Abstract
Bacterial type IV secretion (T4S) systems translocate virulence factors into eukaryotic cells1,2, distribute genetic material between bacteria, and have shown potential as a tool for the genetic modification of human cells3. Given the complex choreography of the substrate through the secretion apparatus4, the molecular mechanism of the T4S system has proven difficult to dissect in the absence of structural data for the entire machinery. Here we use electron microscopy (EM) to reconstruct the T4S system encoded by the Escherichia coli R388 conjugative plasmid. We show that eight proteins assemble in an intricate stoichiometric relationship to form a ~3 megadalton (MDa) nanomachine that spans the entire cell envelope. The structure comprises an outer membrane-associated core complex1 connected by a central stalk to a substantial inner membrane complex that is dominated by a battery of twelve VirB4 ATPase subunits organised as side by side hexameric barrels. Our results show a secretion system with markedly different architecture, and consequently mechanism, to other known bacterial secretion systems1,4-6.
The canonical T4S system comprises 12 proteins, VirB1-11 and VirD4, and forms a large macromolecular complex that spans the cell envelope of Gram-negative bacteria2. The hub protein VirB10 inserts into both the inner and outer membranes and spans the entire width of the periplasm. It is decorated by VirB7 and VirB9 in a 1:1:1 ratio to form a C14 symmetrised outer membrane pore termed the core complex7. The architecture and relative topological positioning of the rest of the T4S system components, particularly at the inner membrane, is unknown. Three ATPases, VirB4, VirB11 and VirD4 energise the secretion apparatus1,2,8. Some T4S systems are known to extend a substantial tubular pilus, comprised of VirB2 supplemented with a minor component VirB5, beyond the cell boundary1,2.
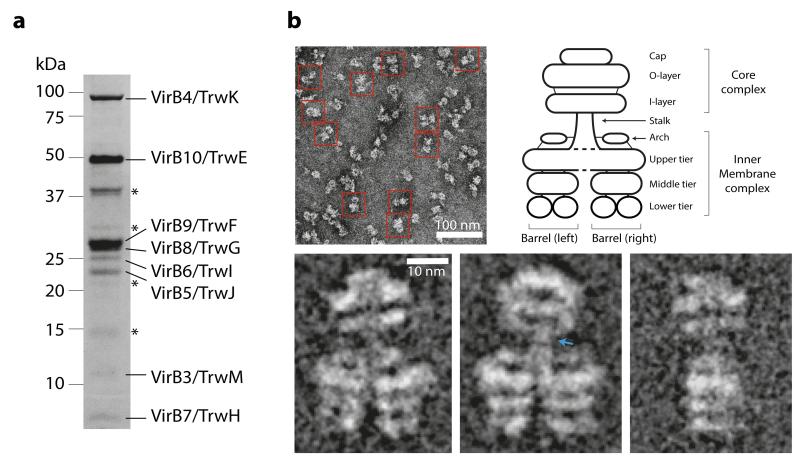
To provide structural insights into an assembled T4S system, the virB/trw operon of the R388 plasmid from virB1/trwN through to virB10/trwE (T4SS1-10) was cloned, over-expressed in E. coli, and the solubilised membrane fraction subjected to affinity chromatography (Extended Data Fig. 1a). Analysis of eluted sample by SDS-PAGE (Fig. 1a), liquid chromatography mass spectrometry (LC-MS) and Western blot (Extended Data Fig. 1b) reproducibly showed a purified complex consisting of VirB3/TrwM, VirB4/TrwK, VirB5/TrwJ, VirB6/TrwI, VirB7/TrwH, VirB8/TrwG, VirB9/TrwF, and VirB10/TrwE (termed the T4SS3-10 complex). Over-expression of virB1/trwN through to virB11/trwD with virB10/trwE tagged as in the T4SS1-10 clone yielded the same T4SS3-10 complex.
Figure 1. Purification of the R388 encoded T4SS3-10 complex.
a) SDS-PAGE analysis of the T4SS3-10 complex. * indicates minor contaminants (from top to bottom: OmpF/OmpA, Dihydrolipoyl dehydrogenase (DLD), single-stranded DNA-binding protein (SSB), and lysozyme). b) Overview negative stain EM image and representative characteristic views (class averages) of the T4SS3-10 complex with a schematic describing the nomenclature of observed structure components. Blue arrow indicates region of high flexibility.
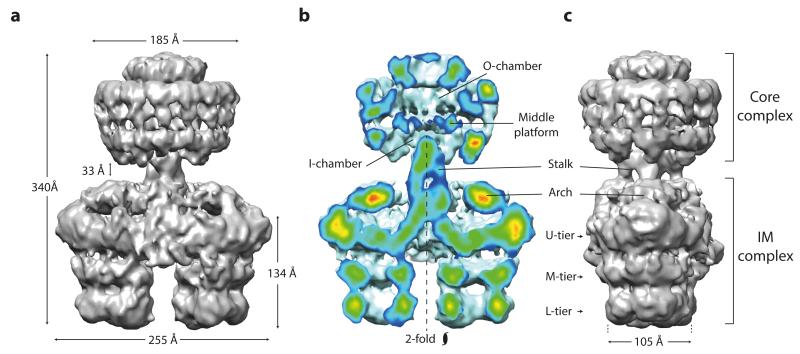
In order to characterise the three-dimensional (3D) architecture of the T4SS3-10 complex, a negative stain EM dataset was collected. Characteristic views of the complex (class averages) were obtained that typically showed double barrel-like densities connected by a thin central stalk to a structure clearly reminiscent of the pKM101 plasmid core complex7 (Fig. 1b). Indeed, separate purification of R388 VirB7/TrwH, VirB9/TrwF and VirB10/TrwE, and negative stain EM analysis of the resultant complex confirmed formation of a pKM101-like core complex complete with 14-fold symmetry (Extended Data Fig. 2). In the T4SS3-10 assembly, those densities outside of the obvious core complex are broadly termed the inner membrane complex (IMC) (Fig. 1b). Significant flexibility between IMC and the core complex was observed (Extended Data Fig. 3a to c), which prompted two discrete 3D reconstructions to be generated for the two complexes (Extended Data Fig. 3d). Final resolutions of 18 Å and 23 Å were achieved respectively (Extended Data Fig. 4a), with flexibility within the IMC accounting for the difference in resolutions. Ultimately, these reconstructions were merged to generate a composite map of the entire T4SS3-10 complex (Fig. 2, 3a, and Extended Data Fig. 4b to e). Final maps were generated with no symmetry applied. However, C14 symmetry is clearly observed in the core complex and overall C2 symmetry in the IMC (Fig. 2b).
Figure 2. Asymmetric composite structure of the T4SS3-10 complex.
a) Front view. The map is a composite generated by merging independently processed core complex and IMC reconstructions. b) Cut-away front view. Electron density is color-coded ranging from red to blue indicating regions of strong to weak density, respectively. The IMC has pseudo 2-fold symmetry around the particle long axis. c) Side view. U, M and L tier substitute for upper, middle and lower tier, respectively.
Figure 3. Segmentation of the T4SS3-10 complex reconstruction.
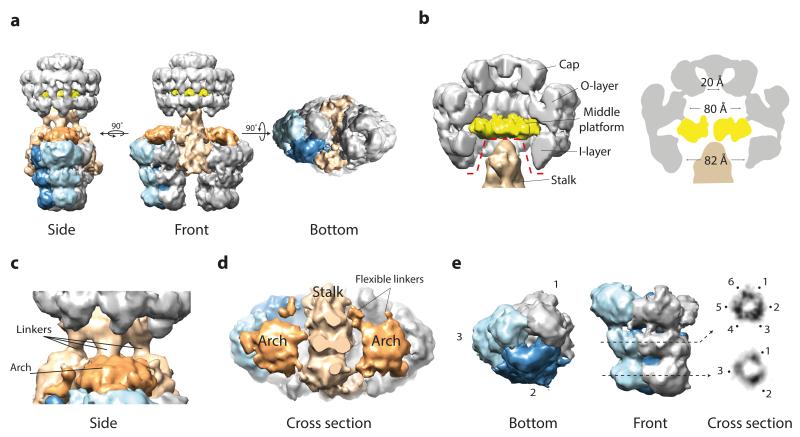
a) Side, front and bottom views. Of the two barrels, only the left one is segmented. The colour scheme used is upheld in all panels. b) Zoom cut-away view of the core complex and stalk. Dotted red line delineates the border at which the separate core complex and IMC reconstructions were merged (left). Central cross section schematic of the core complex from this study (right). c) Zoom side view of the stalk. Some of the linkers between the core complex and IMC are flexible and were therefore poorly resolved. d) Cut-away top view of the stalk and arches. e) Each barrel-like density consists of three dimeric elongated segments. Cross-sections of the lower and middle tiers (right panel) show 3-fold symmetry with a trimer of dimeric densities present in the middle tier.
The whole T4SS3-10 complex is 340 Å in length and comprises a core complex connected by a central stalk to an IMC that is 255 Å at its widest (Fig. 2). The observed core complex is similar to that of the well-characterised pKM101 plasmid7,9, with a diameter of 185 Å and rotationally symmetric inner (I-) and outer (O-) layers integrated to form a chamber that is divided by a central middle platform (Fig. 3b). The core complex cap is in an overall closed conformation as in the cryo-EM reconstruction of the pKM101 core complex9, with any central constriction7 likely occluded by the negative stain of bound detergent or lipid, or both.
In contrast to previous T4S system core complex reconstructions7,9,10, the inner (I-) chamber is partially occluded by the central stalk (Fig. 3, b to d), which extends distally from the core complex to form a central binding hub within the IMC. The stalk does not exhibit any clear symmetry, and in the composite T4SS3-10 complex model makes several limited contacts with the walls of the I-chamber. The top and middle parts of the stalk are linked by four connections that varied in intensity during refinement with the central pair being most stable (Fig. 3c and Extended Data Fig. 5a). Other connections may exist that are not observed due to marked flexibility in this region (Extended Data Fig. 3b and c).
Connected to the stalk distal end are two barrel-like densities related by C2 symmetry with length 134 Å and minimum diameter 105 Å (Fig. 2). Each barrel comprises three tiers with the lower and middle tiers constituting rings with 3- fold symmetry oriented around a central channel. In addition, a dimeric arrangement of densities around each 3- fold axis is observed in the middle tier (Fig. 3e). The upper tier is more substantial and any central channel appears closed. Overall, each barrel is comprised of three basic elongated segments that are each made up of two subunits (Extended Data Fig. 5b).
Directly above the upper tier of each barrel lies a remarkable structure termed the arch, which inter-connects between barrel subunits and the central stalk (Fig. 2 and 3d). Each arch comprises a substantial central density that contacts the underlying barrel via up to six thin linkers (Extended Data Fig. 5a). Note that not all linkers were consistently resolved during refinement and often only a dominant triad of links was observed (positions 1, 3 and 5 in Extended Data Fig. 5a).
To ensure that dry preparation of samples did not affect the integrity of the particles, a tilt series experiment using cryo negative stain (CNS) EM was carried out. It confirms the features and dimensions of the T4SS3-10 complex reconstruction described above (Extended Data Fig. 6).
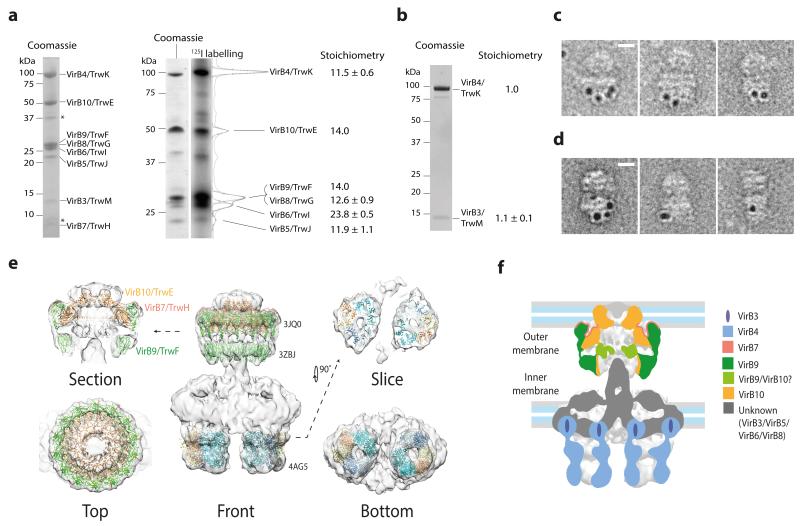
In order to begin dissecting the internal organisation of the T4SS3-10 complex, the stoichiometry of its constituents was determined using radioiodination11. A T4SS3-10 complex clone variant was used in which a His-tag was incorporated at the N-terminus of VirB6/TrwI (T4SS3-10/His6-B6). The T4SS3-10/His6-B6 complex exhibited improved biochemical stability and was equivalent to the T4SS3-10 complex when analysed by EM (Extended Data Fig. 7a). Using the known 14-fold copy number of VirB10/TrwE7,12 for calibration, the relative stoichiometry of VirB4/TrwK, VirB6/TrwI and VirB5/TrwJ are 11.5:23.8:11.9 (Fig. 4a). Deconvolution of overlapping 125Iodine signal for VirB9/TrwF and VirB8/TrwG, based on a copy number of 14 for VirB9/TrwF7,12, suggests there are 12.6 copies of VirB8/TrwG (Fig. 4a). VirB7/TrwH has no tyrosine residue but its stoichiometry within the core complex is known and is the same as VirB10/TrwE. As radioiodination was unsuitable for VirB3/TrwM due to low 125Iodine signal, VirB4/TrwK and VirB3/TrwM were co-expressed independently and purified as a distinct complex from the membrane fraction. Gel filtration and SDS-PAGE analysis showed that these proteins co-elute (Fig. 4b and Extended Data Fig. 7b). Quantification of the respective bands revealed a stoichiometry of 1:1 (Fig. 4b). Strikingly, it therefore appears that all constituents of the T4SS3-10 complex, excluding those of the core complex, essentially exist in multiples of 12. The combined stoichiometry data suggests the T4SS3-10 complex is ~3.4 MDa, which correlates well with the calculated mass estimation derived from the map volume (~3.0 MDa).
Figure 4. Stoichiometric analysis and localisation of various macromolecular components within the T4SS3-10 complex reconstruction.
a) 125Iodine labelling of T4SS3-10/His6-B6 complex constituent proteins. Left: SDS-PAGE of the T4SS3-10/His6-B6 complex. * indicates minor contaminants (OmpF/OmpA and Lpp). Right: SDS-PAGE analysis of 125I-labelled proteins in left lane (Coomassie) and corresponding radiograph in right lane (125I labelling). Relative stoichiometry was calculated by integration of band intensity and is shown at right. Reported means and corresponding standard deviations are from two separate labelling experiments on four independent purifications. b) SDS-PAGE of the VirB4/TrwK and VirB3/TrwM complex stained with SYPRO Ruby. Relative stoichiometry was calculated by integration of band intensity and is shown at right. Reported means and corresponding standard deviations are from two independent purifications. c) 5 nm gold labelling of VirB4/TrwK clusters around the IMC barrels. Scale bar equals 10 nm. d) 5 nm gold labelling of VirB6/TrwI shows a similar localisation pattern to that of VirB4/TrwK (see c). Scale bar equals 10 nm. e) Fit of the VirB4 ATPase domain from T. pseudethanolicus (4AG5), the pKM101 outer membrane complex (3JQ0), and in silico model of VirB9/TraO (3ZBJ). f) Summary schematic showing the localisation of known components and position of cell membranes. For clarity only VirB nomenclature is used in the colour key.
The stacked 3-tier architecture of the IMC barrels is reminiscent of the hexameric EM reconstruction of VirB4/TrwK13. Indeed, gold labelling of the T4SS3-10 complex with a His-tag incorporated at the C-terminus of VirB4/TrwK (T4SS3-10/ B4-His6) shows localisation of label to both IMC barrels (Fig. 4c and Extended Data Fig. 8a and c). Negative stain EM studies of VirB4/TrwK purified from the membrane fraction or as an N-terminal MBP fusion reveals an elongated and flexible monomer with approximate length 105 Å (Extended Data Fig. 7, c to e), which is consistent with the internal length of an IMC barrel. Such data, combined with the VirB4/TrwK stoichiometry and the observed trimer of dimer symmetry within each IMC barrel, supports a model in which each barrel contains six VirB4/TrwK monomers. Gold labelling of VirB6/TrwI (T4SS3-10/His6-B6 complex) shows a similar localisation pattern to that of VirB4/TrwK, which suggests the VirB4/TrwK C-terminus and VirB6/TrwI N-terminus are in the same region within the complex (Fig. 4d and Extended Data Fig. 8b and c).
In order to deduce the position of the inner membrane, the relationship between VirB4/TrwK and the inner membrane was investigated. During T4SS3-10/B4-His6 complex purification, sequential washing of the membrane fraction with 6M urea, high pH, and high salt solutions, failed to separate VirB4/TrwK (Extended Data Fig. 7f), which suggests robust insertion of VirB4/TrwK within the inner membrane. Incorporating data from previous studies13,14, it is proposed that the lower and middle tiers of each VirB4/TrwK barrel house the cytoplasmic VirB4/TrwK C-terminus, whilst the upper tier is either partially or wholly inserted within the inner membrane and houses the VirB4/TrwK N-terminus. Importantly, the soluble VirB4 C-terminal ATPase domain from Thermoanaerobacter pseudethanolicus fits optimally as a trimer of dimer arrangement within the lower and middle tiers (Fig. 4e)10.
The compelling fit of the pKM101 outer membrane complex structure12 and in silico VirB9/TraO I-layer model9 into the core complex of the T4SS3-10 reconstruction (Fig. 4e) highlights the unexpected distance (~40 Å) between the I-layer base and the proposed position of the inner membrane (Fig. 4f). The VirB10/TrwE N-termini, extending distally from the base of the I-layer, either as part of the stalk or as distinct strands, must span this space in order to insert into the inner membrane (Extended Data Fig. 9a). This architecture suggests that substrate could also access the core complex secretion chamber from within the periplasm. Such a model is compatible with some T4S systems thought to utilise a two-step process15,16. Substrate would first cross the inner membrane using either the VirD4/TrwB coupling protein, VirB4/TrwK, or the Sec machinery in some other T4S systems. Then secondarily, substrate would be channelled into the core complex and be extruded (Extended Data Fig. 9b). Evidence is also mounting for interaction between VirD4/TrwB, VirB4/TrwK and VirB11/TrwD17. VirB4/TrwK hexamers could recruit, or even be replaced by, VirB11/TrwD or VirD4/TrwB, which would allow for different enzymatic and functional gearing of the T4S machinery during the secretion cycle.
When compared with the Type III secretion (T3S) system18 (the only known structure for an assembled secretion system; Extended Data Fig. 10), the T4S system exhibits a fundamentally different architecture. While T3S systems are organised as a series of integrated ring-like structures that form a continuous tubular conduit, T4S systems display a more modular design with a 2-fold symmetric inner-membrane complex conjoined via a central stalk to a concentric outer membrane channel. Thus T4S systems represent a radically different evolutionary approach, design and mechanism for the translocation of substrate across the bacterial cell envelope.
Methods
Molecular Biology
Cloning of the R388 plasmid from virB1/trwN through to virB10/trwE (T4SS1-10)
The trw operon region of E. coli R388 plasmid from virB1/trwN through to virB10/trwE (T4SS1-10) was amplified by PCR and cloned into both IBA-GO pASK3c (pASK3c:T4SS1-10) and pBAD-M11 (pBAD:T4SS1-10) expression vectors using a commercial kit based on homologous recombination (Clontech In-Fusion). In both constructs, a sequence encoding the StrepII tag was incorporated at the 3′ terminus of virB10/trwE. The clone pASK3c:T4SS1-10 was used for the over-expression and purification of the T4SS3-10 complex, whilst pBAD:T4SS1-10 was used for the purification of the T4SS3-10/B4-His6 (used for gold labelling experiments) and T4SS3-10/His6-B6 (used for 125I labelling and gold labelling experiments) complexes once the respective tags had been added (see below).
His-, Flag-, and Maltose-binding protein (MBP) derivations of the T4SS1-10 clones
For 125I labelling and gold labelling, a His6 tag was incorporated at the N-terminus of VirB6/TrwI (pBAD:T4SS1-10/His6-B6) or the C-terminus of VirB4/TrwK (pBAD:T4SS1-10/B4-His6) using the Stratagene Quickchange strategy or Clontech In-Fusion kit. A MBP insert from E. coli was similarly added to the N-terminus of VirB4/TrwK within pASK3c:T4SS1-10 to generate pASK3c:T4SS1-10/MBP-B4. For Western blots that tested the presence of specific Trw proteins within the T4SS3-10 complex, Flag and His6 tags were incorporated into pASK3c:T4SS1-10 as specified in the main text and Extended Data Fig. 1b.
Cloning of the virB3/trwM-virB4/trwK complex
virB3/trwM and virB4/trwK insert amplified from pASK3c:T4SS1-10, and generated with BsaI restriction enzyme sites at each end, was digested with BsaI and ligated into similarly cut pASK3c vector. A sequence encoding the strepII tag was therefore located at the 3′ terminus of virB4/trwK (pASK3c:T4SS3-4).
Cloning of virB7/trwH-virB10/trwE
The trw operon region composed of virB7/trwH, virB8/trwG, virB9/trwF and virB10/trwE from R388 plasmid was cloned into pASK3c as for the cloning of the pKM101 traN to traF gene cluster1. A sequence encoding the StrepII tag was therefore located at the 3′ end of virB10/trwE. This plasmid (pASK3c:T4SS7-10) was used for the expression of the R388 core complex.
Protein over-expression and purification
Purification of the T4SS3-10 complex
pASK3c:T4SS1-10 was transformed into E. coli BL21 Star (DE3) strain (Invitrogen), and grown at 35°C in synthetic M9 minimal media supplemented with glucose, vitamins and amino acids (except methionine) (Molecular Dimensions). L-methionine was added separately (0.1 g/litre). At OD600=0.6, cells were induced using anhydrotetracycline (AHT, 0.2 mg/litre) and the temperature dropped to 19°C. Cells were harvested approximately 15 hours later and were used immediately. Pellets were resuspended in cooled 50 mM Tris-HCl pH 8.0, treated with DNase I, lysozyme, and EDTA-free protease inhibitor tablets (Roche), and sonicated on ice. After cells were opened, 1 mM DTT and 1 mM EDTA was added, and the lysate clarified by centrifugation at 38,500 g for 20 minutes. The membrane fraction was then collected by centrifugation at 98,000 g for 45 minutes. Membranes were mechanically homogenised and solubilised in 50 mM Tris-HCl pH 8.0, 200 mM NaCl, 0.5 % w/v n-Dodecyl-β-D-Maltopyranoside (DDM, Anatrace), 0.75 % w/v Decyl Maltose Neopentyl Glycol (DM-NPG, Anatrace), 0.1 % w/v digitonin (Sigma-Aldrich), 1 mM DTT and 1mM EDTA at room temperature for 40 minutes. The suspension was clarified by centrifugation at 98,000 g for 20 minutes. The supernatant was loaded onto a 1ml StrepTrap HP (GE Healthcare) column and then washed with 50 mM Tris-HCl pH 8.0, 200 mM NaCl, 0.06 % w/v DM-NPG, 0.1 % w/v digitonin, 1 mM DTT and 1mM EDTA at 4°C. The purified T4SS3-10 complex was eluted in the equivalent wash buffer supplemented with 2.5 mM desthiobiotin (IBA). The sample was used immediately for EM analysis.
Purification of the R388 core complex
The core complex purification using pASK3c:T4SS7-10 was as described in 7 with minor modification. BL21 (DE3) (Invitrogen) cells were grown at 37°C until induced with AHT at OD600 = 0.65. After approximately 15 hours at 16°C, cells were harvested by centrifugation and opened using an emulsiflex-C5 (Avestin). The cooled lysate was clarified by initial centrifugation at 38,500 g for 15 minutes, then the membrane fraction was collected by centrifugation at 98,000 g for 45 minutes. This membrane fraction was solubilised in 50 mM Tris-HCl pH 8.0, 50 mM NaCl, 1 mM EDTA, 10 mM N,N-Dimethyl-1-Dodecanamine-N-Oxide (LDAO, Anatrace), and 1 % w/v DDM at room temperature for 1 hour. Insoluble material was removed by centrifugation at 98,500 g for 15 minutes. The supernatant was loaded onto a pre-equilibrated StrepTrap HP and washed with 50 mM Tris-HCl pH 8.0, 200 mM NaCl, 10 mM LDAO at 4°C. Elution was with the same buffer supplemented with 2.5 mM desthiobiotin. Peak fractions were pooled, concentrated, and loaded onto a Superose 6 10/300 column (GE Healthcare) equilibrated in 50 mM Tris-HCl pH 8.0, 600 mM NaCl, 10 mM LDAO. The fractions containing the core complex were pooled and concentrated.
Purification of the T4SS3-10/His6-B6 and T4SS3-10/B4-His6 complexes
TOP10 strain (Invitrogen) was used to express pBAD:T4SS1-10/His6-B6 or pBAD:T4SS1-10/B4-His6 in LB media. Transformation and pre-culture was performed in the presence of 0.4 % w/w glucose to minimize leaking expression. To eliminate traces of glucose from the pre-culture, cells were washed by centrifugation and resuspended in fresh LB before starting the large-scale culture. At OD600 = 0.6, cells were incubated at 16°C, and at OD600 = 0.9, cells were induced with addition of 0.08 % w/v arabinose and harvested after approximately 15 hours. Purification of each complex then proceeded as for the T4SS3-10 complex. However, after elution from the streptactin column, the eluate was passed through a 1 ml HisTrap affinity column (GE Healthcare), washed with 50 mM Tris-HCl pH 8.0, 200 mM NaCl, 0.06 % w/v DM-NPG, 0.1 % w/v digitonin, 1 mM DTT and 1mM EDTA supplemented with 50 mM imidazole, and eluted in the presence of 350 mM imidazole.
Purification of the VirB3/TrwM-VirB4/TrwK complex
TOP10 cells transformed with pASK3c:T4SS3-4 were grown in LB broth at 37°C until OD600= 0.65. Cells were then induced using AHT (0.2 mg/litre) and the temperature dropped to 18°C. Cells were harvested approximately 15 hours later and used immediately. Pellets were resuspended in cooled 50 mM Tris-HCl pH 8.0, were treated with DNase I, lysozyme, and EDTA-free protease inhibitor tablets, and sonicated on ice. After cells were opened, the lysate was clarified by centrifugation at 38,500 g for 20 minutes. The membrane fraction was then collected by centrifugation at 98,000 g for 45 minutes. Membranes were mechanically homogenised and solubilised in 50 mM Tris-HCl pH 7.2, 150 mM NaCl, 1 mM EDTA, 1 mM DTT, 10 mM LDAO, 1 % w/v DDM and EDTA-free protease inhibitor tablets at 4°C for 45 minutes. The suspension was clarified by centrifugation at 38,500 g for 15 minutes. The supernatant was loaded onto a StrepTrap HP equilibrated in 50 mM Tris-HCl pH 7.2, 150 mM NaCl, 1 mM LDAO, 1 mM EDTA, 1 mM DTT. The column was first washed and then eluted with the same buffer supplemented with 2.5 mM desthiobiotin. Fractions containing VirB3/TrwM and VirB4/TrwK were pooled together and loaded onto a HiTrapQ HP (GE Healthcare) in order to remove bound DNA. After washing with 50 mM Tris-HCl pH 7.2, 150 mM NaCl, 1 mM LDAO, 1 mM EDTA, and 1 mM DTT, samples were eluted with a linear gradient of the same buffer supplemented with 1 M NaCl. Fractions containing VirB3/TrwM and VirB4/TrwK were loaded onto a Superose 6 10/300 equilibrated in 50 mM Tris-HCl pH 7.2, 150 mM NaCl, 1 mM LDAO, 1 mM EDTA, 1 mM DTT. The gel filtration profile consisted of two characteristic peaks-one corresponding to the oligomeric complex of VirB3/TrwM-VirB4/TrwK, and the other containing only VirB4/TrwK. In order to identify the molar ratio of VirB3/TrwM-VirB4/TrwK in the oligomeric peak, corresponding fractions were analysed by SDS-PAGE and the gels stained with SYPRO Ruby (Bio-Rad). Gel bands were scanned using a Fujifilm FLA-300 scanner and the analysis of band intensity was done with the ImageJ software27.
Purification of VirB4/TrwK
BL21(DE3) cells transformed with pASK3c:T4SS3-4 were used for over-expression and purification of VirB4/TrwK from the membrane fraction. Note that BL21(DE3) cells did not express VirB3/TrwM although they did express VirB4/TrwK. Purification of VirB4/TrwK was performed as described for the VirB3/TrwM-VirB4/TrwK complex including some minor modifications. After membrane solubilisation and clarification of the solubilised fraction by centrifugation, the supernatant was loaded onto a StrepTrap HP equilibrated in 50 mM Tris-HCl pH 7.2, 150 mM NaCl, 1 mM LDAO, 1 mM EDTA, 1 mM DTT. The eluted sample was then loaded onto a Superose 6 10/300 equilibrated in the same buffer. The only peak on the gel filtration profile corresponded to VirB4/TrwK, and the calculation of its molecular weight was done using a Superdex 200 (GE Healthcare) calibrated with high molecular weight standards (Bio-Rad). The purified monomeric VirB4/TrwK was used for subsequent electron microscopy analysis.
Purification of N-terminal MBP-VirB4/TrwK fusion protein
BL21 Star (DE3) cells transformed with pASK3c:T4SS1-10/MBP-B4 were grown at 35°C in synthetic M9 minimal media supplemented with glucose, vitamins and amino acids. L-methionine was added separately (100 mg/litre). At OD600= 0.4, cells were induced using AHT (0.2 mg/litre) and the temperature dropped to 19°C. Cells were harvested approximately 15 hours later. Pellets were resuspended in cooled buffer containing 50 mM Tris-HCl pH 8.0, 200 mM NaCl, were treated with DNase I, lysozyme, and EDTA-free protease inhibitor tablets (Roche), and sonicated on ice. After cells were opened, the lysate was clarified by initial centrifugation at 38,500 g for 20 minutes, then subjected to centrifugation at 98,000 g for 45 minutes. The resulting supernatant was loaded onto a pre-equilibrated column consisting of amylose resin (New England Biolabs). The column was washed with 50 mM Tris-HCl pH6.5, 150 mM NaCl, 50 mM potassium acetate, 5 % glycerol at 4°C. Peak fractions were concentrated using a Vivaspin concentrator with 50 kDa molecular weight cutoff and then loaded onto Superose 6 10/300 gel filtration column equilibrated in 20mM PIPES sodium salt-HCl pH6.5, 100 mM NaCl, 75 mM potassium acetate, 10 mM magnesium acetate, 5 % glycerol, 0.1 mM EDTA. These buffers are derived from the VirB4/TrwK purification strategy described in 13. After gel filtration there were still trace contaminants so peak fractions were diluted in 50 mM Tris-HCl pH8.0, 25 mM NaCl, 1 mM EDTA, 1 mM DTT and loaded onto a HiTrap Q HP (GE Healthcare). Elution of a single purified peak was by gradient against the same buffer supplemented with 1 M NaCl.
Electron microscopy (EM) and image processing
T4SS3-10 complex structure determination strategy
Due to high contrast of negative stain (NS) EM images over those obtained from unstained vitrified samples, negative stain EM was selected as the method of choice. High contrast images allowed particle quality to be judged before manual picking and aided the alignment of the picked particles during processing steps. The determination of the de novo T4SS3-10 complex structure by negative stain EM and a thorough understanding of general features and local symmetry were deemed requisite before future succession to cryo EM. Concerns over the possibility of structural distortion due to the effect of particle drying and flattening during the negative stain procedure were eliminated by validating NS T4SS3-10 complex images against the equivalent obtained using a limited cryo negative stain tilt series.
Negative stain EM structure of the T4SS3-10 complex
3μl of suitably diluted (~0.01 mg/ml) T4SS3-10 complex sample was applied to glow-discharged carbon-coated copper grids (Agar Scientific). After a 30 second incubation, the sample was blotted, washed with 3 drops of water and then stained with 2 % uranyl acetate. Images were recorded on a UltraScan Gatan 4000 4K CCD camera (Gatan, UK) with low electron dose (~25 e/Å2) on a Tecnai F20 FEG microscope operating at a voltage of 200 kV, a magnification of circa 45,500 (3.3 Å/pixel) and a defocus range of 0.8-2.0 μm. A total of 21,680 particles, judged to be evenly stained and non-aggregated, were manually selected from 693 frames and extracted within boxes of 192×192 pixels using EMAN/BOXER19. The contrast transfer function (CTF) was estimated using CTFFIND320 and correction was done by phase flipping using BSOFT21. Images of particles were normalised, band-pass filtered, centred, subjected to reference-free multivariate statistical analysis (MSA)22, and classified with approximately 20 images per class. The particle images were further aligned and classified by several rounds of multi-reference alignment (MRA)22, where the best classes representing characteristic views were used as new references, followed by MSA. A subset of representative class averages (with low variance within classes) were assigned Euler angles by angular reconstitution22 using the reconstruction of the pKM101 core complex bound to VirB4/TraB (EMD-2136 and EMD-2137)10 as a starting model (in this model, VirB4/TraB confers asymmetry to the core complex base). An initial asymmetric (no symmetry imposed) 3D reconstruction of the entire T4SS3-10 complex was then generated. This initial map was improved using anchor set refinement interspersed with rounds of MRA and classification to produce a 3D reconstruction of the entire T4SS3-10 complex. For image processing, MSA, determination of angular orientations by angular reconstitution, and 3D reconstruction and structure refinement were performed using IMAGIC23 whilst image alignments were performed using SPIDER24. Flexibility between the core complex and the IMC prevented further improvement of the reconstruction, restricting particle alignment and ultimately the final resolution of the 3D reconstruction (Extended Data Fig. 3a-c). Thus, a separate analysis for the core complex and the IMC was carried out. Here, the 3D reconstruction of the entire complex was shifted down or up so that either the core complex/stalk sub-region or the IMC sub-region became centred, respectively. These sub-regions were then individually spherically masked to create two sub-structures that were used as starting models to generate higher resolution reconstructions of the two sub-regions as described below.
Core complex/stalk sub-region reconstruction refinement
The aligned image stack was shifted using the same downward shift that was applied to centre the low resolution core complex sub-structure (see above), and a circular mask applied. Refinement of the core complex sub-region was then carried out by the iterative procedure of MRA and classification reducing the number of images per class to between 3 and 4 followed by refinement of angular orientation using the reconstruction from the previous iteration. C14 symmetry was periodically applied during reconstruction although this was restricted to the O-layer and cap regions of the core complex, in order that asymmetric details in the I-layer and stalk should be preserved. To assess map resolution, the classes were divided into two groups (even and odd). No symmetry was applied during the final steps of the reconstruction refinement, which achieved a resolution of 18 Å as determined by Fourier shell correlation at a threshold of 0.5 correlation using 2,500 class averages (Extended Data Fig. 4a).
IMC sub-region reconstruction refinement
The original aligned stack was shifted but this time using the upward shift used to centre the low-resolution IMC sub-region reconstruction (described above), and a circular mask applied. Refinement of the IMC sub-region was then carried out in the same way as described for the core complex sub-region. The first asymmetrical reconstruction clearly demonstrated features corresponding to C2 symmetry with a self cross-correlation in the non-symmetrised volume of ~80 %. C2 symmetry was applied periodically during processing of data. Towards the end of refinement, SPIDER was used for projection matching of class averages instead of angular reconstitution, along with 3D reconstruction. Projection matching and in particular the SPIDER back projection algorithm yielded marginally improved resolution. As above, the classes were divided into two groups (even and odd) to assess map resolution. No symmetry was applied to the final reconstruction, which achieved a resolution of 23 Å as determined by Fourier shell correlation at a threshold of 0.5 correlation (Extended Data Fig. 4a) using 803 class averages.
Generation of the composite T4SS3-10 map
The two discrete maps of the core complex/stalk and IMC were normalised to have equivalent standard deviation (σ) of densities, and then overlaid after applying in reverse the shifts initially applied to the core or IMC sub-regions (see above). Local alignment of the two structures was then optimised using the “Fit in Map” function in Chimera28. Redundant regions of overlap between the two reconstructions were then masked as delineated in Fig. 3b to yield the final composite map. The density threshold for surface rendering of the composite T4SS3-10 map was chosen based on the fit of the outer membrane complex crystal structure12 within the core complex, which corresponds to a threshold level of 0.64σ. Based on this threshold and a volume/mass conversion of 0.81 Da/Å3, the T4SS3-10 map has a calculated mass of ~3.0 MDa.
Assessment of the flexibility between the core complex and the IMC in the T4SS3-10 complex
The angular variation between the core complex and the IMC was assessed using class averages that represent the ‘front’ view of the entire complex. T4SS3-10 complex class averages with the core complex masked out were aligned to a typical IMC ‘front’ view. For each of the top 15 representative unmasked, aligned ‘front’ views (as assessed by the cross-correlation), the angle between the core complex and the IMC was measured by projection matching against back projections of a reference core complex structure. The distribution plot for these angles is shown in Extended Data Fig. 3c.
Cryo negative staining of the T4SS3-10 complex
Fresh T4SS3-10 complex sample (3 μl at ~0.01 mg/ml) was applied for 2 minutes to glow-discharged holey carbon-coated copper grids (Quantifoil) covered with a thin film of carbon, and stained with ammonium molybdate (pH 7.5)29 at saturated concentration prior to vitrification. 15 tilt series (each consisting of 3 tilt angles (−40°, 0°, 40°)) were recorded under low dose conditions (~10 e/Å2) with a FEI Tecnai F20 FEG microscope operating at a voltage of 200 kV, at a magnification of 68,200 (2.2 Å/pixel) and a defocus range of 2.5-3.5 μm. 141 evenly stained and non-aggregated particles were selected and tracked over the 3 tilt angles. Boxed images were normalised, band-pass filtered and centered. Two representative particles with their long axis coinciding with the tilt axis are shown in Extended Data Fig. 6. All images from different tilt angles were also combined in one stack and processed as described above to generate a limited set of class averages (5 images per class), some of them shown in Extended Data Fig. 6.
Crystal structure fitting
The docking of the outer membrane complex crystal structure was performed by manual fitting with automated local refinement in Chimera. Segmentation of the T4SS3-10 complex was performed as part of the Segger package within Chimera. For the docking of the VirB4 C-terminal ATPase domain from Thermoanaerobacter pseudethanolicus two monomers were first superimposed onto two neighbouring subunits of the related TrwB hexameric crystal structure (1GL6) to create a dimer. Three such dimers per segmented IMC barrel were then manually fitted with automated local refinement in Chimera.
Electron microscopy and image processing of the R388 core complex
3 μl of the purified complex diluted to 0.04 mg/ml was applied to glow-discharged carbon-coated grids (Agar Scientific). Triton X-100 was added at a final concentration of 0.1 % in order to increase the number of side views. The sample was negatively stained with 2 % uranyl acetate and visualized in a FEI Tecnai F20 FEG microscope operating at a voltage of 200 kV under low dose conditions (~25 e/Å2). Images were recorded as described above at a magnification of 68,200 (2.2 Å/pixel) and a defocus range of 0.8-2.0 μm. Images were corrected for the CTF effect as described above and 1981 particles selected manually using BOXER. Images were normalised, band-pass filtered and centered using EMAN. They were then subjected to a reference-free classification using EMAN. The images corresponding to end and side views were extracted into two separate subsets containing 638 and 368 single particle images, respectively. End views were further aligned and classified using MLalign2D (XMIPP)30. The final classes were checked for symmetry by calculating their rotational auto-correlation function as described previously7. A plot of the rotational auto-correlation function revealed 14 peaks reflecting the C14 symmetry of the core complex (Extended Data Fig. 2d).
Electron microscopy and image processing of the T4SS3-10/His6-B6 complex
Fresh purified complex (4 μl at ~0.01 mg/ml) was applied to glow-discharged carbon-coated grids (CF-400, Electron Microscopy Sciences). The sample was negatively stained with 2 % uranyl acetate and visualised in a FEI Tecnai T12 BioTWIN LaB6 microscope operating at a voltage of 120 kV. Images were recorded on a FEI Eagle 4K×4K CCD camera under low dose conditions (~25 e/Å2) at a magnification of circa 44,000 (3.4 Å/pixel) and a defocus range of 1-2.5 μm. The CTF parameters were assessed from entire image frames using CTFFIND3. Phase flipping was carried out using SPIDER and applied to entire frames. A total of 1284 particles were selected manually from CTF-corrected micrographs using BOXER (EMAN2). Boxed images were normalised, band-pass filtered and centered. They were then subjected to a reference-free classification. Iterations of MRA using representative views of the complex and MSA were performed in IMAGIC until the classification of the dataset stabilized. The final classes were generated with approximately 20 images per class.
Electron microscopy and image processing of VirB4/TrwK and MBP-VirB4/TrwK fusion protein
3 μl of the purified VirB4/TrwK or MBP-VirB4/TrwK fusion protein diluted to 0.01 mg/ml were applied to glow-discharged carbon-coated grids (Agar Scientific). The sample was negatively stained with 2 % uranyl acetate and observed in a FEI Tecnai F20 FEG microscope operating at a voltage of 200 kV. Images were recorded on a Gatan CCD camera (as for the T4SS3-10 complex) under low dose conditions (~10 e/Å2) at a magnification of 104,167 (1.44 Å/pixel) and a defocus range of 1-2.5 μm. Images were CTF-corrected as described above, and 3100 (VirB4/TrwK) or 4569 (MBP-VirB4/TrwK) particles were selected manually using BOXER. Images were normalised, band pass filtered and centred, subjected to a reference-free classification using EMAN, and then classified using MLalign2D (XMIPP) and/or MRA with MSA (IMAGIC).
Gold labelling, stoichiometry and membrane washing experiments
VirB4/TrwK and VirB6/TrwI gold labelling
T4SS3-10/B4-His6 or T4SS3-10/His6-B6 complexes were applied onto a glow-discharged carbon coated grid (CF-400, Electron Microscopy Sciences). After 1 minute, excess of liquid was blotted, and the grid was washed on a drop of cold purification buffer (50 mM Tris pH8, 200 mM NaCl, 1 mM TCEP, 0.1 % w/v digitonin, 0.06 % w/v DM-NPG) containing 50 mM imidazole, quickly blotted and deposited on a second drop of the same buffer in the presence of 5 nM nanogold beads (Nanoprobes). After 2 minutes, the grid was rinsed sequentially for 20 seconds with: 1 drop of purification buffer, 1 drop of the same buffer without detergent and 3 drops of 2 % uranyl acetate. Incubation was for 1 minute. Images were collected on a Tecnai T12 BioTWIN LaB6 microscope operating at a voltage of 120 kV. Particles were selected manually using EMAN.
Stoichiometry determination by tyrosine labelling
Stoichiometry measurement was performed as in 11 with modifications. T4SS3-10/His6-B6 complex at a concentration ranging from 0.1 mg/ml to 0.5 mg/ml was denatured by heating at 95 °C for 10 minutes in the presence of 1 % SDS (final concentration). Stock solutions of 2 mg/ml chloramine-T and 3 mg/ml sodium metabisulfite were prepared in 20 mM triethylamine-HCl (TEA) pH 7.5. To 52 μl of denatured protein, 1 μl of Na 125I (at 100 μCi/μl) and 10 μl of 2 mg/ml chloramine-T were added and incubated for 2 minutes at room temperature under a hood. Reaction was quenched with 20 μl of sodium metabisulfite. Reaction mixture was dialyzed at room temperature, overnight against 50 mM Tris pH 8, 200 mM NaCl, 1 % SDS to remove excess 125I. The percentage of 125I incorporation was measured in a gamma counter to be about 10 %. SDS-PAGE was performed in Tricine 10 % gels. About 3×106 cpm were loaded on each lane. The gel was fixed in 30 % ethanol, 10 % acetic acid solution overnight, then hydrated in 30 % glycerol for 1 hour and dried (Model 583 Biorad gel dryer) for 1 h at 60 °C on Whatman filter paper. Dried gels were exposed in a PhosphorImager (Molecular Dynamics) cassette for 1 - 3 hours. Signal was analysed in a Storm 860 Molecular Imager with a pixel size of 50. For each lane, the density of each band was measured using Totallab Quant software (automatic band detection without background subtraction). Based on Western blot and mass spectrometry results, we assigned each band to a VirB/Trw protein (Fig. 4). The signal obtained for each protein was then normalized according to the number of tyrosine residues it contains. VirB10/TrwE (stoichiometry of 14 copies per core complex) was used as a reference. The stoichiometry of the other proteins was determined by comparing their normalized signals with the one obtained for VirB10/TrwE. Reported means and corresponding standard deviations are from two separate labelling experiments carried out on four independent preparations of the complex.
Membrane washing
Membranes from TOP10 cells transformed with pBAD:T4SS3-10/B4-His6 complex were isolated as described above. The membrane pellet was resuspended in 6M urea and an aliquot was collected for SDS-PAGE analysis (lane 1, Extended Data Fig. 7f). Then membranes were collected by centrifugation at 98,000 g for 45 minutes. The same procedure (pellet resuspension, aliquot collection and centrifugation) was repeated, sequentially using 6M urea, 100 mM sodium carbonate pH 11 and finally 2 M KCl as resuspension buffers (lane 2-4, Extended Data Fig. 7f). The last pellet was resuspended in buffer 50 mM Tris pH8.0, 200 mM NaCl and collected for SDS-PAGE analysis (lane 5, Extended Data Fig. 7f). The aliquots from each step were then analysed by Western blot on nitrocellulose membrane. Anti-His6 tag antibodies were used to detect the presence of VirB4/TrwK subunit in the membrane or soluble fractions.
Supplementary Material
Acknowledgements
This work was funded by grant 098302 from the Wellcome Trust to GW and by equipment grant 079605 from the Wellcome Trust. RF and AD were funded by Institut Pasteur and the CNRS. FG was the recipient of “Bourse Roux” from Institut Pasteur. We would like to thank Gérard Péhau-Arnaudet for support with the electron microscopes at Institut Pasteur.
Footnotes
The EM composite map of the T4SS3-10 complex has been deposited in the EMD data bank (http://www.ebi.ac.uk/pdbe/) under accession code EMD-2567.
Reprints and permissions information are available at www.nature.com/reprints.
The authors declare no competing financial interests.
References
- 1.Fronzes R, Christie PJ, Waksman G. The structural biology of type IV secretion systems. Nat Rev Microbiol. 2009;7:703–714. doi: 10.1038/nrmicro2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schröder G, Lanka E. The mating pair formation system of conjugative plasmids - A versatile secretion machinery for transfer of proteins and DNA. Plasmid. 2005;54:1–25. doi: 10.1016/j.plasmid.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 3.Schröder G, Schuelein R, Quebatte M, Dehio C. Conjugative DNA transfer into human cells by the VirB/VirD4 type IV secretion system of the bacterial pathogen Bartonella henselae. Proc Natl Acad Sci USA. 2011;108:14643–14648. doi: 10.1073/pnas.1019074108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cascales E, Christie PJ. Definition of a bacterial type IV secretion pathway for a DNA substrate. Science. 2004;304:1170–1173. doi: 10.1126/science.1095211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson TL, Abendroth J, Hol WG, Sandkvist M. Type II secretion: from structure to function. FEMS microbiology letters. 2006;255:175–186. doi: 10.1111/j.1574-6968.2006.00102.x. [DOI] [PubMed] [Google Scholar]
- 6.Marlovits TC, et al. Structural insights into the assembly of the type III secretion needle complex. Science. 2004;306:1040–1042. doi: 10.1126/science.1102610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fronzes R, et al. Structure of a type IV secretion system core complex. Science. 2009;323:266–268. doi: 10.1126/science.1166101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kerr JE, Christie PJ. Evidence for VirB4-mediated dislocation of membrane-integrated VirB2 pilin during biogenesis of the Agrobacterium VirB/VirD4 type IV secretion system. J Bacteriol. 2010;192:4923–4934. doi: 10.1128/JB.00557-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rivera-Calzada A, et al. Structure of a bacterial type IV secretion core complex at subnanometre resolution. Embo J. 2013;32:1195–1204. doi: 10.1038/emboj.2013.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wallden K, et al. Structure of the VirB4 ATPase, alone and bound to the core complex of a type IV secretion system. Proc Natl Acad Sci USA. 2012;109:11348–11353. doi: 10.1073/pnas.1201428109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Passmore LA, et al. Structural analysis of the anaphase-promoting complex reveals multiple active sites and insights into polyubiquitylation. Molecular cell. 2005;20:855–866. doi: 10.1016/j.molcel.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 12.Chandran V, et al. Structure of the outer membrane complex of a type IV secretion system. Nature. 2009;462:1011–1015. doi: 10.1038/nature08588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peña A, et al. The hexameric structure of a conjugative VirB4 protein ATPase provides new insights for a functional and phylogenetic relationship with DNA translocases. J Biol Chem. 2012;287:39925–39932. doi: 10.1074/jbc.M112.413849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Durand E, Oomen C, Waksman G. Biochemical dissection of the ATPase TraB, the VirB4 homologue of the Escherichia coli pKM101 conjugation machinery. J Bacteriol. 2010;192:2315–2323. doi: 10.1128/JB.01384-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Locht C, Coutte L, Mielcarek N. The ins and outs of pertussis toxin. Febs J. 2011;278:4668–4682. doi: 10.1111/j.1742-4658.2011.08237.x. [DOI] [PubMed] [Google Scholar]
- 16.Pantoja M, Chen LS, Chen YC, Nester EW. Agrobacterium type IV secretion is a two-step process in which export substrates associate with the virulence protein VirJ in the periplasm. Mol Microbiol. 2002;45:1325–1335. doi: 10.1046/j.1365-2958.2002.03098.x. [DOI] [PubMed] [Google Scholar]
- 17.Ripoll-Rozada J, Zunzunegui S, de la Cruz F, Arechaga I, Cabezon E. Functional Interactions of VirB11 traffic ATPases with VirB4 and VirD4 molecular motors in type IV secretion systems. J Bacteriol. 2013;195:4195–4201. doi: 10.1128/JB.00437-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schraidt O, Marlovits TC. Three-dimensional model of Salmonella’s needle complex at subnanometer resolution. Science. 2011;331:1192–1195. doi: 10.1126/science.1199358. [DOI] [PubMed] [Google Scholar]
- 19.Ludtke SJ, Baldwin PR, Chiu W. EMAN: Semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 20.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 21.Heymann JB. Bsoft: Image and molecular processing in electron microscopy. J Struct Biol. 2001;133:156–169. doi: 10.1006/jsbi.2001.4339. [DOI] [PubMed] [Google Scholar]
- 22.van Heel M, et al. Single-particle electron cryo-microscopy: towards atomic resolution. Quarterly reviews of biophysics. 2000;33:307–369. doi: 10.1017/s0033583500003644. [DOI] [PubMed] [Google Scholar]
- 23.van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M. A new generation of the IMAGIC image processing system. J Struct Biol. 1996;116:17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- 24.Frank J, et al. SPIDER and WEB: Processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 25.Orlov SS. Theory of three-dimensional reconstruction. 1. Conditions of a complete set of projections. Sov. Phys. Crystallogr. 1975;20:312–314. [Google Scholar]
- 26.Kudryashev M, et al. In situ structural analysis of the Yersinia enterocolitica injectisome. eLife. 2013;2:e00792. doi: 10.7554/eLife.00792. [DOI] [PMC free article] [PubMed] [Google Scholar]
Supplementary references
- 27.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. Journal of computational chemistry. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 29.De Carlo S, Boisset N, Hoenger A. High-resolution single-particle 3D analysis on GroEL prepared by cryo-negative staining. Micron. 2008;39:934–943. doi: 10.1016/j.micron.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 30.Scheres SHW, Nunez-Ramirez R, Sorzano COS, Carazo JM, Marabini R. Image processing for electron microscopy single-particle analysis using XMIPP. Nat Protoc. 2008;3:977–990. doi: 10.1038/nprot.2008.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.