Abstract
Purpose
We undertook this study to determine the prevalence of estrogen receptor (ER) α (ESR1) mutations throughout the natural history of hormone dependent breast cancer and to delineate the functional roles of the most commonly detected alterations.
Experimental Design
We studied a total of 249 tumor specimens from 208 patients. The specimens include 134 ER positive (ER+/HER2–) and, as controls, 115 ER negative (ER−) tumors. The ER+ samples consist of 58 primary breast cancers and 76 metastatic samples. All tumors were sequenced to high unique coverage using next generation sequencing targeting the coding sequence of the estrogen receptor and an additional 182 cancer-related genes.
Results
Recurring somatic mutations in codons 537 and 538 within the ligand-binding domain of ER were detected in ER+ metastatic disease. Overall, the frequency of these mutations was 12% (9/76, 95% CI 6%-21%) in metastatic tumors and in a subgroup of patients who received an average of 7 lines of treatment the frequency was 20% (5/25, 95% CI 7%-41%). These mutations were not detected in primary or treatment naïve ER+ cancer or in any stage of ER− disease. Functional studies in cell line models demonstrate that these mutations render estrogen receptor constitutive activity and confer partial resistance to currently available endocrine treatments.
Conclusions
In this study we show evidence for the temporal selection of functional ESR1 mutations as potential drivers of endocrine resistance during the progression of ER positive breast cancer.
Keywords: breast cancer, estrogen receptor, ESR1 mutations, endocrine resistance
Introduction
ERα is a nuclear transcription factor that drives proliferation and growth of luminal type breast cancers and is the target of endocrine therapies in this disease. Although such anti-estrogen treatments are highly effective, a major clinical limitation is the development of acquired resistance to these therapies. Despite the fact that approximately 50% of patients with ER+ breast cancer benefit from adjuvant endocrine treatment, a significant fraction of them recur with metastatic disease (1-3). Furthermore, all patients with ER+ metastatic breast cancer who respond to endocrine treatment will eventually progress with antiestrogen resistant, hormone-independent disease.
In most cases of acquired endocrine resistance, ER continues to be expressed. Proposed mechanisms of resistance include; activation of growth factor receptor, cell survival and cell cycle signaling pathways as well as stress induced pathways (4, 5). Additionally, aberrant expression of ER co-activators and co-repressors has been implicated in endocrine resistance (6, 7). ER mutations have also been explored as a potential mechanism of drug resistance and initially it was postulated that loss of function mutations could contribute to resistance. However, several pre-clinical studies analyzed the effects of point mutations in ER and found a number of mutations that can actually enhance ER function similar to androgen receptor mutations in castrate resistant prostate cancer (8-10). One such functional mutation in ESR1 leads to the substitution of tyrosine by serine or alanine at position 537 in the ligand-binding domain (LBD) and results in ligand independent ER transcriptional activity that does not respond to endocrine manipulation (11-13). Despite these preclinical findings, the frequency of ER mutations in primary breast cancers was found to be extremely rare. In one early study of 118 ER positive and 70 ER negative primary breast cancers, only two ESR1 mutations of unknown significance were found and both were in ER− breast cancers (14). More recently, several studies describing next generation sequencing (NGS) in hundreds of primary breast cancer samples detected multiple significant somatic alterations, but none were detected in ESR1 (15-17). Two small studies conducted in the 1990s detected mutations in the ER LBD with a frequency of 3-10% in metastatic breast cancer samples, suggesting that the frequency of ER mutations in metastatic disease may be higher (18, 19). However, these studies were small and had not been validated with more sensitive sequencing technology and/or integrated with clinical correlations. Therefore, in this study we sought to comprehensively study the frequency and functional significance of ER mutations in both primary and metastatic breast cancer using targeted NGS.
Methods
Breast Cancer Human Tissue Specimens and Clinical Data
Paraffin embedded blocks from formalin fixed ER positive/HER2 negative primary and in–breast local recurrence/metastatic breast cancer specimens were obtained from the Pathology Departments of three institutions (MD Anderson Cancer Center, Houston, TX; Beth Israel Deaconess Medical Center, Boston, MA; Hospital Clínico Universitario in Valencia, Spain), including samples from the NCT00780676 trial (a phase II trial for patients with advanced metastatic breast cancer treated with dasatinib or selumetinib based on the expression profile of their metastatic tumor). ER+/HER2+ tumors were excluded as they are known to be resistant to endocrine therapy.
Samples were all stained for ER, progesterone receptor (PR) and HER2 and reviewed by a pathologist at each institution and were not centrally reviewed. ER positivity was defined as more than 1% of cells with strong staining. Anti-ERα antibodies were from Thermo Scientific and Ventana (clone SP1). HER 2 was defined as positive either by IHC of 3+ or FISH HER2/CEP17 ratio of greater than 2.2. The anti-HER2 antibody was purchased from Dako or Ventana. Clinical data were also collected. For comparison, additional ER− (either ER−/HER2− or ER−/HER2+) primary breast cancer samples were obtained (MD Anderson, and Instituto Nacional de Enfermedades Neoplásicas), as well as a small number of specimens from ER− metastatic disease (MD Anderson). Brief descriptions of all studied cohorts can be found in Table 1 and more detailed information about the ER+ metastatic cohort is in supplemental table 1. All tissue collections were done with the approval of the corresponding institutional review boards.
Table 1.
Patient cohorts.
| Patient Cohort | # of specimens | Average # of treatments* prior to biopsy | |
|---|---|---|---|
| LM+ | Metastases from patients with advanced ER+ disease that were heavily pre-treated prior to biopsy (participants in “Personalized Treatment Selection for Metastatic Breast Cancer” trial NCT00780676) | 25 | 7 |
| EM+ | Metastases from patients with early metastatic ER+ disease | 51 | 1-2 |
| P+ | Primary ER+ tumors | 58 | NA |
| M− | Metastases from patients with ER-disease | 11 | NA |
| P− | Primary ER- primary breast cancer disease | 104 | NA |
Abbreviations: LM+, late metastatic ER positive breast cancer, EM+, early metastatic ER positive disease, P+, ER positive primary breast cancer, M−, ER negative metastatic disease and P−, ER negative primary breast cancer.
Including endocrine treatments and chemotherapy regimens.
Sequencing and primary sequence data analysis
Tumor tissue sections of 40 μm were macrodissected using hematoxylin and eosinophil staining to identify areas of at least 80% cellularity. Genomic DNA was extracted using the Maxwell 16 FFPE Plus LEV DNA Purification kit (Promega) and quantified using a PicoGreen fluorescence assay (Invitrogen); ≥50ng and up to 200ng of extracted DNA was sheared to ~100-400 bp by sonication, followed by end-repair, dA-addition and ligation of indexed, Illumina sequencing adaptors. Sequencing libraries were hybridization captured using RNA-based baits (Agilent), targeting a total of 3,320 exons of 182 cancer-related genes (most commonly altered in cancer, from http://www.sanger.ac.uk/genetics/CGP/cosmic/) plus 37 introns from 14 genes often rearranged in cancer (Table S1). Paired end sequencing (49 × 49 cycles) was performed using the HiSeq2000 (Illumina). Sequence data from gDNA were mapped to the reference human genome (hg19) using the BWA aligner (20). PCR duplicate read removal and sequence metric collection was performed using Picard (http://picard.sourceforge.net) and SAMtools (21). Local alignment optimization was performed using GATK (22).
Genomic alteration detection
Base substitution detection was performed using a Bayesian methodology, which allows detection of novel somatic mutations at low mutation annotation format and increased sensitivity for mutations at hotspot sites (23) through the incorporation of tissue-specific prior expectation: P(Mutation present| Read data “R”) =P(Frequency of mutation “F” > 0|R) ∝ 1 - P(R|F= 0) P (F= 0), where P(R|F) is evaluated with a multinomial distribution of the observed allele counts using empirically observed error rates and P(F = 0) is the prior expectation of mutation in the tumor type. To detect indels, de-novo local assembly in each targeted exon was performed using the de-Bruijn approach (24). Candidate calls are filtered using a series of quality metrics, including strand bias, read location bias and a custom database of sequencing artifacts derived from normal controls. Germline alterations are identified and filtered using dbSNP (version 135, www.ncbi.nlm.nih.gov/projects/SNP/) and subsequently annotated for known and likely somatic mutations using the COSMIC database (version 62, http://cancer.sanger.ac.uk/cancergenome/projects/cosmic/). Detection of copy-number alterations (CNAs) was performed by obtaining a log-ratio profile of the sample by normalizing the sequence coverage obtained at all exons against a process-matched normal control. The profile is segmented and interpreted using allele frequencies of ~1,800 additional genome-wide SNPs to estimate tumor purity and copy number based on established methods (25-27) by fitting parameters of the equation copy , where lrseg, Cseg numbers at each segment and sample purity respectively. Focal mplifications are called at segments with ≥6 copies (7-8 in high aneuploidy samples) and homozygous deletions at 0 copies, in samples with purity >20%.
Cell and tissue culture
293T and MCF7 cells (purchased from ATCC and early passage used) were maintained in DMEM supplemented with 10% Fetal Bovine Serum and Pen/Strep. Hormone depletion was carried out for 48 hours using Phenol-red free DMEM (Cellgro) + 10% Charcoal Dextran Treated FBS (CDT, Omega Scientific) and was followed by estradiol (E2 10nM) stimulation. Transient transfections were accomplished using Lipofectamin 2000 (Life Technologies), as per the manufacturer's protocol.
Luciferase Assays
Luciferase reporter assay system (Promega) was used to monitor luciferase activity in 293T cells as per the manufacturer's recommendations, using a single tube luminometer (BD Monolight 2010). Briefly, 1×105 293T cells were plated in 6-well plate in hormone-depleted medium. Transient transfections were accomplished using Lipofectamin 2000 (Life Technologies) with the following vectors: pcDNA-wt/mut ER (0.1μg/well), ERE-TK-Luc (2 copies of Estrogen Response Element upstream Luciferase reporter, minimal TK promoter, 1μg/well), pCMV- βGal (internal control normalization vector, 0.1μg/well), pcDNA3.1 (empty vector, 0.8μg/well). Forty eight hours post-transfection, cells were treated with E2 or vehicle. For the dose response studies doses ranged between 100 to 1500 nM for the 4-hydroxytamoxifen (Sigma-Aldrich, catalog # H7904) and between 50 to 2500 nM for fulvestrant (AstraZeneca). All transfection studies were performed in triplicates and luciferase results are reported as relative light units (RLU) and normalized with β Galactosidase activity using Mammalian β-gal assay kit (Thermo Scientific).
Site-directed Mutagenesis
The GeneArt Site-Directed mutagenesis system (Life Technologies) was used to generate Y537N, Y537C and D538G mutations within ER ligand binding domain. Wild-type ER expression vector (pcDNA-ER) was used as a template with the following mutagenesis primers: Y537N: Forward, 5’- AACGTGGTGCCCCTCAATGACCTGCTGCTGGAGA T -3’ Reverse, 5’- ATCTCCAGCAGCAGGTCATTGAGGGGCACCACGTT -3’ Y537C: Forward, 5’- AACGTGGTGCCCCTCTGTGACCTGCTGCTGGAGAT -3’ Reverse, 5’- ATCTCCAGCAGCAGGTCACAGAGGGGCACCACGTT -3’ D538G: Forward, 5’- AACGTGGTGCCCCTCTATGGCCTGCTGCTGGAGAT -3’ Reverse, 5’- ATCTCCAGCAGCAGGCCATAGAGGGGCACCACGTT -3’.
Western-blot and Real-time PCR
The following antibodies were used for ER (sc-543, Santa Cruz) and β-Actin (A5441, Sigma) protein detection. For mRNA level measurement, total RNA was prepared using Trizol reagent (Life Technologies) and cDNA were synthesized using High capacity cDNA reverse transcription kit (ABI). The following primers were used for GREB1, pS2/TFF1, PR, CA12, β-Actin mRNAs detection using ABI 7300 Real-time PCR system in combination with Power SYBR Green PCR Master Mix (Life Technologies): GREB1 mRNA: Forward, 5’- CTG CCC CAG AAT GGT TTT TA -3’ Reverse, 5’- GGA CTG CAG AGT CCA GAA GC -3’ pS2/TFF1 mRNA: Forward, 5’-CCGAGCTCTGGGACTAATCA -3’ Reverse, 5’- TTG TGG TTT TCC TGG TGT CA -3’ ; β-Actin mRNA: Forward, 5’- CAC ACG CAG CTC ATT GTA GA -3 Reverse, 5’- GGC ATG GGT CAG AAG GAT T -3’ ; PR mRNA: Forward: 5'-AGC CAG AGC CCA CAA TAC AG-3', Reverse: 5'-GAC CTT ACA GCT CCC ACA GG-3' ; CA12 mRNA: Forward: 5'-GTG GTG TCC ATT TGG CTT TT-3', Reverse: 5'-GTG TCG CAA GTG TCC AGA GA -3'
Results
Sequencing Studies
To examine the prevalence and clinical implications of ER mutations in human breast cancer, we assembled a dataset of 249 specimens, obtained from 208 patients, representing both ER+/HER2− and ER− disease (Table 1). The cohorts selected enabled the analysis of the different stages of breast cancer, ranging from diagnostic biopsies of early breast cancers to samples from metastatic recurrences used for biomarker assessment before enrollment in clinical trials. DNA sequencing was performed for 3230 exons of 182 cancer-related genes plus 37 introns of 14 genes commonly fused on indexed hybridization-captured libraries to an average unique coverage of > 500x.
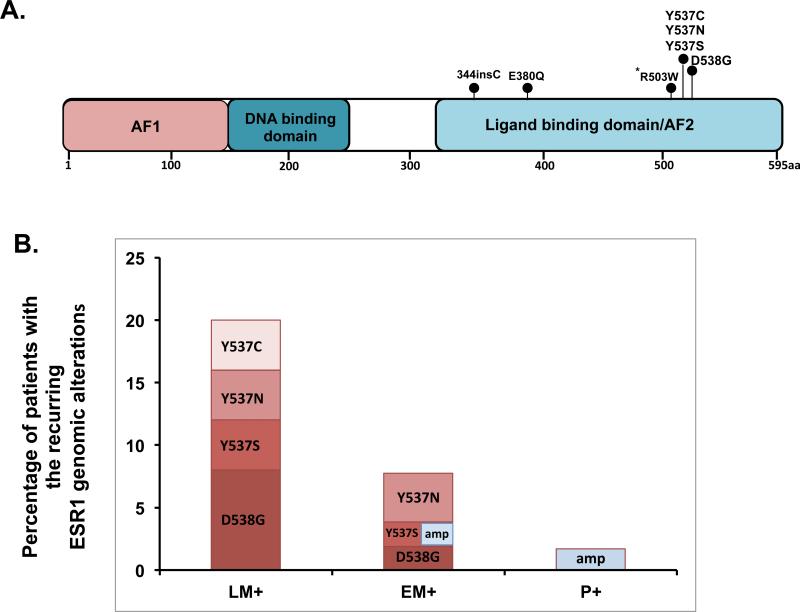
Across the 249 specimens, a total of 16 ESR1 point mutations (substitutions or indels) and 2 instances of locus focal amplification were observed. Although matched normal specimens were unavailable for definitive somatic status determination in our study, a computational assessment of variant allele frequencies supported the germ-line hypothesis for 4/16 observed events (see methods, Supplementary Table 1). Of the remaining 12 somatic variants (Table 2), 9 (75%) occurred at the known recurrent mutation sites in the ER LBD – codons 537/538- while the rest were elsewhere in the LBD (Figure 1A).
Table 2.
Detailed description of the detected somatic ESR1 variants.
| Cohort/Sample Id | Primary/Metastasis | ER Status | HER2 Status | Biopsy Site | Transcript Change | Protein Change | Known Somatic Variant (literature) | Predicted Variant Zygosity In Tumor | Endocrine Treatment Prior To Biopsy |
|---|---|---|---|---|---|---|---|---|---|
| LM3 | Metastasis | + | _ | liver | 1033_1034 insGCT |
344insC | no | clonal homozygos |
anastrozole tamoxifen exemestane fulvestrant |
| EM29 | Metastasis | +* | _* | bone | 1138G>C | E380Q | no | unknown^ | anastrazole |
| P371 | Primary | _ | + | 1507C>T | R503W | no | Subclonal | none | |
| LM17 | Metastasis | + | _ | lymph node | 1609T>A | Y537N | yes | subclonal | tamoxifen exemestane fulvestrant letrozole megestrol |
| EM43 | Metastasis | + | _ | lymph node | 1609T>A | Y537N | yes | clonal heterozygous |
tamoxifen |
| LM12 | Metastasis | + | _ | bone | 1610A>C | Y537C | yes | clonal heterozygous |
Tamoxifen goserelin anastrozole |
| EM11 | Metastasis | + | _ | lung | 1610A>C | Y537C | yes | subclonal | anastrazole |
| LM4 | Metastasis | + | _ | liver | 1610A>C | Y537S | yes | clonal heterozygous |
tamoxifen goserelin letrozole exemestane fulvestrant |
| EM51 | Metastasis | + | _ | liver | 1610A>C | Y537S | yes | subclonal | tamoxifen |
| LM19 | Metastasis | + | _ | liver | 1613A>G | D538G | yes | clonal homozygos |
tamoxifen anastrozole fulvestrant |
| LM2 | Metastasis | + | _ | bone | 1613A>G | D538G | yes | clonal heterozygous |
anastrozole exemestane fulvestrant |
| EM8 | Metastasis | + | _ | skin | 1613A>G | D538G | yes | unknown | anastrazole |
Inferred (assessed on another specimen from patient)
Tumor purity or coverage too low for assessment.
Fig. 1. Location of the ER mutations and frequencies per cohort.
(A) Diagram of ER with detected predicted somatic point mutations designated with a circle at the representative protein position. * Denotes a mutation found in a primary ER negative breast cancer.
(B) Frequency of the 538/537 substitution mutations and ER amplifications per cohort in ER positive tumors showing an increase in frequency with the progression and increase in number of treatment lines for ER positive breast cancer. Each bar represents the percentage of patients with a mutation and the different colors within the bars represent the frequency of the indicated alteration. In the EM+ cohort one patient had both an ER amplification and an Y537S mutation.
Focusing on the recurrent codon 537/538 LBD mutations, we observed a striking enrichment of these variants in ER+ metastatic disease (Figure 1B): Of the 76 ER+ metastatic specimens profiled, the recurring ER LBD mutations were found in 9 (12%) cases. The specific mutations include: Y537N (33%), D538G (33%), Y537S (22%) and Y537C (11%). The prevalence was even higher with a frequency of 20% (5/25 patients) in heavily pre-treated patients who received an average of seven lines of treatment including at least two endocrine treatments for metastatic disease. The mutations were not found in any of the 58 ER+ primary treatment-naïve tumors or the 115 ER− tumors.
Our study included 37 matched primary and metastatic pairs. In two of these pairs, EM8 and EM11 the point mutations Y537C and D538G were detected in the metastatic samples but not in the matching untreated primary tumors. For one patient with a mutation (sample EM43), we were able to sequence tissue from an earlier metastatic specimen. Both biopsies were from the same metastatic site but were attained at two time points from a patient who did not receive adjuvant hormonal treatment. The first metastatic lesion, without a detectable ESR1 mutation, was obtained prior to the initiation of tamoxifen and the second one, found to have an Y537C mutation, was obtained at the time of disease progression after 8 years of treatment. Finally, in 3 specimens, codon 537/538 mutations exhibited allele frequencies consistent with sub-clonal cell populations. Collectively, these findings suggest a clonal selection of an endocrine resistant phenotype.
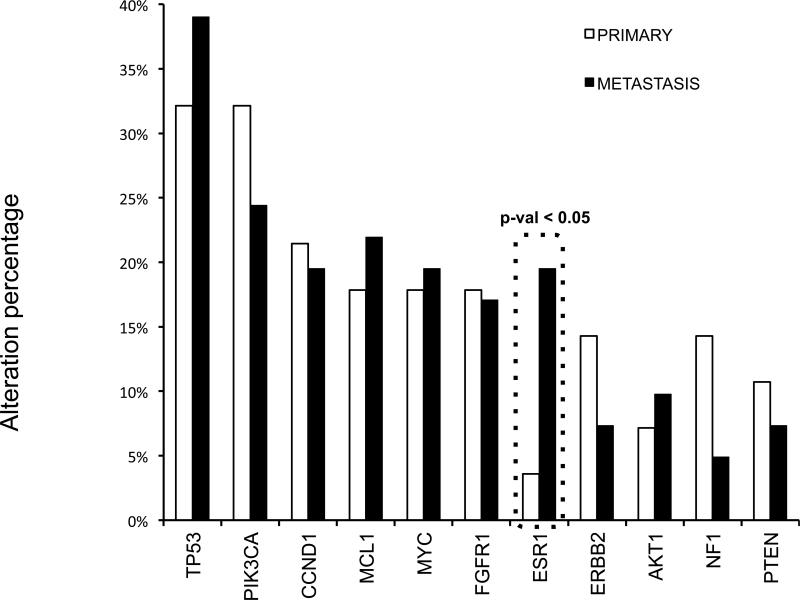
We then broadened our analysis to the genomic context of ER alterations by examining the full spectrum of genomic alterations in primary and metastatic ER+ disease. The list of the 182 genes that were sequenced can be found in supplementary Table 3. All 25 advanced metastatic, as well as 46 early metastatic and primary specimens were considered in this analysis. A similar analysis of the other 63 ER positive specimens has been presented previously(28). ER alterations were not mutually exclusive with any other commonly altered gene in ER+ breast cancer (P53, PIK3CA, CCND1, MYC, FGFR1, MCL1), although HER2 copy gains could not be assessed in our data set as clinical ER+/HER2+ cases were excluded. Nonetheless, we believe more samples will be needed to verify this observation (supplemental Figure 1). Of the most frequently altered genes (altered in >5 samples), all but ESR1 alterations displayed similar frequencies across primary and metastatic specimens, suggesting a role for aberrant ER in the development of recurrent disease (Figure 2).
Fig. 2. Genetic alteration in primary versus metastatic breast cancer.
The frequencies of the common genomic alterations found in our cohorts comparing primary tumors versus metastatic samples shows a significant increase in the ESR1 alterations in metastatic samples. P-value calculated using the Fisher's exact test.
Functional Studies
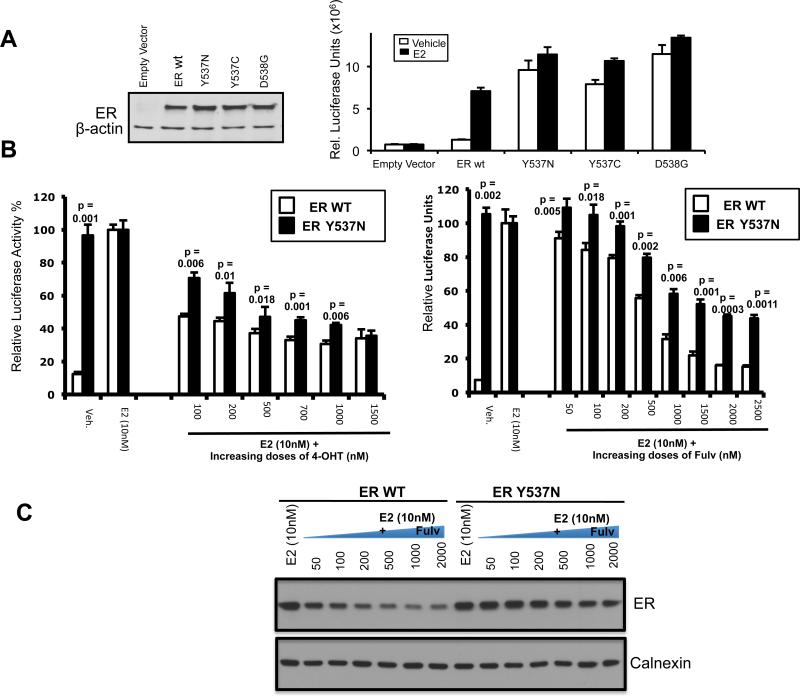
To delineate the functional roles of the mutations in codons 537/538 found in metastatic breast cancer patient samples, we performed site-directed mutagenesis to prepare three ER expression constructs containing 2 different amino acid substitutions at position 537 (Y537C and Y537N) and one amino acid substitution at position D538 (D538G). All ER mutations were confirmed by sequencing. The constitutive activity of the Y537N and Y537S mutations has been established previously11, 19. To assess the transcriptional activity of the Y537C, Y537N and D538G mutants we transiently transfected ER negative 293T cells with wild type (WT) ER or the mutant ER constructs and an estrogen-responsive promoter-reporter construct, ERE-TK-Luc (Figure 3). Cells were treated with either control vehicle or estradiol (E2) and luciferase activity was measured. As expected, the WT-ER construct exhibited activity only with E2 stimulation. In contrast, all three ER mutants possessed constitutive activity in the absence of E2 stimulation whereas the addition of E2 did not increase the reporter activity significantly. In addition, both the ligand and nonligand dependent activity in the mutants was higher than the ligand stimulated activity of the WT-ER. We next treated the cells with increasing doses of 4-hydroxytamoxifen or fulvestrant with or without E2. Cells expressing the WT-ER responded to 4-hydroxytamoxifen and fulvestrant with a significant and dose dependent reduction in luciferase activity. The ER mutants also responded to 4-hydroxytamoxifen and fulvestrant in a dose dependent manner, however, the mutant ER displayed a significantly reduced response to all doses of anti-estrogens except for the highest dose of 4-hydroxytamoxifen. Mutant ER exhibited decreased response to 4-hydroxytamoxifen and fulvestrant and higher doses of these anti-estrogens were required to achieve the level of inhibition seen in the WT receptor. To confirm the decreased response of the mutant ER to fulvestrant we examined ER degradation after 24 hours with increasing doses of fulvestrant. As we expected, mutant ER degradation compared to WT-ER was less pronounced at all doses of fulvestrant tested (Figure 3C). In addition, since it has been shown that endocrine resistant breast cancer can respond to estrogen treatment (29, 30), we tested the response of the Y537N and D538G mutants to a wide range of E2 levels but did not detect a significant change in the luciferase activity (supplementary figure 2). These results suggest that the ER mutants would be relatively resistant to established clinical doses of tamoxifen and fulvestrant as well as estrogen treatment and higher doses of SERMS or newer selective estrogen receptor degraders (SERDS) with improved pharmacokinetics may be required to inhibit mutant-ER activity.
Fig. 3. Analysis of the transcriptional activity of the recurring mutant ER and dose response to tamoxifen and fulvestrant.
(A) Transient transfection of 293T cells with wild-type (WT) or mutant ER expression vectors in addition to ERE-TK-Luc reporter vector. ER expression levels were determined to be equivalent by western-blot. β-actin expression level is shown as an internal loading control (left panel). Luciferase activity comparison of transfected 293 cells then treated for 24 hours with 10nM estrogen (E2) or vehicle (ethanol) for 24hours. Experiments were done in triplicates and repeated three times. Data represent mean +/−SD and are normalized by Beta-galactosidase internal control activity (left panel).
(B) Luciferase activity comparison of 293T cells transfected with WT or mutant ER expression vectors in addition to ERE-TK-Luc reporter vector, then treated with vehicle (ethanol) or indicated dose of E2 and increasing doses of 4-hydroxytamoxifen (4-OHT) or fulvestrant (Fulv.) for 24hours. Response to 4-OHT and fulvestrant is normalized to the response to E2 stimulation. Data represent mean +/−SD and are normalized by Beta-galactosidase internal control activity. Unpaired two-tail t-test was used to examine the statistical differences between the WT and Y537N mutant and p-values are shown.
(C) Luciferase activity levels of 293T cells transfected with WT or mutant ER expression vectors in addition to ERE-TK-Luc reporter vector after treatment with a wide range of E2 doses. Activity levels were not altered by high doses of E2.
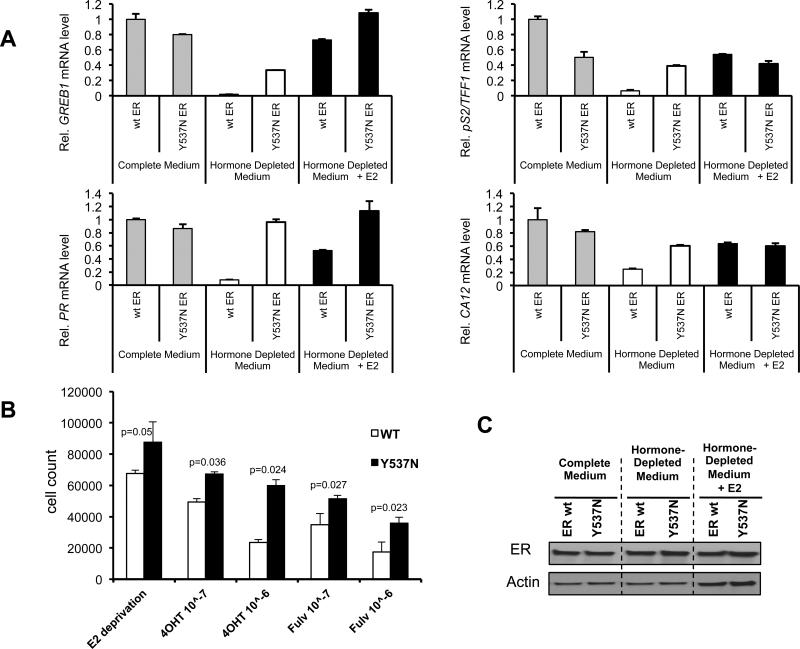
In order to study the transcriptional activity of the ER mutant in ER+ breast cancer cells we transfected a WT-ER construct and the Y537N construct in MCF7 cells. WT and mutant ER signaling was measured by determining the expression levels of ER regulated transcripts with or without E2. As shown in Figure 4A, PR, GREB1, CA12 and PS2 transcript levels were comparable between the WT and ER mutant in full media whereas in E2 deprived conditions these ER regulated genes are induced by the Y537N mutant but not the WT-ER. In addition, when we examined cell proliferation in estrogen-deprived conditions, which mimic aromatase inhibitor treatment, as well as treatment with 4-hydroxytamoxifen or fulvestrant, we found a significant growth advantage for the Y537N mutant (Figure 4B). These results demonstrate that cells expressing the Y537N mutant exhibit constitutive transcriptional activity of ER target genes and an abrogated growth inhibitory response to E2 deprivation, tamoxifen or fulvestrant treatment.
Fig.4. Transcriptional activity and growth of mutant ER in a breast cancer cell line.
(A) Transient transfection of MCF7 cells with empty vector (E.V.), wild-type (wt) or mutant Y537N ER expression vectors, then grown in complete medium (grey bars), hormone depleted medium (white bars) or hormone depleted medium with 10nM E2 (black bars). Relative mRNA levels of ER target genes GREB1 and PR (left panel) and pS2/TFF1 and CA12 (right panel) were determined by real-time PCR. Bars represent fold change +/− SD of two independent experiments.
(B) MCF7 cells were transfected with WT or Y537N ER. Cells were grown in E2 deprived conditions alone, with 4-OHT or fulvestrant and monitored for 5 days. Histograms depict cell count at day 5. Unpaired two-tail t-test was used to examine the statistical difference and p-value is shown.
(C) Western blot of ER levels in MCF7 cells after transfection of either WT-ER or Y537N mutant ER showing similar expression levels.
Discussion
The advances in sequencing technologies over the past few years have led to a new paradigm in the understanding of the mutational landscape of breast cancer and tumor heterogeneity. In this study we performed targeted next generation sequencing, which enables high levels of sequencing coverage to detect low frequency mutations within limited cellular populations, in order to study ER mutations in breast cancer. In this study, which is an international effort, we assembled a large sample set of breast cancers comprised primarily of ER+/HER2− metastatic samples and associated treatment histories, as well as ER+ primary tumors and control ER− tumors. The majority of the ER+ metastatic samples were obtained from patients who received at least one form of endocrine treatment, in order to focus on endocrine resistant tumors.
In this report, we detected in 11 of the 76 (14%) metastatic tumors ESR1 mutations in the LBD. Nine of these mutations are substitution mutations affecting Y537 and D538. In addition we identified two mutations, 344insC and E380Q, that were not described in the literature previously and further studies are needed to test their functional significance. In line with our data, other groups have recently identified the Y537 and D538 mutations and additional other mutations (31-34). The mutations detected in these studies are clustered in the LBD in a substantial number of metastatic ER+ cancers Among these three studies, the largest cohorts consisted of 36 and 44 metastatic tumors with a mutation frequency of 25% and 11%, respectively, supporting the significance of these mutations in advanced disease. We did not detect LBD ESR1 mutations in 58 primary ER+ tumors and is consistent with the findings of The Cancer Genome Atlas of over 450 cases(35). In contrast, LBD mutations were detected in 3% of the primary tumors from the BOLERO 2 study, where patients with ER+ metastatic breast cancer that had progressed in the metastatic setting were randomized to exemestane ± the TORC1 inhibitor everolimus (32, 36). This discrepancy may be due to the fact that the primary tumors in the BOLERO 2 represent a selective group of patients who all developed resistance to endocrine treatment, among them approximately 20% developed resistance early after adjuvant hormonal adjuvant treatment. Thus, the frequency of the recurrent ESR1 LBD mutations in primary remains an open and important question, since the existence of these mutations in early disease has important implications for the selection of adjuvant treatment for thousands of patients annually.
The recurring LBD mutations in positions Y537 and D538 are at the start of helix 12, a highly conserved α-helix that undergoes conformational changes during ER activation. Therefore it is not surprising that mutations in these positions would have functional implications. Similar to previous studies that focused on the Y537N, Y537S and D538A mutations (11, 19, 37), our functional studies indicate that the Y537N, Y537C and D538G mutations lead to ligand independent activity that is relatively resistant to tamoxifen, fulvestrant and estrogen deprivation. Studies led by Katzenellenbogen and colleagues have shed light on the mechanisms of these findings by demonstrating that the ER mutants that confer constitutive activity adopt a conformation that resembles the WT ligand-bound ER and the Y537S mutant has a decreased affinity to E2 with increased binding to co-activators (12, 38). The mutant ER affinity to tamoxifen and fulvestrant has not been tested, however, we hypothesize that it is also reduced and together with increased co-activator binding leads to the relative resistance to these drugs. Our studies show that higher doses of tamoxifen or fulvestrant are able to decrease partially the mutant transcriptional activity, suggesting the possibility that higher doses of these agents could be a strategy to circumvent this relative resistance. This might explain in part the results of the CONFIRM trial in which a higher dose of fulvestrant improved progression free survival (39). Further experiments to understand the detailed mechanism of the endocrine resistance of these mutants and test this hypothesis are underway.
Previous studies in xenograft models of MCF7 cells demonstrated the emergence of a D351Y LBD mutation in a tamoxifen resistant and stimulated tumor (40, 41) In this study, we analyzed tumor specimens obtained at different stages of ER positive breast cancer including; primary treatment naïve primary disease, local recurrent and/or metastatic disease with an average of two lines of treatment for metastatic disease and metastatic disease biopsied after an average of seven treatments. The frequencies of the recurring ER-LBD mutations in these three cohorts were 0%, 8% and 20%, respectively, demonstrating a correlation between the increase in the frequency of these mutations and tumor progression. In addition, in a case from which serial biopsies were available we were able to detect the emergence of the LBD-mutation as the patient developed resistance to tamoxifen treatment. Taken together, these findings suggest a temporal evolution of ER mutations with emergence of an endocrine resistant phenotype.
In our study we detected ESR1 amplifications in a primary and metastatic tumor. Multiple studies have investigated the frequency of ESR1 amplifications in ER+ primary breast cancer though the data have been controversial. Two large studies found ESR1 amplifications in approximately 20% of cancers, which correlated with an increased response to tamoxifen, whereas in other studies amplifications were detected in just 1-3% of tumors (42-45). Our study is consistent with the latter studies as we observed an ESR1 amplification rate of 1.7% (1/58) in primary ER+ tumors. Similarly, the frequency of amplifications in metastatic samples was also very low (1.3%, 1/76). This is the first study to report the frequency of ESR1 amplifications in metastatic breast cancer.
In summary, our study demonstrates an overall 14% frequency of somatic ESR1 mutations in a large cohort of metastatic ER+ breast cancers though the retrospective nature of our study limits our ability to precisely determine the frequency. The absence of mutations in the primary tumors suggests clonal evolution as the mechanism of rdetectable mutations in the primary tumors suggests clonal evolution as the mechanism of resistance. Thus, we propose that these mutations are a genomic mechanism of endocrine resistance. As such, the detection of these mutations can assist in clinical decision making as disease progresses underscoring the value of serial biopsies and profiling of metastatic recurrences. In addition, since the frequencies of these mutations are substantial when sensitive testing methods are used in patients with ER+ breast cancer, studies to identify alternative dosing schedules of currently approved antiestrogens and novel therapeutics that can overcome this resistance are warranted.
Supplementary Material
Translational Relevance.
This study demonstrates an overall 12% frequency of somatic ESR1 mutations in metastatic ER+ breast cancer and absence of detectable mutations in the primary tumors. In pre-clinical models we show that these mutations render constitutive activity and relative resistance to endocrine therapy. Taken together, these results suggest clonal evolution as the mechanism of resistance. Thus, we propose that these mutations are biomarkers and drivers of endocrine resistance in ER+ metastatic breast cancer. As such, these mutations can assist in clinical decision making as disease progresses underscoring the value of serial biopsies and profiling of metastatic recurrences. In addition, further studies are warranted to investigate therapeutic strategies to circumvent the resistance conferred by these mutations.
Acknowledgments
Financial Support:
This study was supported in part by grants from Susan G. Komen for the Cure (to MB), the NCI (P01 CA080111 to MB) and NIDDK (R01 DK074967 to MB).
Footnotes
Disclaimers:
Roman Yelensky is an employee and stockholder of Foundation Medicine. Garrett Frampton is an employee and stockholder of Foundation Medicine. Philip Stephens is an employee and stockholder of Foundation Medicine. Vincent A. Miller is an employee and stockholder of Foundation Medicine. Massimo Cristofanilli receives honorarium from Foundation Medicine. Addie Dvir is an employee of Teva Pharmaceuticals. Lior Soussan-Gutman is an employee of Teva Pharmaceuticals. Maureen T Cronin is an employee and stockholder of Foundation Medicine. Mirna Jarosz is an employee and stockholder of Foundation Medicine. Geoff Otto is an employee and stockholder of Foundation Medicine. Jeffrey S Ross is an employee and stockholder of Foundation Medicine.
REFERENCES
- 1.Bonneterre J, Thurlimann B, Robertson JF, Krzakowski M, Mauriac L, Koralewski P, et al. Anastrozole versus tamoxifen as first-line therapy for advanced breast cancer in 668 postmenopausal women: results of the Tamoxifen or Arimidex Randomized Group Efficacy and Tolerability study. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2000;18(22):3748–57. doi: 10.1200/JCO.2000.18.22.3748. Epub 2000/11/15. PubMed PMID: 11078487. [DOI] [PubMed] [Google Scholar]
- 2.Nabholtz JM, Buzdar A, Pollak M, Harwin W, Burton G, Mangalik A, et al. Anastrozole is superior to tamoxifen as first-line therapy for advanced breast cancer in postmenopausal women: results of a North American multicenter randomized trial. Arimidex Study Group. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2000;18(22):3758–67. doi: 10.1200/JCO.2000.18.22.3758. Epub 2000/11/15. PubMed PMID: 11078488. [DOI] [PubMed] [Google Scholar]
- 3.Baum M, Budzar AU, Cuzick J, Forbes J, Houghton JH, Klijn JG, et al. Anastrozole alone or in combination with tamoxifen versus tamoxifen alone for adjuvant treatment of postmenopausal women with early breast cancer: first results of the ATAC randomised trial. Lancet. 2002;359(9324):2131–9. doi: 10.1016/s0140-6736(02)09088-8. Epub 2002/07/02. PubMed PMID: 12090977. [DOI] [PubMed] [Google Scholar]
- 4.Osborne CK, Schiff R. Mechanisms of endocrine resistance in breast cancer. Annual review of medicine. 2011;62:233–47. doi: 10.1146/annurev-med-070909-182917. Epub 2010/10/05. doi: 10.1146/annurev-med-070909-182917. PubMed PMID: 20887199; PubMed Central PMCID: PMC3656649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garcia-Becerra R, Santos N, Diaz L, Camacho J. Mechanisms of Resistance to Endocrine Therapy in Breast Cancer: Focus on Signaling Pathways, miRNAs and Genetically Based Resistance. International journal of molecular sciences. 2012;14(1):108–45. doi: 10.3390/ijms14010108. Epub 2013/01/25. doi: 10.3390/ijms14010108. PubMed PMID: 23344024; PubMed Central PMCID: PMC3565254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Osborne CK, Bardou V, Hopp TA, Chamness GC, Hilsenbeck SG, Fuqua SA, et al. Role of the estrogen receptor coactivator AIB1 (SRC-3) and HER-2/neu in tamoxifen resistance in breast cancer. Journal of the National Cancer Institute. 2003;95(5):353–61. doi: 10.1093/jnci/95.5.353. Epub 2003/03/06. PubMed PMID: 12618500. [DOI] [PubMed] [Google Scholar]
- 7.Lavinsky RM, Jepsen K, Heinzel T, Torchia J, Mullen TM, Schiff R, et al. Diverse signaling pathways modulate nuclear receptor recruitment of N-CoR and SMRT complexes. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(6):2920–5. doi: 10.1073/pnas.95.6.2920. Epub 1998/04/18. PubMed PMID: 9501191; PubMed Central PMCID: PMC19670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Veldscholte J, Ris-Stalpers C, Kuiper GG, Jenster G, Berrevoets C, Claassen E, et al. A mutation in the ligand binding domain of the androgen receptor of human LNCaP cells affects steroid binding characteristics and response to anti-androgens. Biochemical and biophysical research communications. 1990;173(2):534–40. doi: 10.1016/s0006-291x(05)80067-1. Epub 1990/12/14. PubMed PMID: 2260966. [DOI] [PubMed] [Google Scholar]
- 9.Zhao XY, Malloy PJ, Krishnan AV, Swami S, Navone NM, Peehl DM, et al. Glucocorticoids can promote androgen-independent growth of prostate cancer cells through a mutated androgen receptor. Nature medicine. 2000;6(6):703–6. doi: 10.1038/76287. Epub 2000/06/03. doi: 10.1038/76287. PubMed PMID: 10835690. [DOI] [PubMed] [Google Scholar]
- 10.Herynk MH, Fuqua SA. Estrogen receptor mutations in human disease. Endocrine reviews. 2004;25(6):869–98. doi: 10.1210/er.2003-0010. Epub 2004/12/08. doi: 10.1210/er.2003-0010. PubMed PMID: 15583021. [DOI] [PubMed] [Google Scholar]
- 11.Weis KE, Ekena K, Thomas JA, Lazennec G, Katzenellenbogen BS. Constitutively active human estrogen receptors containing amino acid substitutions for tyrosine 537 in the receptor protein. Mol Endocrinol. 1996;10(11):1388–98. doi: 10.1210/mend.10.11.8923465. Epub 1996/11/01. PubMed PMID: 8923465. [DOI] [PubMed] [Google Scholar]
- 12.Carlson KE, Choi I, Gee A, Katzenellenbogen BS, Katzenellenbogen JA. Altered ligand binding properties and enhanced stability of a constitutively active estrogen receptor: evidence that an open pocket conformation is required for ligand interaction. Biochemistry. 1997;36(48):14897–905. doi: 10.1021/bi971746l. Epub 1997/12/16. doi: 10.1021/bi971746l. PubMed PMID: 9398213. [DOI] [PubMed] [Google Scholar]
- 13.White R, Sjoberg M, Kalkhoven E, Parker MG. Ligand-independent activation of the oestrogen receptor by mutation of a conserved tyrosine. The EMBO journal. 1997;16(6):1427–35. doi: 10.1093/emboj/16.6.1427. Epub 1997/03/17. doi: 10.1093/emboj/16.6.1427. PubMed PMID: 9135157; PubMed Central PMCID: PMC1169739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roodi N, Bailey LR, Kao WY, Verrier CS, Yee CJ, Dupont WD, et al. Estrogen receptor gene analysis in estrogen receptor-positive and receptor-negative primary breast cancer. Journal of the National Cancer Institute. 1995;87(6):446–51. doi: 10.1093/jnci/87.6.446. Epub 1995/03/15. PubMed PMID: 7861463. [DOI] [PubMed] [Google Scholar]
- 15.Ellis MJ, Ding L, Shen D, Luo J, Suman VJ, Wallis JW, et al. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature. 2012;486(7403):353–60. doi: 10.1038/nature11143. Epub 2012/06/23. doi: 10.1038/nature11143. PubMed PMID: 22722193; PubMed Central PMCID: PMC3383766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stephens PJ, Tarpey PS, Davies H, Van Loo P, Greenman C, Wedge DC, et al. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486(7403):400–4. doi: 10.1038/nature11017. Epub 2012/06/23. doi: 10.1038/nature11017. PubMed PMID: 22722201; PubMed Central PMCID: PMC3428862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Banerji S, Cibulskis K, Rangel-Escareno C, Brown KK, Carter SL, Frederick AM, et al. Sequence analysis of mutations and translocations across breast cancer subtypes. Nature. 2012;486(7403):405–9. doi: 10.1038/nature11154. Epub 2012/06/23. doi: 10.1038/nature11154. PubMed PMID: 22722202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karnik PS, Kulkarni S, Liu XP, Budd GT, Bukowski RM. Estrogen receptor mutations in tamoxifen-resistant breast cancer. Cancer research. 1994;54(2):349–53. Epub 1994/01/15. PubMed PMID: 8275466. [PubMed] [Google Scholar]
- 19.Zhang QX, Borg A, Wolf DM, Oesterreich S, Fuqua SA. An estrogen receptor mutant with strong hormone-independent activity from a metastatic breast cancer. Cancer research. 1997;57(7):1244–9. Epub 1997/04/01. PubMed PMID: 9102207. [PubMed] [Google Scholar]
- 20.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–60. doi: 10.1093/bioinformatics/btp324. Epub 2009/05/20. doi: btp324 [pii] 10.1093/bioinformatics/btp324. PubMed PMID: 19451168; PubMed Central PMCID: PMC2705234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–9. doi: 10.1093/bioinformatics/btp352. Epub 2009/06/10. doi: btp352 [pii] 10.1093/bioinformatics/btp352. PubMed PMID: 19505943; PubMed Central PMCID: PMC2723002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491–8. doi: 10.1038/ng.806. Epub 2011/04/12. doi: 10.1038/ng.806 ng.806 [pii]. PubMed PMID: 21478889; PubMed Central PMCID: PMC3083463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39(Database issue):D945–50. doi: 10.1093/nar/gkq929. Epub 2010/10/19. doi: gkq929 [pii] 10.1093/nar/gkq929. PubMed PMID: 20952405; PubMed Central PMCID: PMC3013785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Compeau PE, Pevzner PA, Tesler G. How to apply de Bruijn graphs to genome assembly. Nat Biotechnol. 2011;29(11):987–91. doi: 10.1038/nbt.2023. Epub 2011/11/10. doi: 10.1038/nbt.2023 nbt.2023 [pii]. PubMed PMID: 22068540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carter SL, Cibulskis K, Helman E, McKenna A, Shen H, Zack T, et al. Absolute quantification of somatic DNA alterations in human cancer. Nature biotechnology. 2012;30(5):413–21. doi: 10.1038/nbt.2203. Epub 2012/05/01. doi: 10.1038/nbt.2203. PubMed PMID: 22544022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Loo P, Nordgard SH, Lingjaerde OC, Russnes HG, Rye IH, Sun W, et al. Allelespecific copy number analysis of tumors. Proc Natl Acad Sci U S A. 2010;107(39):16910–5. doi: 10.1073/pnas.1009843107. Epub 2010/09/15. doi: 10.1073/pnas.1009843107 1009843107 [pii]. PubMed PMID: 20837533; PubMed Central PMCID: PMC2947907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yau C, Mouradov D, Jorissen RN, Colella S, Mirza G, Steers G, et al. A statistical approach for detecting genomic aberrations in heterogeneous tumor samples from single nucleotide polymorphism genotyping data. Genome Biol. 2010;11(9):R92. doi: 10.1186/gb-2010-11-9-r92. Epub 2010/09/23. doi: 10.1186/gb-2010-11-9-r92 gb-2010-11-9-r92 [pii]. PubMed PMID: 20858232; PubMed Central PMCID: PMC2965384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meric-Bernstam F FG, Ferrer-Lozano J, Yelensky R. Cancer gene profile of metastatic breast cancer. JCO. 2012:30. [Google Scholar]
- 29.Swaby RF, Jordan VC. Low-dose estrogen therapy to reverse acquired antihormonal resistance in the treatment of breast cancer. Clinical breast cancer. 2008;8(2):124–33. doi: 10.3816/CBC.2008.n.012. Epub 2008/07/16. doi: 10.3816/CBC.2008.n.012. PubMed PMID: 18621608. [DOI] [PubMed] [Google Scholar]
- 30.Ellis MJ, Gao F, Dehdashti F, Jeffe DB, Marcom PK, Carey LA, et al. Lower-dose vs high-dose oral estradiol therapy of hormone receptor-positive, aromatase inhibitor-resistant advanced breast cancer: a phase 2 randomized study. JAMA : the journal of the American Medical Association. 2009;302(7):774–80. doi: 10.1001/jama.2009.1204. Epub 2009/08/20. doi: 10.1001/jama.2009.1204. PubMed PMID: 19690310; PubMed Central PMCID: PMC3460383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li S, Shen D, Shao J, Crowder R, Liu W, Prat A, et al. Endocrine-therapy-resistant ESR1 variants revealed by genomic characterization of breast-cancer-derived xenografts. Cell reports. 2013;4(6):1116–30. doi: 10.1016/j.celrep.2013.08.022. Epub 2013/09/24. doi: 10.1016/j.celrep.2013.08.022. PubMed PMID: 24055055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Toy W, Shen Y, Won H, Green B, Sakr RA, Will M, et al. ESR1 ligand-binding domain mutations in hormone-resistant breast cancer. Nature genetics. 2013 doi: 10.1038/ng.2822. Epub 2013/11/05. doi: 10.1038/ng.2822. PubMed PMID: 24185512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robinson DR, Wu YM, Vats P, Su F, Lonigro RJ, Cao X, et al. Activating ESR1 mutations in hormone-resistant metastatic breast cancer. Nature genetics. 2013 doi: 10.1038/ng.2823. Epub 2013/11/05. doi: 10.1038/ng.2823. PubMed PMID: 24185510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Merenbakh-Lamin K, Ben-Baruch N, Yeheskel A, Dvir A, Soussan-Gutman L, Jeselsohn R, et al. D538G Mutation in Estrogen Receptor-alpha: A Novel Mechanism for Acquired Endocrine Resistance in Breast Cancer. Cancer research. 2013;73(23):6856–64. doi: 10.1158/0008-5472.CAN-13-1197. Epub 2013/11/13. doi: 10.1158/0008-5472.CAN-13-1197. PubMed PMID: 24217577. [DOI] [PubMed] [Google Scholar]
- 35.Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. Epub 2012/09/25. doi: 10.1038/nature11412. PubMed PMID: 23000897; PubMed Central PMCID: PMC3465532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baselga J, Campone M, Piccart M, Burris HA, 3rd, Rugo HS, Sahmoud T, et al. Everolimus in postmenopausal hormone-receptor-positive advanced breast cancer. The New England journal of medicine. 2012;366(6):520–9. doi: 10.1056/NEJMoa1109653. Epub 2011/12/14. doi: 10.1056/NEJMoa1109653. PubMed PMID: 22149876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pearce ST, Liu H, Jordan VC. Modulation of estrogen receptor alpha function and stability by tamoxifen and a critical amino acid (Asp-538) in helix 12. The Journal of biological chemistry. 2003;278(9):7630–8. doi: 10.1074/jbc.M211129200. Epub 2002/12/24. doi: 10.1074/jbc.M211129200. PubMed PMID: 12496244. [DOI] [PubMed] [Google Scholar]
- 38.Lazennec G, Ediger TR, Petz LN, Nardulli AM, Katzenellenbogen BS. Mechanistic aspects of estrogen receptor activation probed with constitutively active estrogen receptors: correlations with DNA and coregulator interactions and receptor conformational changes. Mol Endocrinol. 1997;11(9):1375–86. doi: 10.1210/mend.11.9.9983. Epub 1997/08/01. PubMed PMID: 9259327. [DOI] [PubMed] [Google Scholar]
- 39.Di Leo A, Jerusalem G, Petruzelka L, Torres R, Bondarenko IN, Khasanov R, et al. Results of the CONFIRM phase III trial comparing fulvestrant 250 mg with fulvestrant 500 mg in postmenopausal women with estrogen receptor-positive advanced breast cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2010;28(30):4594–600. doi: 10.1200/JCO.2010.28.8415. Epub 2010/09/22. doi: 10.1200/JCO.2010.28.8415. PubMed PMID: 20855825. [DOI] [PubMed] [Google Scholar]
- 40.Wolf DM, Jordan VC. The estrogen receptor from a tamoxifen stimulated MCF-7 tumor variant contains a point mutation in the ligand binding domain. Breast cancer research and treatment. 1994;31(1):129–38. doi: 10.1007/BF00689683. Epub 1994/01/01. PubMed PMID: 7981453. [DOI] [PubMed] [Google Scholar]
- 41.Wolf DM, Jordan VC. Characterization of tamoxifen stimulated MCF-7 tumor variants grown in athymic mice. Breast cancer research and treatment. 1994;31(1):117–27. doi: 10.1007/BF00689682. Epub 1994/01/01. PubMed PMID: 7981452. [DOI] [PubMed] [Google Scholar]
- 42.Holst F, Stahl PR, Ruiz C, Hellwinkel O, Jehan Z, Wendland M, et al. Estrogen receptor alpha (ESR1) gene amplification is frequent in breast cancer. Nature genetics. 2007;39(5):655–60. doi: 10.1038/ng2006. Epub 2007/04/10. doi: 10.1038/ng2006. PubMed PMID: 17417639. [DOI] [PubMed] [Google Scholar]
- 43.Tomita S, Zhang Z, Nakano M, Ibusuki M, Kawazoe T, Yamamoto Y, et al. Estrogen receptor alpha gene ESR1 amplification may predict endocrine therapy responsiveness in breast cancer patients. Cancer science. 2009;100(6):1012–7. doi: 10.1111/j.1349-7006.2009.01145.x. Epub 2009/03/27. doi: 10.1111/j.1349-7006.2009.01145.x. PubMed PMID: 19320640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Adelaide J, Finetti P, Charafe-Jauffret E, Wicinski J, Jacquemier J, Sotiriou C, et al. Absence of ESR1 amplification in a series of breast cancers. International journal of cancer Journal international du cancer. 2008;123(12):2970–2. doi: 10.1002/ijc.23786. Epub 2008/09/26. doi: 10.1002/ijc.23786. PubMed PMID: 18816632. [DOI] [PubMed] [Google Scholar]
- 45.Brown LA, Hoog J, Chin SF, Tao Y, Zayed AA, Chin K, et al. ESR1 gene amplification in breast cancer: a common phenomenon? Nature genetics. 2008;40(7):806–7. doi: 10.1038/ng0708-806. author reply 10-2. Epub 2008/06/28. doi: 10.1038/ng0708-806. PubMed PMID: 18583964; PubMed Central PMCID: PMC2846830. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.