrial fusion does not depend on OPA1 processing. This is in agreement with previous OPA1 knockdown experiments in HeLa cells (Ishihara et al., 2006) and the observation that noncleavable L-OPA1 is sufficient to promote stress-induced mitochondrial hyperfusion (SIMH) in Opa1−/− cells (Tondera et al., 2009). Consistently, we observed SIMH in DKO cells (Fig. S2, D and E).
Long OPA1 forms preserve cristae morphology and apoptotic resistance
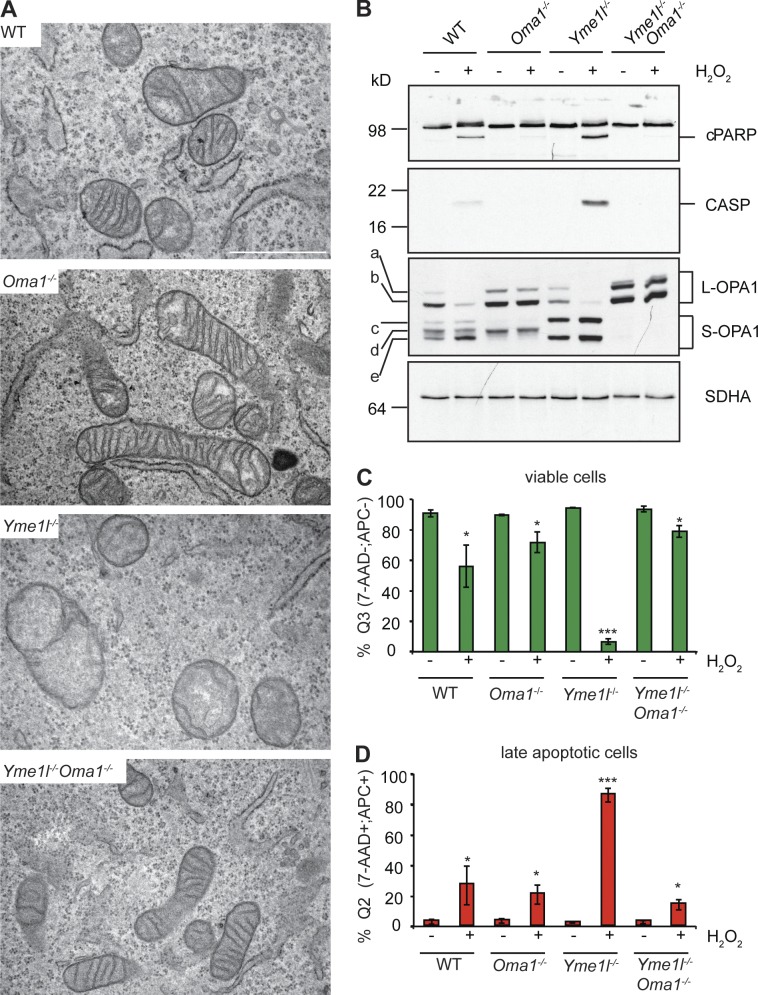
We examined the ultrastructure of mitochondria in DKO cells by transmission electron microscopy, as cristae morphology is known to depend on OPA1 (or Mgm1 in yeast; Olichon et al., 2003; Meeusen et al., 2006). We observed normally shaped cristae in Oma1−/− cells but severely disorganized and swollen cristae in Yme1l−/− cells (Fig. 3 A), as previously observed in YME1L-depleted HEK293T cells (Stiburek et al., 2012). Strikingly, the mitochondrial ultrastructure was completely restored in DKO cells (Fig. 3 A), demonstrating that L-OPA1 is sufficient to maintain the tubular, sheet-like structure of mitochondrial cristae.
Figure 3.
Loss of OMA1 restores cristae morphology and apoptotic resistance of Yme1l−/− cells. (A) WT, Oma1−/−, Yme1l−/−, and Yme1l−/−Oma1−/− MEFs were analyzed by transmission electron microscopy. Bar, 1 µm. (B–D) WT and protease-deficient MEFs were exposed to H2O2 to induce apoptosis. (B) Immunoblot analysis of apoptotic marker proteins (cleaved PARP, cPARP; caspase 3, CASP). (C and D) Flow cytometry analysis of viable cells (Q3: 7-AAD−;APC−) and late apoptotic cells (Q2: 7-AAD+; APC+). *, P ≤ 0.05; ***, P ≤ 0.001. Error bars indicate mean ± SD.
Apoptosis is associated with the reshaping of mitochondrial cristae controlled by OPA1 (Olichon et al., 2003; Frezza et al., 2006; Merkwirth et al., 2008; Yamaguchi et al., 2008). Overexpression of OPA1 confers apoptotic resistance (Frezza et al., 2006), whereas loss of OPA1 (Cipolat et al., 2006) or the selective loss of L-OPA1 alone (Merkwirth et al., 2008) increases the apoptotic sensitivity of cells. We therefore assessed the susceptibility of cells lacking YME1L, OMA1, or both to H2O2-induced apoptosis, monitoring the accumulation of apoptotic markers (Fig. 3, B–D). Cells depleted of YME1L were reported to be more susceptible to apoptosis, whereas loss of OMA1 protects cells against apoptosis (Quirós et al., 2012; Stiburek et al., 2012). Consistently, cleaved poly ADP-ribose polymerase (cPARP) and activated caspase 3 accumulated in H2O2-treated Yme1l−/− cells but not in Oma1−/− cells, whose apoptotic sensitivity was decreased when compared with WT cells (Fig. 3, B–D). Deletion of Oma1 protected Yme1l−/− cells against apoptosis (Fig. 3, B–D). We therefore conclude that L-OPA1 confers apoptotic resistance to cells.
Short OPA1 forms trigger mitochondrial fragmentation in Yme1l−/− cells and colocalize with the fission machinery
Our findings raise the question of why fragmented mitochondria accumulate in Yme1l−/− cells that contain similar levels of L-OPA1 as WT cells (Fig. 1 C). We noted a striking correlation of the accumulation of S-OPA1 with mitochondrial fragmentation and apoptosis. Although a tubular network is preserved in Oma1−/− cells lacking S-OPA1 forms c and e, mitochondrial fragmentation in Yme1l−/− cells is associated with an accumulation of these S-OPA1 forms generated by OMA1 (Figs. 1 C and 3 B). This effect is even more pronounced upon H2O2-induced apoptosis, which destabilizes L-OPA1 and leads to the formation of S-OPA1 by OMA1 in WT and Yme1l−/− cells (Fig. 3 B). We therefore reasoned that the accumulation of S-OPA1 may trigger mitochondrial fragmentation in the absence of YME1L, either by inhibiting fusion or by stimulating fission.
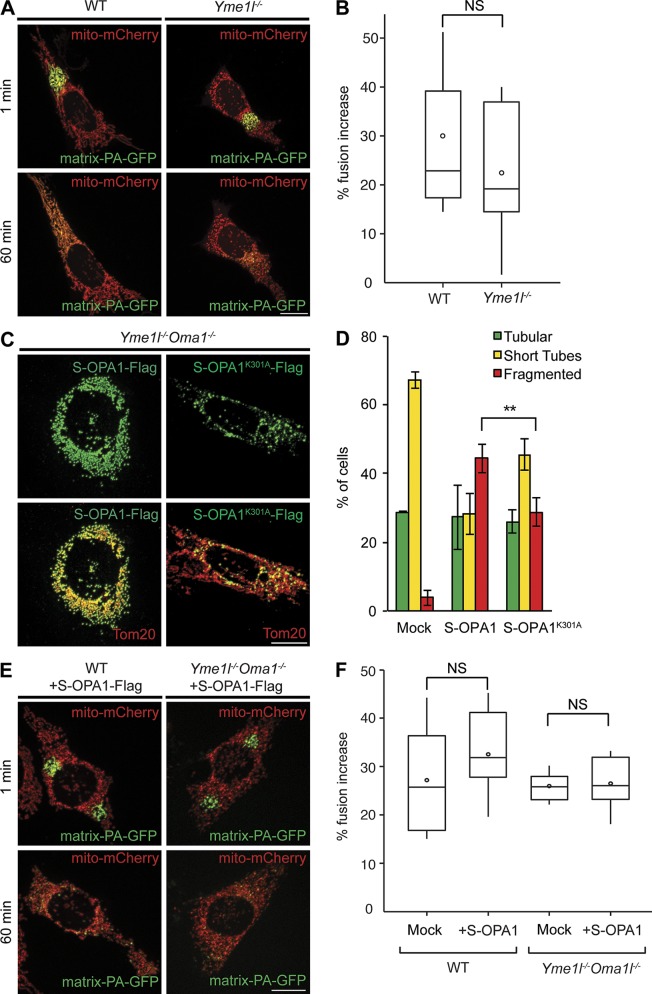
Mitochondrial fusion proceeded normally in Yme1l−/− cells (Fig. 4, A and B), which is consistent with recent YME1L knockdown experiments (Ruan et al., 2013). To examine if S-OPA1 can induce mitochondrial fission, we ectopically expressed a hybrid protein of S-OPA1 and the amino-terminal region of apoptosis-inducing factor (AIF) containing the mitochondrial targeting signal (Ishihara et al., 2006). A significant proportion of WT and DKO cells expressing S-OPA1 showed mitochondrial fragmentation (Fig. 4, C and D; and Fig. S3 A). S-OPA1 forms c and d fragment mitochondria, which indicates a similar activity of different S-OPA1 forms (Fig. S3 A). Moreover, mitochondrial fragmentation was observed upon expression of S-OPA1 in HEK293T cells (Fig. S3, B–E). Thus, although loss of OMA1, and concomitantly S-OPA1 forms c and e, restored a tubular network in the absence of YME1L, reintroduction of S-OPA1 in DKO cells triggered mitochondrial fragmentation. Notably, this is not a canonical stress response, as fragmented mitochondria in DKO cells expressing S-OPA1 can fuse (Fig. 4, E and F) and retain normal membrane potential (Fig. S3, F and G). These experiments therefore strongly suggest that mitochondrial fragmentation in Yme1l−/− cells is caused by an unbalanced processing of OPA1 and indicate that S-OPA1 is functionally linked to mitochondrial fission.
Figure 4.
The function of short OPA1 forms is linked to mitochondrial fission. (A) Mitochondria fuse in Yme1−/− cells. Mitochondrial fusion was analyzed as in Fig. 2 using matrix-PA-GFP. (B) Quantification of mitochondrial fusion in Yme1l−/− cells after 60 min. Results are represented in the form of a box plot (see Fig. 2). WT versus Yme1l−/− cells, P = 0.14. (C) Expression of S-OPA1 forms in Yme1l−/−Oma1−/− cells induces mitochondrial fragmentation. Mitochondrial morphology was assessed in Yme1l−/−Oma1−/− MEFs expressing Flag-tagged variants of rat S-OPA1-Flag or S-OPA1K301A-Flag (bottom) harboring a mutation in the GTPase domain of OPA1. (D) Quantification of three independent experiments (error bars indicate mean ± SD). n ≥ 100; **, P ≥ 0.01. (E) Mitochondrial fusion occurs in fragmented mitochondria of WT and Yme1l−/−Oma1−/− cells expressing S-OPA1-Flag. (F) Quantification of mitochondrial fusion after 60 min. Results are shown in the form of a box plot (see Fig. 2). Shown are WT/Yme1l−/−Oma1−/− cells versus WT/Yme1l−/−Oma1−/− cells expressing rat S-OPA1. Bars, 15 µm.
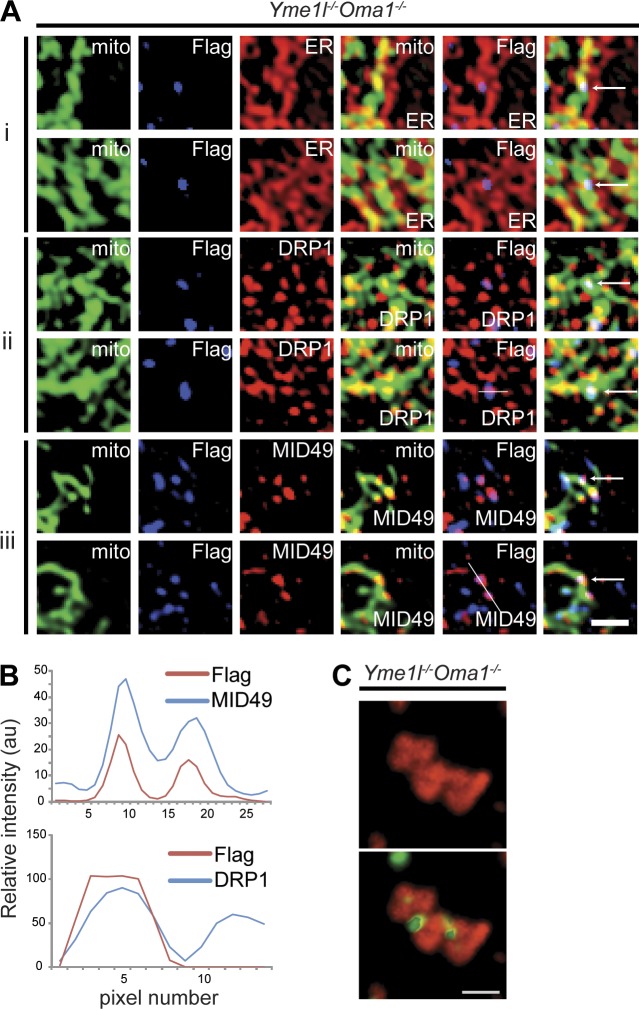
We expressed mitochondrially targeted S-OPA1K301A, harboring a mutation in the GTPase domain of OPA1, in DKO cells. Tubular mitochondria were preserved in a significantly increased proportion of cells when compared with DKO cells expressing S-OPA1 (Fig. 4, C and D; and Fig. S3, B and C). Mitochondrial fragmentation upon overexpression of S-OPA1K301A may reflect intermolecular complementation of the GTPase activity by endogenous L-OPA1 within an OPA1 oligomer, as has been described previously for yeast Mgm1 (DeVay et al., 2009; Zick et al., 2009). Interestingly, we observed that S-OPA1K301A assembled into punctate structures in mitochondria, as opposed to S-OPA1 or noncleavable L-OPA1K301A, which were uniformly dispersed within the organelle (Figs. 4 C and S3 A). A similar punctate distribution has been observed for the mitochondrial fission machinery, which is recruited to mitochondrial fission sites marked by the ER (Friedman et al., 2011; Murley et al., 2013). We therefore visualized ER–mitochondria contact sites by fluorescence microscopy of DKO cells and assessed the degree of colocalization of S-OPA1K301A punctae with these sites. Strikingly, ∼50% of these punctae localized at ER–mitochondria contact sites (Fig. 5 A), where fission of mitochondrial membranes occurs. A punctate distribution at mitochondrial division sites has also been observed for DRP1 (Sesaki and Jensen, 1999; Smirnova et al., 2001) and the DRP1 recruitment factor MID49 (Palmer et al., 2011; Zhao et al., 2011). Fluorescence microscopy of DKO cells expressing S-OPA1K301A revealed colocalization of ∼15% or ∼30% of the S-OPA1K301A containing punctae with DRP1 or MID49 spots, respectively (Fig. 5, A and B). Moreover, S-OPA1K301A punctae are conspicuously present at sites of mitochondrial constriction (Fig. 5 C).
Figure 5.
GTPase-inactive short OPA1 forms colocalize with sites of mitochondrial division. (A) S-OPA1K301A-Flag expressed in Yme1l−/−Oma1−/− cells assembles into punctae partially colocalizing with ER–mitochondria contact sites (i; a total of 90 spots in five different cells), endogenous DRP1 (ii; a total of 230 spots in five different cells), and endogenous MID49 (iii; a total of 186 spots in six different cells). Bars, 2 µm. (B) Line scan of ii and iii along the lines indicated in A. (C) S-OPA1K301A-Flag punctae coalesce at sites of mitochondria constriction. Mitochondria are visualized using mito-mCherry, the ER using GRP78-specific antibodies. Bar, 2 µm.
Collectively, several lines of evidence support a stimulatory role of S-OPA1 for mitochondrial fission: First, we observed a striking correlation between the accumulation of S-OPA1 and mitochondrial fragmentation in cells lacking YME1L, although fusion of mitochondrial membranes proceeds normally in these cells. Second, ectopic expression of S-OPA1 in DKO cells induces fragmentation of the mitochondrial network, which appears to occur in a GTPase-dependent manner and can be mediated by different S-OPA1 forms. Third, S-OPA1K301A assembles into punctate structures that partially colocalize with ER–mitochondria contact sites, mitochondrial constriction sites, and foci containing DRP1 or MID49. How S-OPA1 affects mitochondrial fission remains to be elucidated. Mitochondrial division occurs in Opa1−/− cells and thus does not depend on S-OPA1. However, S-OPA1 may stimulate membrane fission and contribute to the coordination of mitochondrial fusion and fission. Indeed, an increased probability for membrane fission has been observed after a fusion event in various cell types (Twig et al., 2008; Wang et al., 2012; Cagalinec et al., 2013). It is conceivable that S-OPA1 interacts with the fission machinery at the OM or is involved in IM fission. A stimulatory role of S-OPA1 in mitochondrial fission may be crucial under stress conditions or upon induction of apoptosis and/or mitophagy and is therefore likely to be of particular importance for mitochondrial quality control (Youle and van der Bliek, 2012). In agreement with an active role of S-OPA1 for stress-induced mitochondrial fission, OMA1 is activated under stress, resulting in the increased formation of S-OPA1 forms c and/or e (e.g., in apoptotic cells; Fig. 3 B). Inhibition of stress-induced OPA1 processing in cells lacking OMA1 is thus likely to affect both mitochondrial fusion and fission: while L-OPA1 is stabilized and fusion preserved, the formation of S-OPA1 forms c and e is prevented, resulting in the maintenance of a tubular mitochondrial network and protection against apoptosis. Loss of YME1L, however, impairs constitutive OPA1 processing at S2 but leads to the OMA1-dependent accumulation of S-OPA1 forms c and e and mitochondrial fragmentation. Our analyses of YME1L- and OMA1-deficient cells thus revealed an unexpected role of OPA1 processing in mitochondrial fusion–fission dynamics, which determine the shape, size, and distribution of these organelles within a cell and thus are paramount to cell physiology.
Materials and methods
Generation and culture of cell lines
Yme1lloxP/loxP and Oma1loxP/loxP conditional mice were generated by conventional ESC targeting using pRapidFlirt constructs designed to insert intronic LoxP sites flanking exon 3 of either Yme1l or Oma1. Germline chimeras were generated on a C57Bl6/N background and subsequently intercrossed to generate Yme1lloxP/loxPOma1loxP/loxP animals. Primary fibroblasts were isolated from Yme1lloxP/loxP, Oma1loxP/loxP, and Yme1lloxP/loxPOma1loxP/loxP embryonic day 13.5 embryos and immortalized using a plasmid encoding SV40 large T antigen. Immortalized fibroblasts were transduced with recombinant His-TAT-NLS-Cre (HTNC) fusion protein (4 µM), incubated for 20 h, washed with PBS, and supplemented with growth medium. Individual clones were sorted by flow cytometry into 96-well plates (FACS Aria III; BD) using the FASC DIVA software (BD), expanded, and subsequently genotyped for deletion of Yme1l and/or Oma1 by PCR and immunoblotting. MEFs were grown in DMEM + GlutaMAX (Life Technologies) supplemented with sodium pyruvate (Gibco), nonessential amino acids (Gibco), and 10% fetal calf serum (Life Technologies). Cells were cultured at 37°C in a humidified incubator with 5% CO2. Respiring media consisted of DMEM without glucose supplemented with 10% FBS, nonessential amino acids, and galactose (10 mM). Opa1−/− cells were grown in glucose-containing media supplemented with 50 µg/ml uridine. S-OPA1 (1–97 amino acids of rat AIF fused to 230–997 amino acids of rat OPA1) and the mutant variant S-OPA1K301A thereof were stably expressed in Flp-In T-Rex HEK293T cells (Invitrogen) under the control of a tetracycline-regulatable promoter according to the manufacturer’s instructions.
Confocal fluorescence microscopy
To monitor mitochondrial morphology by immunofluorescence microscopy, cells were stained with antibodies directed against cytochrome c (anti–mouse 1:500; BD) or TOM20 (anti–rabbit 1:500; Santa Cruz Biotechnology, Inc.) or directly visualized with mito-mCherry. The ER was visualized with antibodies directed against GRP78 (anti–mouse 1:200; Abcam), DRP1 foci with DRP1-specific antibodies (anti–mouse 1:200; BD), MID49 foci with MID49-specific antibody (anti–rabbit 1:50; Proteintech), and transiently expressed OPA1 variants using antibodies directed against the Flag epitope (anti–mouse 1:500; Sigma-Aldrich). Cells were fixed with paraformaldehyde (4% [wt/vol] in PBS) for 15 min and permeabilized using 0.15% (vol/vol) Triton X-100. 3% (wt/vol) bovine serum albumin in PBS was used for blocking and to dilute primary and secondary antibodies. Fluorescently coupled secondary antibodies Alexa Fluor 488 (goat anti–mouse and goat anti–rabbit), Alexa Fluor 568 (goat anti–mouse and goat anti–rabbit), and Alexa Fluor 633 (goat anti–mouse) were used at 1:1,000 dilution (Invitrogen). Images were acquired using an UltraVIEW VoX spinning disc (CSU-X1; Yokogawa Corporation of America) confocal microscope (PerkinElmer) on a microscope (Ti; Nikon) equipped with a 60× (Apochromat total internal reflection fluorescence 60× oil, NA 1.49) objective lens and camera (EMCCD C9100-50; CamLink). Live cells were imaged in DMEM medium at 37°C with 5% CO2. Standard image processing was performed using Volocity (PerkinElmer) and ImageJ (Schneider et al., 2012). No image manipulation was performed, except a background subtraction in Fig. 1 (ImageJ, rolling ball with 10 pixel radius). Images in Fig. 5 were deconvolved using Huygens Professional with the “Classical Maximum Likelihood Estimation (CMLE) algorithm” and measured point spread functions of 488-, 546-, and 634-nm channels (Scientific Volume Imaging) before manual analysis for colocalization. For colocalization analyses, S-OPA1 punctae near the periphery of the cell were scored for their 3D coincidence with DRP1, MID49, or ER–mitochondria contact sites using Imaris (Bitplane). 3D visualization in Fig. 5 C was performed in Volocity (PerkinElmer) using “3D opacity view.”
Assessing mitochondrial fusion
5 × 104 cells were transfected with 1 µg matrix-PA-GFP and 0.5 µg mito-mCherry, and imaged after 24 h. Approximately 23 ± 12% of mitochondria were photoactivated using 5% power of a 405-nm laser for one cycle (30 iterations). Z stack images of PA-GFP and mito-mCherry using a step size of 0.3 µm were acquired using 488-nm and 568-nm lasers immediately after photoactivation (1 min delay) and after 20, 40, or 60 min. We thank O. Shirihai (Boston University, Boston, MA) for the plasmid encoding IM-PA-GFP and A. Kumar Kondadi (University of Cologne, Cologne, Germany) for the plasmid encoding mito-mCherry.
Mitochondrial fusion was quantified using a similar approach to one described previously (Zunino et al., 2007). The relative amounts of mitochondria containing PA-GFP were compared directly after activation (1 min) and after the indicated time points. The total volume of mitochondria was obtained by thresholding image stacks of mito-mCherry and the volume of mitochondria containing PA-GFP by thresholding the image stacks of PA-GFP. Alterations in the percentage of mitochondria containing PA-GFP were calculated for each time point to determine mitochondrial fusion. Thresholding and image analysis was performed using Volocity (PerkinElmer). Data were represented with boxplots. Boxes represent data between the 25th and 75th percentiles, lines extend between the 10th and 90th percentiles, the horizontal line indicates the median value, and the circle the mean value. Changes in fusion were evaluated by a Student’s t test.
Transmission electron microscopy
MEFs were prepared for transmission electron microscopy analysis as previously described (Wakabayashi et al., 2009). MEFs were fixed for 1 h in prewarmed 2% (vol/vol) glutaraldehyde, 2.5% (wt/vol) sucrose, 3 mM CaCl2, and 100 mM Hepes-KOH, pH 7.4. After washes, MEFs were postfixed using reduced OsO4 (1% [wt/vol] OsO4, 10 mg/ml potassium ferrocyanide, 1.25% [wt/vol] sucrose, and 100 mM sodium cacodylate, pH 7.4) for 1 h on ice. After washes in water, cells were incubated in 2% (wt/vol) uranyl acetate for 30 min. After dehydration using 50, 70, 90, and 100% ethanol, samples were embedded in epon resin. Samples were observed under a transmission electron microscope (EM902; Carl Zeiss) at an acceleration voltage of 80 kV, and micrographs were generated at 20,000×.
Analysis of H2O2-induced apoptosis
To induce apoptosis, cells were incubated in the presence of 1 mM H2O2 for 7 h. Double staining with annexin V (AV) and 7-AAD, as described previously, assessed apoptotic cell death (Rappl et al., 2001). Cells were analyzed by flow cytometry (FACS Aria III) using the FASC DIVA software (BD). The accumulation of apoptotic marker proteins, cPARP, and activated caspase 3 (CASP3) was examined in cell lysates by immunoblotting.
Online supplemental material
Fig. S1 shows that the formation of tubular mitochondria in Yme1l−/−Oma1−/− cells depends on OPA1. Fig. S2 shows mitochondrial fusion in Yme1l−/−Oma1−/− cells. Fig. S3 shows that ectopic S-OPA1 expression induces mitochondrial fragmentation but does not cause general mitochondrial dysfunction. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201308006/DC1.
Supplementary Material
Acknowledgments
We are grateful to Dr. Gunter Rappl for cell sorting, Esther Barth and Gudrun Zimmer for expert technical assistance, and Christian Jüngst for help with the imaging software. We thank Dr. Orian Shirihai for the plasmid encoding IM-PA-GFP, Dr. Katsuyoshi Mihara for the plasmid encoding rat S-OPA1, Arun Kumar Kondadi for the plasmid encoding mito-mCherry, Dr. Luigi Palmieri for the anti-SLC25A13 antibody, and Dr. Jean-Claude Martinou for the Opa1−/− cells.
This work was supported by post-doctoral fellowships by the Human Frontier Science Program (HFSP; LT000887/2010-L) to T. Wai and by the Alexander von Humboldt Foundation to M.J. Baker, and grants of the Deutsche Forschungsgemeinschaft and the European Research Council (AdG No. 233078) to T. Langer.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- AIF
- apoptosis-inducing factor
- cPARP
- cleaved poly ADP-ribose polymerase
- DKO
- double knockout
- IM
- inner membrane
- MEF
- mouse embryonic fibroblast
- OM
- outer membrane
- WT
- wild type
References
- Alexander C., Votruba M., Pesch U.E., Thiselton D.L., Mayer S., Moore A., Rodriguez M., Kellner U., Leo-Kottler B., Auburger G., et al. 2000. OPA1, encoding a dynamin-related GTPase, is mutated in autosomal dominant optic atrophy linked to chromosome 3q28. Nat. Genet. 26:211–215 10.1038/79944 [DOI] [PubMed] [Google Scholar]
- Baricault L., Ségui B., Guégand L., Olichon A., Valette A., Larminat F., Lenaers G. 2007. OPA1 cleavage depends on decreased mitochondrial ATP level and bivalent metals. Exp. Cell Res. 313:3800–3808 10.1016/j.yexcr.2007.08.008 [DOI] [PubMed] [Google Scholar]
- Cagalinec M., Safiulina D., Liiv M., Liiv J., Choubey V., Wareski P., Veksler V., Kaasik A. 2013. Principles of the mitochondrial fusion and fission cycle in neurons. J. Cell Sci. 126:2187–2197 10.1242/jcs.118844 [DOI] [PubMed] [Google Scholar]
- Chan D.C. 2012. Fusion and fission: interlinked processes critical for mitochondrial health. Annu. Rev. Genet. 46:265–287 10.1146/annurev-genet-110410-132529 [DOI] [PubMed] [Google Scholar]
- Cipolat S., Martins de Brito O., Dal Zilio B., Scorrano L. 2004. OPA1 requires mitofusin 1 to promote mitochondrial fusion. Proc. Natl. Acad. Sci. USA. 101:15927–15932 10.1073/pnas.0407043101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipolat S., Rudka T., Hartmann D., Costa V., Serneels L., Craessaerts K., Metzger K., Frezza C., Annaert W., D’Adamio L., et al. 2006. Mitochondrial rhomboid PARL regulates cytochrome c release during apoptosis via OPA1-dependent cristae remodeling. Cell. 126:163–175 10.1016/j.cell.2006.06.021 [DOI] [PubMed] [Google Scholar]
- Delettre C., Lenaers G., Griffoin J.M., Gigarel N., Lorenzo C., Belenguer P., Pelloquin L., Grosgeorge J., Turc-Carel C., Perret E., et al. 2000. Nuclear gene OPA1, encoding a mitochondrial dynamin-related protein, is mutated in dominant optic atrophy. Nat. Genet. 26:207–210 10.1038/79936 [DOI] [PubMed] [Google Scholar]
- Delettre C., Griffoin J.M., Kaplan J., Dollfus H., Lorenz B., Faivre L., Lenaers G., Belenguer P., Hamel C.P. 2001. Mutation spectrum and splicing variants in the OPA1 gene. Hum. Genet. 109:584–591 10.1007/s00439-001-0633-y [DOI] [PubMed] [Google Scholar]
- DeVay R.M., Dominguez-Ramirez L., Lackner L.L., Hoppins S., Stahlberg H., Nunnari J. 2009. Coassembly of Mgm1 isoforms requires cardiolipin and mediates mitochondrial inner membrane fusion. J. Cell Biol. 186:793–803 10.1083/jcb.200906098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvezin-Caubet S., Jagasia R., Wagener J., Hofmann S., Trifunovic A., Hansson A., Chomyn A., Bauer M.F., Attardi G., Larsson N.G., et al. 2006. Proteolytic processing of OPA1 links mitochondrial dysfunction to alterations in mitochondrial morphology. J. Biol. Chem. 281:37972–37979 10.1074/jbc.M606059200 [DOI] [PubMed] [Google Scholar]
- Ehses S., Raschke I., Mancuso G., Bernacchia A., Geimer S., Tondera D., Martinou J.-C., Westermann B., Rugarli E.I., Langer T. 2009. Regulation of OPA1 processing and mitochondrial fusion by m-AAA protease isoenzymes and OMA1. J. Cell Biol. 187:1023–1036 10.1083/jcb.200906084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar-Henriques M., Anton F. 2013. Mechanistic perspective of mitochondrial fusion: tubulation vs. fragmentation. Biochim. Biophys. Acta. 1833:162–175 10.1016/j.bbamcr.2012.07.016 [DOI] [PubMed] [Google Scholar]
- Frezza C., Cipolat S., Martins de Brito O., Micaroni M., Beznoussenko G.V., Rudka T., Bartoli D., Polishuck R.S., Danial N.N., De Strooper B., Scorrano L. 2006. OPA1 controls apoptotic cristae remodeling independently from mitochondrial fusion. Cell. 126:177–189 10.1016/j.cell.2006.06.025 [DOI] [PubMed] [Google Scholar]
- Friedman J.R., Lackner L.L., West M., DiBenedetto J.R., Nunnari J., Voeltz G.K. 2011. ER tubules mark sites of mitochondrial division. Science. 334:358–362 10.1126/science.1207385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griparic L., Kanazawa T., van der Bliek A.M. 2007. Regulation of the mitochondrial dynamin-like protein Opa1 by proteolytic cleavage. J. Cell Biol. 178:757–764 10.1083/jcb.200704112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillery O., Malka F., Landes T., Guillou E., Blackstone C., Lombès A., Belenguer P., Arnoult D., Rojo M. 2008. Metalloprotease-mediated OPA1 processing is modulated by the mitochondrial membrane potential. Biol. Cell. 100:315–325 10.1042/BC20070110 [DOI] [PubMed] [Google Scholar]
- Head B., Griparic L., Amiri M., Gandre-Babbe S., van der Bliek A.M. 2009. Inducible proteolytic inactivation of OPA1 mediated by the OMA1 protease in mammalian cells. J. Cell Biol. 187:959–966 10.1083/jcb.200906083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara N., Fujita Y., Oka T., Mihara K. 2006. Regulation of mitochondrial morphology through proteolytic cleavage of OPA1. EMBO J. 25:2966–2977 10.1038/sj.emboj.7601184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh K., Nakamura K., Iijima M., Sesaki H. 2013. Mitochondrial dynamics in neurodegeneration. Trends Cell Biol. 23:64–71 10.1016/j.tcb.2012.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott A.B., Bossy-Wetzel E. 2008. Impairing the mitochondrial fission and fusion balance: a new mechanism of neurodegeneration. Ann. N. Y. Acad. Sci. 1147:283–292 10.1196/annals.1427.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.J., Jeong S.Y., Karbowski M., Smith C.L., Youle R.J. 2004. Roles of the mammalian mitochondrial fission and fusion mediators Fis1, Drp1, and Opa1 in apoptosis. Mol. Biol. Cell. 15:5001–5011 10.1091/mbc.E04-04-0294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liesa M., Shirihai O.S. 2013. Mitochondrial dynamics in the regulation of nutrient utilization and energy expenditure. Cell Metab. 17:491–506 10.1016/j.cmet.2013.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Weaver D., Shirihai O., Hajnóczky G. 2009. Mitochondrial ‘kiss-and-run’: interplay between mitochondrial motility and fusion-fission dynamics. EMBO J. 28:3074–3089 10.1038/emboj.2009.255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeusen S., DeVay R., Block J., Cassidy-Stone A., Wayson S., McCaffery J.M., Nunnari J. 2006. Mitochondrial inner-membrane fusion and crista maintenance requires the dynamin-related GTPase Mgm1. Cell. 127:383–395 10.1016/j.cell.2006.09.021 [DOI] [PubMed] [Google Scholar]
- Merkwirth C., Dargazanli S., Tatsuta T., Geimer S., Löwer B., Wunderlich F.T., von Kleist-Retzow J.C., Waisman A., Westermann B., Langer T. 2008. Prohibitins control cell proliferation and apoptosis by regulating OPA1-dependent cristae morphogenesis in mitochondria. Genes Dev. 22:476–488 10.1101/gad.460708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller-Rischart A.K., Pilsl A., Beaudette P., Patra M., Hadian K., Funke M., Peis R., Deinlein A., Schweimer C., Kuhn P.H., et al. 2013. The E3 ligase parkin maintains mitochondrial integrity by increasing linear ubiquitination of NEMO. Mol. Cell. 49:908–921 10.1016/j.molcel.2013.01.036 [DOI] [PubMed] [Google Scholar]
- Murley A., Lackner L.L., Osman C., West M., Voeltz G.K., Walter P., Nunnari J. 2013. ER-associated mitochondrial division links the distribution of mitochondria and mitochondrial DNA in yeast. Elife. 2:e00422 10.7554/eLife.00422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunnari J., Suomalainen A. 2012. Mitochondria: in sickness and in health. Cell. 148:1145–1159 10.1016/j.cell.2012.02.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olichon A., Baricault L., Gas N., Guillou E., Valette A., Belenguer P., Lenaers G. 2003. Loss of OPA1 perturbates the mitochondrial inner membrane structure and integrity, leading to cytochrome c release and apoptosis. J. Biol. Chem. 278:7743–7746 10.1074/jbc.C200677200 [DOI] [PubMed] [Google Scholar]
- Olichon A., Elachouri G., Baricault L., Delettre C., Belenguer P., Lenaers G. 2007. OPA1 alternate splicing uncouples an evolutionary conserved function in mitochondrial fusion from a vertebrate restricted function in apoptosis. Cell Death Differ. 14:682–692 10.1038/sj.cdd.4402048 [DOI] [PubMed] [Google Scholar]
- Otera H., Ishihara N., Mihara K. 2013. New insights into the function and regulation of mitochondrial fission. Biochim. Biophys. Acta. 1833:1256–1268 10.1016/j.bbamcr.2013.02.002 [DOI] [PubMed] [Google Scholar]
- Palmer C.S., Osellame L.D., Laine D., Koutsopoulos O.S., Frazier A.E., Ryan M.T. 2011. MiD49 and MiD51, new components of the mitochondrial fission machinery. EMBO Rep. 12:565–573 10.1038/embor.2011.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quirós P.M., Ramsay A.J., Sala D., Fernández-Vizarra E., Rodríguez F., Peinado J.R., Fernández-García M.S., Vega J.A., Enríquez J.A., Zorzano A., López-Otín C. 2012. Loss of mitochondrial protease OMA1 alters processing of the GTPase OPA1 and causes obesity and defective thermogenesis in mice. EMBO J. 31:2117–2133 10.1038/emboj.2012.70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappl G., Abken H., Hasselmann D.O., Tilgen W., Ugurel S., Reinhold U. 2001. The CD7− subset of CD4+ memory T cells is prone to accelerated apoptosis that is prevented by interleukin-15 (IL-15). Cell Death Differ. 8:395–402 10.1038/sj.cdd.4400825 [DOI] [PubMed] [Google Scholar]
- Ruan Y., Li H., Zhang K., Jian F., Tang J., Song Z. 2013. Loss of Yme1L perturbates mitochondrial dynamics. Cell Death Dis. 4:e896 10.1038/cddis.2013.414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., Eliceiri K.W. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 9:671–675 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sesaki H., Jensen R.E. 1999. Division versus fusion: Dnm1p and Fzo1p antagonistically regulate mitochondrial shape. J. Cell Biol. 147:699–706 10.1083/jcb.147.4.699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova E., Griparic L., Shurland D.L., van der Bliek A.M. 2001. Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol. Biol. Cell. 12:2245–2256 10.1091/mbc.12.8.2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z., Chen H., Fiket M., Alexander C., Chan D.C. 2007. OPA1 processing controls mitochondrial fusion and is regulated by mRNA splicing, membrane potential, and Yme1L. J. Cell Biol. 178:749–755 10.1083/jcb.200704110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiburek L., Cesnekova J., Kostkova O., Fornuskova D., Vinsova K., Wenchich L., Houstek J., Zeman J. 2012. YME1L controls the accumulation of respiratory chain subunits and is required for apoptotic resistance, cristae morphogenesis, and cell proliferation. Mol. Biol. Cell. 23:1010–1023 10.1091/mbc.E11-08-0674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura Y., Itoh K., Sesaki H. 2011. SnapShot: Mitochondrial dynamics. Cell. 145:1158–1158: e1 10.1016/j.cell.2011.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tondera D., Grandemange S., Jourdain A., Karbowski M., Mattenberger Y., Herzig S., Da Cruz S., Clerc P., Raschke I., Merkwirth C., et al. 2009. SLP-2 is required for stress-induced mitochondrial hyperfusion. EMBO J. 28:1589–1600 10.1038/emboj.2009.89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twig G., Elorza A., Molina A.J., Mohamed H., Wikstrom J.D., Walzer G., Stiles L., Haigh S.E., Katz S., Las G., et al. 2008. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 27:433–446 10.1038/sj.emboj.7601963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Bliek A.M., Shen Q., Kawajiri S. 2013. Mechanisms of mitochondrial fission and fusion. Cold Spring Harb. Perspect. Biol. 5:a011072 10.1101/cshperspect.a011072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi J., Zhang Z., Wakabayashi N., Tamura Y., Fukaya M., Kensler T.W., Iijima M., Sesaki H. 2009. The dynamin-related GTPase Drp1 is required for embryonic and brain development in mice. J. Cell Biol. 186:805–816 10.1083/jcb.200903065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Xiao W., Shan S., Jiang C., Chen M., Zhang Y., Lü S., Chen J., Zhang C., Chen Q., Long M. 2012. Multi-patterned dynamics of mitochondrial fission and fusion in a living cell. PLoS ONE. 7:e19879 10.1371/journal.pone.0019879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann B. 2010. Mitochondrial fusion and fission in cell life and death. Nat. Rev. Mol. Cell Biol. 11:872–884 10.1038/nrm3013 [DOI] [PubMed] [Google Scholar]
- Yamaguchi R., Lartigue L., Perkins G., Scott R.T., Dixit A., Kushnareva Y., Kuwana T., Ellisman M.H., Newmeyer D.D. 2008. Opa1-mediated cristae opening is Bax/Bak and BH3 dependent, required for apoptosis, and independent of Bak oligomerization. Mol. Cell. 31:557–569 10.1016/j.molcel.2008.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youle R.J., van der Bliek A.M. 2012. Mitochondrial fission, fusion, and stress. Science. 337:1062–1065 10.1126/science.1219855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Liu T., Jin S., Wang X., Qu M., Uhlén P., Tomilin N., Shupliakov O., Lendahl U., Nistér M. 2011. Human MIEF1 recruits Drp1 to mitochondrial outer membranes and promotes mitochondrial fusion rather than fission. EMBO J. 30:2762–2778 10.1038/emboj.2011.198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zick M., Duvezin-Caubet S., Schäfer A., Vogel F., Neupert W., Reichert A.S. 2009. Distinct roles of the two isoforms of the dynamin-like GTPase Mgm1 in mitochondrial fusion. FEBS Lett. 583:2237–2243 10.1016/j.febslet.2009.05.053 [DOI] [PubMed] [Google Scholar]
- Zunino R., Schauss A., Rippstein P., Andrade-Navarro M., McBride H.M. 2007. The SUMO protease SENP5 is required to maintain mitochondrial morphology and function. J. Cell Sci. 120:1178–1188 10.1242/jcs.03418 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.