Abstract
Through a targeted knockdown (KD) screen of chromatin regulatory genes we identified the EP400 complex components EPC1 and EPC2 as critical oncogenic co-factors in acute myeloid leukemia (AML). EPC1 and EPC2 were required for the clonogenic potential of human AML cells of multiple molecular subtypes. Focusing on MLL-mutated AML as an exemplar, Epc1 or Epc2 KD induced apoptosis of murine MLL-AF9 AML cells and abolished leukemia stem cell potential. By contrast, normal hematopoietic stem and progenitor cells (HSPC) were spared. Similar selectivity was observed for human primary AML cells versus normal CD34+ HSPC. In keeping with these distinct functional consequences, Epc1 or Epc2 KD induced divergent transcriptional consequences in murine MLL-AF9 granulocyte-macrophage progenitor-like (GMP) cells versus normal GMP, with a signature of increased MYC activity in leukemic but not normal cells. This was caused by accumulation of MYC protein and was also observed following KD of other EP400 complex genes. Pharmacological inhibition of MYC:MAX dimerization, or concomitant MYC KD, reduced apoptosis following EPC1 KD, linking the accumulation of MYC to cell death. Therefore EPC1 and EPC2 are components of a complex which directly or indirectly serves to prevent MYC accumulation and AML cell apoptosis, thus sustaining oncogenic potential.
Keywords: AML, EPC1, EPC2, MLL, MYC
Introduction
The discovery of recurrently occurring mutations in genes coding for regulators and components of chromatin, such as MLL, TET2, DNMT3A, ASXL1 and EZH2 among others 1,2 suggests that epigenetic dysfunction is a key feature of the pathology of acute myeloid leukemia (AML) and myelodysplasia. This is further emphasized by the observation of unique DNA methylation patterns in myeloid malignancy 3-5 and the clinical efficacy of azacitidine in selected cases.6,7 Novel approaches that target the structure or function of chromatin may prove beneficial in AML and MDS, diseases which for the most part remain fatal.8,9 Indicative of this, potent and selective molecules that inhibit the activity of the histone methyltransferase DOT1L,10 the histone demethylase LSD1 11 or the epigenetic reader BRD4 12,13 are effective in pre-clinical models of leukemia and are advancing towards or through early phase clinical trials.
Given the importance of disordered epigenetic regulation in AML, we performed a targeted knockdown (KD) screen of chromatin regulatory genes using human THP1 cells as an exemplar AML line, aiming to uncover novel and hitherto unappreciated critical regulators of leukemogenic potential. THP1 cells exhibit a t(9;11) translocation which is the cytogenetic hallmark of MLL-AF9, an oncogenic fusion found in approximately 5% of human AMLs.14,15 The approach was similar to that recently used to uncover a selective dependency of murine MLL-AF9;NRASxG12D AML cells on BRD4 12 and PRC2 complex components,16 although in our study a wider range of chromatin genes was targeted and human cells were used. The screen led to our identification of the homologous Enhancer of Polycomb genes EPC1 and EPC2 as required for the functional potential of AML stem cells, but not normal haematopoietic stem and progenitor cells (HSPC). Surprisingly, KD of EPC1 or EPC2 in AML cells, but not normal HSPC, led to accumulation of MYC protein, a phenomenon which contributed to apoptotic death. Thus, counterintuitively, EPC1 and EPC2 sustain oncogenic potential by contributing to cellular processes that limit the accumulation of MYC.
Materials and methods
Viral vectors, antibodies, Q-PCR primers and reagents
Viral vectors, antibodies and Q-PCR primers and probes are listed in Supplementary Tables 1 and 2. Cycloheximide, puromycin, neomycin, blasticidin, doxycycline, PD184352, U0126 and 10058-F4 were purchased from Sigma (St Louis, MO).
Human Tissue
Use of human tissue was in compliance with the ethical and legal framework of the Human Tissue Act, 2004. Normal CD34+ mobilized HSPC surplus to requirements were from patients undergoing chemotherapy and autologous transplantation for lymphoma and myeloma. Their use was authorized by the Salford and Trafford Research Ethics Committee and, for samples collected since 2006, following the written informed consent of donors. Normal human bone marrow (BM) was collected with informed consent from healthy adult male donors, with the ethical approval of the Yorkshire Independent Research Ethics Committee. Primary human AML blasts were from Manchester Cancer Research Centre’s Tissue Biobank (instituted with approval of the South Manchester Research Ethics Committee). Their use was authorized following ethical review by the Tissue Biobank’s scientific subcommittee, and with the informed consent of donors.
Mice and murine experiments
Experiments were approved by the Cancer Research UK Manchester Institute’s Animal Ethics Committee and performed under a project license issued by the United Kingdom Home Office, in keeping with the Home Office Animal Scientific Procedures Act 1986. C57BL/6 (CD45.2+) mice were purchased from Harlan (Shardlow, UK). B6.SJL-Ptprca Pepcb/BoyJ (CD45.1+) and NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice were purchased from Jackson Laboratories (Bar Harbor, ME) and bred in-house. The cohort of mice with syngeneic MLL-AF9 AML, initiated using a retroviral transduction and transplantation protocol, was generated as described.17,18 For secondary transplantation experiments of either syngeneic MLL-AF9 AML cells or normal HSPC, control or Epc KD cells were FACS sorted for GFP expression 42 hours following lentiviral infection and immediately injected into the tail veins of sub-lethally irradiated (450cGy) mice. For xenogeneic transplantation of primary human MLL-mutated AML cells, control or EPC KD cells were FACS sorted for GFP expression 42 hours following lentiviral infection and then transplanted into neonatal NSG mice. Prior to transplantation, neonatal NSG mice were sublethally irradiated (100cGy) and treated with 50μg anti-CD122 antibody (see Supplementary Information) by intraperitoneal injection, dosing of the latter being repeated at monthly intervals. Cells were injected into the facial vein.
Cell lines, primary KD screen, lentiviral particle manufacture and alamarBlue assays
Details may be found in the Supplementary Information.
Lentiviral and retroviral infection of cell lines and primary cells
Lentiviral and retroviral supernatants were prepared, and normal and leukemic, murine and human cells were infected with viral particles as described.11 For primary cell infections, lentiviral supernatants were on occasion concentrated using polyethylene glycol precipitation. Normal murine KIT+ HSPC were selected from BM, and human CD34+ HSPC from aliquots of cryopreserved peripheral blood stem and progenitor cells, using anti-CD117 or anti-CD34 microbeads respectively and an AutoMACS Pro device (all from Miltenyi Biotec, Bisley, UK), according to manufacturer’s instructions.
Clonogenic and stromal assays
Details may be found in the Supplementary Information.
Flow cytometry, apoptosis and cell cycle analysis
Details may be found in the Supplementary Information.
RNA extraction, Q-PCR, microarray hybridization and data analysis
RNA extraction, Q-PCR, RNA processing for exon array analysis and processing of exon array data to produce gene level summaries for murine protein coding genes with defined human orthologs were all performed as described in detail elsewhere.11 Preranked gene set enrichment analyses were also performed as described.11,19 Exon array data are available in the Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo) under accession number GSE39172 (reviewer access link is: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=rnmvfwqswwkkghg&acc=GSE39172).
Results
EPC1 and EPC2 are required to sustain the functional potential of MLL leukemia stem cells
The lentiviral shRNA screen targeted 272 genes for KD in THP1 AML cells (Supplementary Table 1). The readout was fold change in cell biomass over four days in KD cells by comparison with control cells infected with lentivirus expressing a non-targeting (NTC) hairpin, and this was determined by paired analyses of resorufin fluorescence (Supplementary Figure 1a). In addition to MLL itself and MEN1, which is essential for the oncogenic potential of MLL fusion oncogenes,20 this initial approach identified the homologous Enhancer of Polycomb genes EPC1 and EPC2 as required for AML cell proliferation and/or survival (Supplementary Figure 1b and Supplementary Table 3). There was no overlap in the preliminary screening hits identified by our study and those identified by recent studies.12,16 EPC is conserved and essential in yeast, fly and mouse.21-25 Its gene product forms part of the EP400 chromatin regulatory complex,26 variants of which include the TIP60 (also known as KAT5) histone acetyltransferase (HAT) complex 23,24 and a MYC binding complex.27 There is no information to date as to a role for EPC in normal or malignant hematopoiesis.
Our preliminary findings were confirmed in repeat experiments (Figure 1a), as were KD of EPC1 and EPC2 transcripts and proteins (Figures 1b,c). EPC1 and EPC2 KD cells failed to form colonies in semisolid culture (Figures 1d,e). The observed phenotypes were due to induction of apoptosis rather than cell cycle arrest or differentiation (Figure 1f and Supplementary Figures 1c-e). Importantly, KD of EPC1 and EPC2 in AML cell lines representative of other molecular subtypes also caused apoptosis and loss of clonogenic potential, indicating that the dependency for EPC was not confined to the MLL molecular subtype (Figure 1g and Supplementary Figure 1f). To confirm the requirement for EPC in AML cells from a different species we made use of a retroviral transduction and syngeneic transplantation model of human MLL-mutated AML.11,17,18 Epc1 or Epc2 KD substantially reduced the clonogenic potential of MLL-AF9 AML cells in semisolid culture (Figures 2a-c), again due to induction of apoptosis rather than cell cycle arrest (Figure 2d & Supplementary Figure 2a). Similar results were obtained for murine MLL-ENL and MLL-AF10 AML cells (Supplementary Figure 2b), indicating that the murine phenotype was not confined to MLL-AF9 transformed cells. Forced expression of human EPC1 or EPC2 rescued the clonogenic potential of Epc1 and Epc2 KD AML cells respectively (Figure 2e & Supplementary Figure 2c). Secondary transplantation of 2000 Epc1 or Epc2 KD MLL-AF9 AML cells failed to initiate AML in sub-lethally irradiated syngeneic recipients, in contrast to control cells which induced short latency disease (Figure 2f). In similar experiments, EPC1 and EPC2 KD primary AML cells from patients exhibiting different MLL gene rearrangements failed to form colonies in semisolid culture (Figures 2g,h) and failed to expand in stromal co-culture assays (Supplementary Figure 2d), in contrast to control cells. Likewise, EPC1 and EPC2 KD primary AML cells from two separate patients exhibiting different MLL gene rearrangements failed to initiate leukemic engraftment following transplantation into sub-lethally irradiated immune deficient neonatal mice, in contrast to control cells which engrafted every mouse to variable extent (Figures 2i-k). Together these data demonstrate that EPC1 and EPC2 are required for the clonogenic and proliferative potential of cell lines representing a range of AML molecular subtypes. More specifically, using syngeneic and xenogeneic mouse models of MLL-mutated AML cells, our data demonstrate that EPC1 and EPC2 are required to prevent apoptosis of AML cells, and to maintain their leukemia stem cell (LSC) potential.
Figure 1.
EPC1 and EPC2 are required to prevent apoptosis of human THP1 AML cells. Human THP1 AML cells were infected with lentiviruses targeting EPC for KD, or a non-targeting control (NTC), with puromycin drug resistance as the selectable marker. (a) Bar chart shows mean±SEM change in resorufin signal over four days in EPC KD cells relative to control cells (NTC) (n=3). (b) Bar chart shows mean±SEM expression of EPC1 or EPC2 relative to control cells (n=3), and (c) representative western blots, three days following lentiviral infection and initiation of EPC1 or EPC2 KD. (d) Bar chart shows mean±SEM frequency of colony forming cells (CFC) enumerated following ten days in semisolid culture (n=3) and (e) representative images from (d). For colony assays, viable cells (determined by trypan blue dye exclusion) were plated out three days following initiation of EPC KD. (f) Exemplar FACS plots indicate annexin V and 7-aminoactinomycin D (7-AAD) binding to EPC KD or control THP1 cells on day 7 of liquid culture. (g) Bar chart shows mean±SEM CFC frequencies for control and EPC KD cells from the indicated AML cell lines enumerated after ten days in semisolid culture (n=4).
Figure 2.
Epc1 and Epc2 are required for the functional potential of MLL leukemia stem cells. (a-e) Murine MLL-AF9 leukemic splenocytes were infected with lentiviruses targeting Epc for KD, or a non-targeting control (NTC), with GFP as the selectable marker. Cells were FACS purified 48 hours following lentiviral infection. Bar charts show (a) mean±SEM frequency of AML colony forming cells (CFC) enumerated after five days in semisolid culture (n=7) and (b) mean±SEM expression of Epc1 or Epc2, relative to control AML cells, 48 hours after lentiviral infection and initiation of KD, as determined by Q-PCR (n=3 replicates). (c) Exemplar western blots show expression of the indicated proteins 48 hours after initiation of KD. (d) FACS plots show annexin V and 7-aminoactinomycin D (7-AAD) binding to Epc KD or control cells six days after initiation of KD. Cells were FACS sorted for GFP (the selectable marker) 48 hours following lentiviral infection and then cultured in liquid conditions for four days. (e) Bar chart shows mean±SEM frequency of AML CFC following Epc1 or Epc2 KD, with or without forced expression of human EPC1 or EPC2 (n=3). (f) Survival curves of mice secondarily transplanted with 2000 Epc KD or control cells (n=6 per cohort). (g-k) Primary unfractionated human AML cells were infected with lentiviruses targeting EPC for KD, or a non-targeting control (NTC), with GFP as the selectable marker. (g) Bar chart shows mean frequency of AML colony forming cells (CFC) enumerated after ten days in semisolid culture (error bars refer to SEM of triplicate analyses; BB number is the Manchester Cancer Research Centre Biobank sample identifier; the chromosomal translocation associated with each sample is shown). (h) Representative images of primary human AML colonies. Graphs show percentage human AML cell engraftment in mouse BM (i) 200 days following transplant of 3×105 or (j) 150 days following transplant of 1.25 ×105 FACS purified GFP+ EPC KD or control cells into sub-lethally irradiated neonatal immune deficient mice (n=4 per cohort). (k) Representative FACS plot shows primary human AML engraftment in BM.
EPC1 and EPC2 KD spares normal hematopoietic stem and progenitor cells
The identification of genes and cellular pathways which are required for the proliferation or survival of LSC but which are of less importance in normal hematopoietic stem and progenitor cells (HSPC) is key to the development of novel leukemia-selective therapies. We observed no significant reduction in the clonogenic potential of Epc KD normal murine KIT+ BM HSPCs (Figures 3a,b) nor any increase in apoptosis (Supplementary Figure 3a). This was even though levels of Epc1 and Epc2 expression and efficiencies of KD were broadly similar to those observed in AML cells (Figures 3c,d). Epc1 and Epc2 expression levels relative to Actb in normal HSPC were 74±7% (mean±SEM) and 110±17% respectively of those found in MLL-AF9 AML cells, as determined by quantitative polymerase chain reaction (Q-PCR) (Figure 3c). Epc1 transcripts were reduced to 53±8% of control levels in MLL-AF9 AML cells and 56±4% of control levels in normal HSPC following Epc1 KD. For Epc2 KD the values were 66±2% and 53±11% respectively (Figure 3c). Experiments with normal human CD34+ HSPC gave similar results: the clonogenic and multilineage differentiation potential of EPC KD CD34+ cells was maintained with respect to myeloid lineage colonies, although there was a significant reduction in erythroid burst-forming units (Figure 3e). Of note, EPC1 and EPC2 are expressed in both immature and mature human BM cell populations, although the highest level of expression relative to ACTB was noted for both in the immunophenotypic HSC compartment (Supplementary Figures 3b,c). In keeping with the selective requirement for EPC in leukemic hematopoiesis, it was also of note that Epc KD murine KIT+ BM HSPC could not be immortalized by MLL-AF9 (Supplementary Figure 3d).
Figure 3.
Epc1 or Epc2 KD spares normal hematopoietic stem and progenitor cells. (a-d) Murine HSPCs were infected with lentiviruses targeting Epc for KD, or a non-targeting control (NTC), with GFP as the selectable marker. (a) Bar chart shows mean±SEM frequency of colony forming cells (CFC) following Epc KD in KIT+ BM HSPC, or in control cells (NTC), as determined by semisolid culture for seven days in conditions supporting myeloid progenitor proliferation (n=4). p=NS indicates observed variance is not significant using one way ANOVA. (b) Images show representative colonies from (a). (c) Bar chart shows expression of Epc1 or Epc2, as determined by Q-PCR, in the indicated cell populations relative to expression levels in control MLL-AF9 AML cells, 48 hours following initiation of KD (n=3). (d) Exemplar western blots show expression of the indicated proteins 48 hours after initiation of KD in KIT+ BM HSPC. (e) Bar chart shows mean±SEM colony forming cell (CFC) frequencies, and colony types, of control and EPC KD normal human CD34+ cells grown in semi-solid culture for 14 days relative to control cells (n=4 separate individuals). * indicates p<0.01 using one-way ANOVA followed by Fisher’s least significant difference post hoc test. CFU-GM – colony forming unit granulocyte/macrophage; CFU-M – colony forming unit macrophage; BFU-E – burst-forming unit erythroid. (f) Graph shows donor chimerism in blood four weeks following transplantation of Epc1 KD or control (NTC) KSL cells (n=4 recipients per cohort). (g) Representative FACS plots show donor:recipient chimerism in BM 23 weeks after transplantation of Epc1 KD KSL cells.
To confirm these findings in an in vivo setting, we infected FACS purified murine CD45.2+ BM KSL (KIT+Sca1+Lin−) cells (a population enriched for hematopoietic stem cells and transiently self-renewing multipotent progenitors) with lentiviruses targeting Epc for KD or a control vector, with GFP as a selectable marker. Forty-eight hours later, 4×104 GFP+ cells were transplanted into lethally irradiated CD45.1+ congenic recipients with 2×105 CD45.1+ nucleated BM helper cells. Analysis of blood four weeks later demonstrated similar levels of donor:recipient chimerism in the two transplanted cohorts (Figure 3f & Supplementary Figure 3e). Mice transplanted with Epc1 KD KSL cells exhibited long term lympho-myeloid reconstitution of donor origin (Figure 3g). In additional experiments, BM donor:recipient chimerism six weeks post-transplant was similar in mice receiving Epc2 KD or control KSL cells (Supplementary Figure 3f). Together these data demonstrate that AML cells, as exemplified by our detailed analyses on the MLL-mutated molecular subtype, have a dependency on EPC which, with the partial exception of BFU-E, is not shared by normal HSPC.
Transcriptional consequences of Epc KD in GMP and MLL-AF9 GMP-like cells
To investigate the transcriptional consequences of Epc1 and Epc2 KD in normal and leukemic myeloid cells of equivalent immunophenotype, the transcriptome of KD or control normal GMP was compared to that of KD or control MLL-AF9 GMP-like cells using exon arrays, 42 hours following lentiviral infection (Supplementary Figures 4a-d). As for normal KIT+ HSPC (Figure 3a), the clonogenic potential of Epc KD normal GMP was maintained (Supplementary Figure 4e). Exonic expression values were condensed to gene level summaries and, to facilitate cross species comparison, only 15920 murine protein coding genes with annotated human homologues were considered further (Supplementary Table 4). Comparison of the two control populations with one another revealed, as expected, significantly higher expression of genes previously associated with MLL leukemia, such as Hoxa7, Hoxa9 and Meis1, in MLL-AF9 GMP-like cells versus normal GMP (Supplementary Figure 5a).
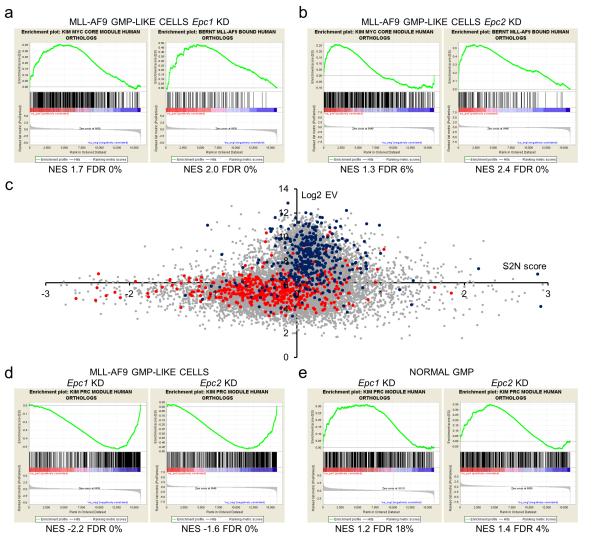
Next we compared the transcriptome of Epc1 KD and Epc2 KD normal GMP and MLL-AF9 GMP-like cells with their respective control populations. As already demonstrated by Q-PCR and western blotting (Figures 3c,d), exon array analysis confirmed that Epc expression, and the proportional extent of Epc KD, were broadly similar in normal and malignant cells (Supplementary Table 4 and Supplementary Figures 5b,c). The first main observation was that in MLL-AF9 GMP-like cells, KD of either Epc1 or Epc2 had similar effects: changes in gene expression following Epc1 KD were highly correlated with those observed following Epc2 KD (Supplementary Figure 5d). Likewise, in normal GMP cells a similar positive correlation was noted (Supplementary Figure 5e). Thus in a given cell type, EPC1 and EPC2 regulate similar transcriptional programs. However, the second main observation was in complete contrast: changes in gene expression following Epc1 KD in MLL-AF9 GMP-like cells exhibited minimal correlation with those following Epc1 KD in normal GMP. The same was true for Epc2 KD (Supplementary Figures 5f,g). Thus, in keeping with the different functional requirements for Epc in leukemic versus normal cells, Epc1 and Epc2 directly or indirectly control distinct transcriptional programs in MLL-AF9 versus normal myeloid progenitor cells. The third key observation was concordant with this: surprisingly, following Epc1 or Epc2 KD in MLL-AF9 GMP-like cells, among the genes whose expression was up regulated, there was significant enrichment of MYC module genes, genes bound by the MLL-AF9 oncoprotein and other gene sets associated with active cellular metabolism (Figures 4a,b and Supplementary Tables 5 & 6). MYC module genes are genes co-occupied by MYC, MAX, MYCN, DMAP1 (an EP400 complex component), E2F1, E2F4 and ZFX in murine embryonic stem cells (ESC).27 No such significant enrichments were seen following Epc1 or Epc2 KD in normal GMP (Supplementary Figures 5h,i). In keeping with the role of MYC as a generalized amplifier of the transcription of expressed genes,28,29 the changes in expression of defined MYC module genes and MLL-AF9 bound genes following Epc1 KD in AML cells occurred in concert with generalized up regulation of expression of active genes (i.e. those with above median array expression level) (Figure 4c). Among the genes whose relative expression was down regulated following Epc1 or Epc2 KD in MLL-AF9 GMP-like cells, there was significant enrichment of Polycomb-related complex (PRC) module genes (Figure 4d). PRC module genes are genes co-occupied by SUZ12, EED, PHC1 and RNF2 in murine ESC and include lineage-specific developmental regulators.27 By contrast, there was modest (Epc1 KD) or substantial (Epc2 KD) enrichment of PRC module genes among those up regulated in Epc1 or Epc2 KD normal GMP (Figure 4e), suggesting divergent action of Epc on PRC module gene expression, either directly or indirectly, in leukemic versus normal myeloid cells.
Figure 4.
A transcriptional signature of MYC activation in Epc KD MLL-AF9 GMP-like cells. GSEA plots show relative up regulation of MYC module genes and MLL-AF9 bound genes in (a) Epc1 and (b) Epc2 KD MLL-AF9 GMP-like cells by comparison with control cells. (c) Scatter plot shows log2 gene level summary array expression values (Log2 EV, reflecting extent of expression) versus signal-to-noise ranking metric scores (S2N score, reflecting change in expression; see Supplementary Table 7) following Epc1 KD in MLL-AF9 GMP-like cells. The x-axis crosses at the median expression value for all protein coding genes. Grey dots represent the 15,920 protein coding genes represented on the array that have annotated human orthologs; blue dots represent MYC module genes; red dots represent PRC module genes. (d) GSEA plots show relative down regulation of PRC module genes in Epc1 and Epc2 KD MLL-AF9 GMP-like cells and (e) up regulation of PRC module genes in Epc1 and Epc2 KD GMP cells by comparison with control cells. NES – normalized enrichment score; FDR – false discovery rate. Gene sets are shown in Supplementary Table 8.
These data demonstrate that the transcriptional consequences of Epc KD in MLL-AF9 GMP-like cells versus normal GMP are distinct. They also raised a question as to whether Epc KD activated MYC in leukemia cells.
Accumulation of MYC protein in AML cells upon KD of EP400 complex components
Given these observations, and the prior observation of EPC1 and EPC2 in complex with MYC, MAX, TRRAP, EP400, DMAP1 and BRD8 in murine ESC,27 we next investigated whether Epc KD altered expression of MYC. While there was no significant difference in Myc transcript expression in Epc KD versus control MLL-AF9 GMP-like cells (Supplementary Table 4 and Supplementary Figures 6a,b), there was a marked accumulation of MYC protein, as shown by western blotting 48 hours following lentiviral infection and initiation of KD (Figure 5a & Supplementary Figure 6c). MYC protein accumulation was not observed in Epc1 or Epc2 KD KIT+ normal murine BM HSPC (Figure 5b & Supplementary Figure 6c), in keeping with the array data. Cycloheximide chase assays in control or Epc1 KD murine MLL-AF9 AML cells demonstrated the expected rapid turnover of MYC in control cells (half-life ~15 minutes). By contrast, in Epc1 KD cells the half-life of MYC was longer (~50 minutes) (Figures 5c), suggesting that EPC directly or indirectly regulates turnover of MYC. Of note, steady state whole cell MYC protein levels did not differ substantially in MLL-AF9 AML cells by comparison with KIT+ normal BM HSPC (Supplementary Figure 6c).
Figure 5.
EPC KD results in accumulation of MYC in AML cells. Western blots show expression of the indicated proteins in (a) murine MLL-AF9 AML cells or (b) normal KIT+ BM HSPC 48 hours following lentiviral infection of cells with Epc KD or non-targeting control (NTC) vectors. Cells were FACS purified for GFP expression 48 hours following lentiviral infection. (c) Western blot and graph show a representative cycloheximide chase experiment for MYC protein levels in control and Epc1 KD murine MLL-AF9 AML cell 48 hours after lentiviral infection. Cycloheximide 20μg/ml was used. Graph data points were derived by semi-quantitative evaluation of western bands using ImageJ analysis. (d-h) Murine MLL-AF9 or human THP1 AML cells were infected with lentiviruses targeting EP400 complex component genes or Myc for KD, or a non-targeting control, with puromycin drug resistance as the selectable marker. Drug selection was applied 24 hours following lentiviral infection. Representative western blots show expression levels of MYC 72 hours following infection of (d) murine MLL-AF9 AML cells or (e) human THP1 AML cells with the indicated KD constructs. Vertical dashed lines on images indicate omission of intervening gel lanes for clarity of presentation. (f) Bar chart shows mean±SEM change in resorufin signal over four days in KD cells relative to control cells for the indicated KD constructs (n=3). (g) Bar chart shows expression of the indicated genes relative to control cells three days following initiation of KD. Values shown are means of triplicate analyses. (h) Bar chart shows mean±SEM percentage apoptotic cells (defined as annexin V+ and/or 7-AAD+) on day seven of liquid culture following KD with the indicated constructs (n=3).
These data raised the question as to whether EPC alone regulates MYC levels in AML cells or, alternatively, whether this is also a property of other EP400 complex components. To address this we induced KD of EP400 complex component genes 26 in murine MLL-AF9 AML cells and human THP1 AML cells. Of those tested, accumulation of MYC was noted in cells following KD of EPC1, EP400, DMAP1, VPS72, RUVBL1, RUVBL2 and MORF4L1 (Figures 5d,e). In THP1 AML cells, KD of DMAP1, VPS72, RUVBL1, RUVBL2 and MORF4L1 also limited proliferation through induction of apoptosis (Figures 5f-h). We also noted accumulation of MYC protein following EPC KD in other human AML cell lines, including Kasumi1, NB4, HL60 and U937 (Supplementary Figure 6d) indicating that the phenomenon was not confined to MLL mutated AML cell lines. Together these data suggest a general role for the EP400 complex in regulating MYC turnover in AML cells, and in preventing leukemia cell apoptosis. They also provide an explanation for the transcriptional changes observed following Epc KD: accumulation of MYC protein.
MYC contributes to AML cell apoptosis following EPC1 KD
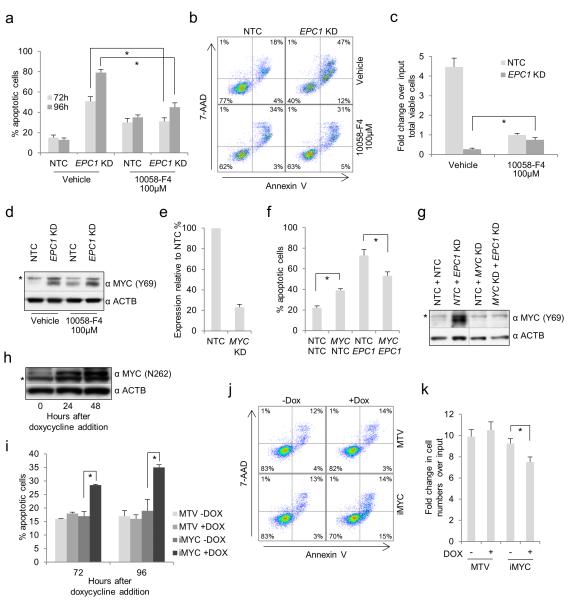
Using THP1 cells as a model system, we next investigated whether the accumulation of MYC following EPC1 KD is MAP kinase dependent, because MYC is stabilized by MAPK1/MAPK3 (ERK2/ERK1) signaling through phosphorylation of serine 62.30 Treatment of control and EPC1 KD cells with U0126 or PD184352, compounds which are selective inhibitors of MAP2K1/2 (MEK1/2), blocked MAPK1/MAPK3 phosphorylation, reduced baseline levels of MYC, largely abolished the accumulation of MYC seen following EPC1 KD (Figure 6a) and induced a G1 cell cycle arrest (Supplementary Figure 7). Importantly, treatment of EPC1 KD cells with MAP2K1/2 inhibitors significantly reduced the proportion of apoptotic cells 96 hours following lentiviral infection and initiation of KD (Figure 6b,c), without inducing substantial apoptosis of control cells, and significantly increased the total number of surviving cells at 120 hours (Figure 6d). Thus EPC1 KD-dependent accumulation of MYC and onset of apoptosis requires MAP2K1/2 activity.
Figure 6.
Accumulation of MYC protein after EPC1 KD is dependent on MAP kinase activity. Human THP1 AML cells were infected with lentiviruses targeting EPC1 for KD, or a non-targeting control (NTC), with puromycin drug resistance as the selectable marker. Drug selection was applied 24 hours following lentiviral infection. (a) Western blot shows expression of the indicated proteins in control and EPC1 KD cells 48 hours following lentiviral infection and 24 hours following addition of the indicated MAP2K1/2 inhibitors. * indicates a non-specific band. (b) Bar chart shows mean±SEM percentage of apoptotic control and EPC1 KD cells (defined as annexin V+ and/or 7-AAD+) at the indicated time points following lentiviral infection and in the indicated conditions (n=5). * indicates p<0.01 for comparison of vehicle-treated with inhibitor-treated EPC1 KD cells using one-way ANOVA and Fisher’s least significant difference post hoc test. (c) Representative FACS plots from (b). (d) Bar chart shows mean±SEM fold change over input in viable cell numbers, as determined by trypan dye exclusion and hemocytometer counting, of control or EPC1 KD cells 120 hours following lentiviral infection (n=3). * indicates p<0.001 for comparison of vehicle-treated with drug-treated EPC1 KD cells using one-way ANOVA and Fisher’s least significant difference post hoc test.
To determine directly whether MYC contributes to apoptosis following EPC1 KD, we treated control and KD THP1 AML cells with 10058-F4, a small molecule inhibitor of MYC:MAX dimerization.31 In the presence of 100μM 10058-F4, there was a modest increase in apoptosis of control THP1 cells (Figure 7a) together with a G1 cell cycle arrest (Supplementary Figure 7), observations which are in keeping with the requirement for MYC in AML cells and a previous study of the effects of this compound in human AML cell lines.32 In complete contrast, in EPC1 KD cells 10058-F4 treatment significantly reduced the proportion of apoptotic cells 72 and 96 hours following initiation of KD (Figures 7a,b), and significantly increased the total number of surviving cells at 120 hours (Figure 7c). Of note, 10058-F4 did not block EPC1 KD-induced accumulation of MYC, although baseline levels of the protein were higher in 10058-F4 treated versus vehicle treated control cells (Figure 7d). Similar results were obtained using a double KD approach. Whereas MYC KD to 23±3% of control levels (Figure 7e) significantly increased apoptosis compared with control cells (Figure 7f), concomitant MYC and EPC1 KD both blocked the accumulation of MYC protein (Figure 7g) and reduced the proportion of apoptotic cells by comparison with EPC1 KD alone (Figure 7f). Together these data demonstrate that apoptosis of AML cells following EPC1 KD is dependent, at least in part, on MYC.
Figure 7.
MYC and apoptosis in THP1 AML cells. Human THP1 AML cells were infected with lentiviruses targeting EPC1 for KD, or a non-targeting control (NTC), with puromycin drug resistance as the selectable marker. Drug selection was applied 24 hours following lentiviral infection. (a) Bar chart shows the percentage of apoptotic control and EPC1 KD cells (defined as annexin V+ and/or 7-AAD+) at the indicated time points following lentiviral infection and in the indicated conditions. * indicates p<0.001 for comparison of vehicle-treated with inhibitor-treated EPC1 KD cells using an unpaired t-test. (b) Representative FACS plots from (a). (c) Bar chart shows mean±SEM fold change over input in viable cell numbers of control or EPC1 KD cells 120 hours following lentiviral infection (n=6). * indicates p<0.01 for comparison of vehicle-treated with drug-treated EPC1 KD cells using an unpaired t-test. (d) Western blot shows expression of the indicated proteins in the indicated conditions in control and EPC1 KD cells 48 hours following lentiviral infection and 24 hours following addition of 10058-F4. * indicates a non-specific band. (e-g) THP1 cells were infected with pairwise combinations of lentiviruses targeting EPC1 and/or MYC for KD, or non-targeting controls (NTC), with puromycin drug resistance and GFP as the selectable markers. Bar charts show (e) mean±SEM expression of MYC in MYC KD cells relative to control cells, 72 hours following lentiviral infection (n=3) and (f) mean±SEM percentage of apoptotic cells 96 hours following double infection with the indicated combinations of KD or control vectors. (g) Representative western blot from (f). * indicates a non-specific band. (h-k). Myc expression was induced in human THP1 AML cells using a doxycycline-regulated system. (h) Western blot shows induction of MYC. * indicates a non-specific band. (i) Bar chart shows mean±SEM percentage of apoptotic THP1 cells at the indicated time points following addition of doxycycline (n=3). MTV = empty vector; iMYC = inducible Myc; DOX = doxycycline. * indicates p<0.01 for comparison of vehicle-treated versus DOX-treated iMYC cells using an unpaired t-test. (j) Representative FACS plots from (i). (k) Bar chart shows mean±SEM fold change in viable cell numbers over five days for the indicated THP1 lines treated with doxycycline or vehicle (n=5). * indicates p<0.05 for comparison of vehicle-treated versus DOX-treated iMYC cells using an unpaired t-test.
Finally, we tested the effect of acutely increasing cellular MYC levels in THP1 cells using a doxycycline-inducible system. Induction of MYC (Figure 7h) led to a significant and persistent increase in apoptotic cells (Figure 7i,j) and a consequent modest but significant reduction in proliferation (Figure 7k). However, the extent of the reduction in proliferation following MYC induction did not approach that observed following EPC1 KD indicating that while MYC contributes to the onset of apoptosis following EPC1 KD, EPC1 likely serves additional critical roles in AML cells over and above regulation of MYC.
Discussion
EPC1 and EPC2 are components of an essential chromatin regulatory complex which is conserved from yeast to man.24 The yeast homolog Epl1 is near exclusively found in two complexes: a three protein complex (termed Piccolo NuA4) which additionally contains yeast homologues of mammalian ING3 and the HAT TIP60, and a larger complex, NuA4 (for nucleosome acetyltransferase of H4), which in addition to Piccolo NuA4 components includes yeast homologues of mammalian TRRAP, EP400 and DMAP1, among others.23,24 A region at the N-terminal of Epl1 tethers Piccolo NuA4 to nucleosome core particles.33 Deletion of Epl1 in yeast causes accumulation of cells in G2M, increased sensitivity to DNA damaging agents, defective telomeric silencing and global loss of histone H2A and H4 acetylation.23 Genetic studies in Drosophila suggest an important role in gene silencing: in heterozygous condition E(Pc) mutations markedly enhance the homeotic transformations characteristic of the Polycomb phenotype 21 and are strong suppressors of position effect variegation.34 In mammalian biology, while a role for EPC1 in regulating skeletal muscle differentiation has been reported, and Epc1−/− mice die in early gestation,25 little is known of any role for EPC in normal or leukemic hematopoiesis. Interestingly, EPC1 is mutated by chromosomal translocation in endometrial stromal sarcoma, resulting in a novel EPC1-PHF1 fusion gene,35 and in adult T-cell leukemia/lymphoma SO4 cells resulting in a novel EPC1-ASXL2 fusion gene,36 although the functional consequences of these fusions are not clear. More broadly the TIP60 complex has roles in transcriptional control, cellular growth control and DNA double strand break repair.37
EPC is found in several protein complex settings in mammalian cells. In murine embryonic stem cells, as in yeast, both DMAP1 and TIP60 immunoprecipitations pull down most members of the conserved TIP60 complex including EPC1 and EPC2. However, in contrast to yeast and reflecting the evolutionary development of MYC, MYC’s immunoprecipitation pulled down MAX, TRRAP, EP400, DMAP1, BRD8, EPC1 and EPC2, but not TIP60.27 A similar complex which included RUVBL1 and RUVBL2 was identified following EP400 pulldown in HeLa cells.26 This indicates that, at least in ESC and carcinoma cells, association with an EP400 complex that includes EPC, but not necessarily TIP60, represents the predominant protein:protein interaction made by MYC MAX dimers. The functional importance of the association of MYC with EP400 complex components is reflected by prior observations that TRRAP and RUVBL1, both of which directly interact with MYC, are required for the in vitro transformation of rat embryonic fibroblasts by MYC and HRAS G12V.38,39 EPC1 also interacts with E2F6 and the H3K27 methyltransferase EZH2 in proliferating but not quiescent human diploid fibroblast TIG3 cells.40
Our studies have uncovered a critical role in leukemic hematopoiesis for EPC1 and EPC2 in particular, and EP400 complex components in general: AML cells representing multiple molecular subtypes, but not normal HSPC, are selectively sensitive to EPC1 and EPC2 KD. This is of significance because there are enzymatic activities within the EP400 complex which could be targeted therapeutically, such as the ATPase or DNA helicase activities of RUVBL1, RUVBL2 or EP400. Indeed, RUVBL2 has also recently been reported to be a candidate therapeutic target in AML41 and inhibitors for homologs of the bromodomain protein BRD8 have been developed.12,13 Remarkably, apoptosis of AML cells was due, at least in part, to acute accumulation of the oncoprotein MYC. This accumulation was observed following KD of multiple EP400 complex component genes and in multiple AML cell lines, indicating that it was not associated with any one particular transforming oncogene. Deregulated MYC expression is associated with both oncogenic transformation and apoptosis, although the reasons for these different biological outcomes are not fully understood. In fact, MYC-induced apoptosis has been suggested to represent an important natural tumor suppressor mechanism.42 While low level deregulated expression of Myc in vivo induces proliferation in pancreatic islet cells and is potently tumorigenic in collaboration with other genetic lesions such as RAS G12D, higher level expression induces apoptosis (although this too may be overcome to oncogenic effect by concomitant forced expression of Bcl2).43,44 Apoptosis associated with high level MYC expression is also observed in serum-deprived MYC-expressing Rat-1 fibroblasts.45 In keeping both with these observations and the requirement for MYC in AML,12 we observed that in THP1 AML cells both KD (Figure 7f) and forced expression (Figure 7i) of MYC induced apoptosis, suggesting that an optimal cellular level of MYC is required. The accumulation of MYC following EPC1 KD contributed to apoptosis, as evidenced by pharmacological and genetic KD approaches, although its extent was much greater than in cells in which Myc expression had been induced without EPC1 KD. This suggests that EPC1 has additional critical functions in AML cells over and above regulation of MYC. Given the important role of the TIP60 complex in DNA damage repair 37 and that EPC may function to tether the complex to chromatin 33 one potential explanation for our observations is that the phenotype is the consequence of the combined effect of impaired DNA damage repair and accumulation of MYC.
It remains unclear why MYC accumulates following EPC KD in AML cells. Cellular levels are normally controlled by tight regulation of the synthesis and destruction of both transcript and protein.46,47 Proteolysis is regulated by physical interactions with other proteins and post-translational modifications such as phosphorylation, acetylation and ubiquitylation.46-48 Indeed a prolonged MYC half-life has been implicated as an oncogenic mechanism in Burkitt’s Lymphoma with P57S or T58A MYC mutations 49 or precursor T-acute lymphoblastic leukemia with FBXW7 mutations.50 Our observation of an increased cellular half-life of MYC following EPC KD as well as the known binding of the EP400 complex to MYC:MAX dimers 27 suggests that the complex may directly promote turnover of MYC in AML cells by, for example, preventing its stabilization by HATs such as GCN5L2, PCAF, TIP60, CREBBP and EP300, preventing its phosphorylation by MAP kinase pathway effectors or promoting its degradation by E3 ligases.46,47,51-54 Interestingly, we have recently reported that depletion of EP300 or its co-factor TTC5 in AML cells also results in accumulation of MYC protein,55 suggesting a complex regulation of MYC levels in haematopoietic cells by multiple factors active genome wide at promoters. These observations prompt further speculation that the recurrently occurring mutations in TRRAP, EP400 and HATs identified in solid and liquid tumors by high throughput sequencing efforts 56 may alter cellular levels of MYC as a mechanism of oncogenic transformation. Likewise, it also remains unclear why there is a differential requirement for EPC in normal versus leukemic cells. Elaboration of the binding pattern of EP400 complex components to chromatin in normal HSPC versus leukemic cells and, more generally, enhanced understanding of the differences in the structure and function of chromatin between these two cell types may provide an explanation for our observations. Indeed, understanding the ways in which the chromatin architecture of the two cell types differ is an important approach to the identification of candidate therapeutic targets and strategies.
Supplementary Material
Acknowledgements
We thank Jodie Whitaker, Gillian Newton, Morgan Blaylock, Jeff Barry, Michael Hughes, Gail Bruder, Angela Cooke and Deepti Wilks for technical support; William Harris for assistance with plasmid preparations; Chris Womack and staff at the Clinical Pharmacology Unit at Astra Zeneca, Alderley Park, UK for access to normal subjects for bone marrow collection; and Georges Lacaud, Nullin Divecha and Valerie Kouskoff for a critical reading of the manuscript. This work was supported by Cancer Research UK grant number C5759/A12328.
Funding source: Cancer Research UK grant number C5759/A12328
Footnotes
Conflict of Interest The authors report no conflict of interest.
Supplementary Information Supplementary information is available at Leukemia’s website.
References
- 1.The Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013 May 30;368(22):2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Boer J, Walf-Vorderwulbecke V, Williams O. In focus: MLL-rearranged leukemia. Leukemia. 2013 Jun;27(6):1224–1228. doi: 10.1038/leu.2013.78. [DOI] [PubMed] [Google Scholar]
- 3.Figueroa ME, Lugthart S, Li Y, Erpelinck-Verschueren C, Deng X, Christos PJ, et al. DNA methylation signatures identify biologically distinct subtypes in acute myeloid leukemia. Cancer Cell. 2010 Jan 19;17(1):13–27. doi: 10.1016/j.ccr.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Figueroa ME, Skrabanek L, Li Y, Jiemjit A, Fandy TE, Paietta E, et al. MDS and secondary AML display unique patterns and abundance of aberrant DNA methylation. Blood. 2009 Oct 15;114(16):3448–3458. doi: 10.1182/blood-2009-01-200519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schoofs T, Berdel WE, Muller-Tidow C. Origins of aberrant DNA methylation in acute myeloid leukemia. Leukemia. 2013 Aug 20; doi: 10.1038/leu.2013.242. [DOI] [PubMed] [Google Scholar]
- 6.Fenaux P, Mufti GJ, Hellstrom-Lindberg E, Santini V, Finelli C, Giagounidis A, et al. Efficacy of azacitidine compared with that of conventional care regimens in the treatment of higher-risk myelodysplastic syndromes: a randomised, open-label, phase III study. Lancet Oncol. 2009 Mar;10(3):223–232. doi: 10.1016/S1470-2045(09)70003-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Estey EH. Epigenetics in clinical practice: the examples of azacitidine and decitabine in myelodysplasia and acute myeloid leukemia. Leukemia. 2013 Sep;27(9):1803–1812. doi: 10.1038/leu.2013.173. [DOI] [PubMed] [Google Scholar]
- 8.Burnett AK, Hills RK, Milligan DW, Goldstone AH, Prentice AG, McMullin MF, et al. Attempts to optimize induction and consolidation treatment in acute myeloid leukemia: results of the MRC AML12 trial. J Clin Oncol. 2010 Feb 1;28(4):586–595. doi: 10.1200/JCO.2009.22.9088. [DOI] [PubMed] [Google Scholar]
- 9.Wiseman DH, Greystoke BF, Somervaille TC. The variety of leukemic stem cells in myeloid malignancy. Oncogene. 2013 Jul 8; doi: 10.1038/onc.2013.269. [DOI] [PubMed] [Google Scholar]
- 10.Daigle SR, Olhava EJ, Therkelsen CA, Majer CR, Sneeringer CJ, Song J, et al. Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer Cell. 2011 Jul 12;20(1):53–65. doi: 10.1016/j.ccr.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harris WJ, Huang X, Lynch JT, Spencer GJ, Hitchin JR, Li Y, et al. The histone demethylase KDM1A sustains the oncogenic potential of MLL-AF9 leukemia stem cells. Cancer Cell. 2012;21(4):473–87. doi: 10.1016/j.ccr.2012.03.014. [DOI] [PubMed] [Google Scholar]
- 12.Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478(7370):524–8. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dawson MA, Prinjha RK, Dittmann A, Giotopoulos G, Bantscheff M, Chan WI, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011;478(7370):529–33. doi: 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Balgobind BV, Zwaan CM, Pieters R, Van den Heuvel-Eibrink MM. The heterogeneity of pediatric MLL-rearranged acute myeloid leukemia. Leukemia. 2011 Aug;25(8):1239–1248. doi: 10.1038/leu.2011.90. [DOI] [PubMed] [Google Scholar]
- 15.Meyer C, Hofmann J, Burmeister T, Groger D, Park TS, Emerenciano M, et al. The MLL recombinome of acute leukemias in 2013. Leukemia. 2013 Apr 30; doi: 10.1038/leu.2013.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi J, Wang E, Zuber J, Rappaport A, Taylor M, Johns C, et al. The Polycomb complex PRC2 supports aberrant self-renewal in a mouse model of MLL-AF9;Nras(G12D) acute myeloid leukemia. Oncogene. 2013 Feb 14;32(7):930–938. doi: 10.1038/onc.2012.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Somervaille TC, Cleary ML. Identification and characterization of leukemia stem cells in murine MLL AF9 acute myeloid leukemia. Cancer Cell. 2006;10(4):257–68. doi: 10.1016/j.ccr.2006.08.020. [DOI] [PubMed] [Google Scholar]
- 18.Somervaille TC, Matheny CJ, Spencer GJ, Iwasaki M, Rinn JL, Witten DM, et al. Hierarchical maintenance of MLL myeloid leukemia stem cells employs a transcriptional program shared with embryonic rather than adult stem cells. Cell Stem Cell. 2009;4(2):129–40. doi: 10.1016/j.stem.2008.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yokoyama A, Somervaille TC, Smith KS, Rozenblatt-Rosen O, Meyerson M, Cleary ML. The menin tumor suppressor protein is an essential oncogenic cofactor for MLL-associated leukemogenesis. Cell. 2005;123(2):207–18. doi: 10.1016/j.cell.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 21.Sato T, Russell MA, Denell RE. Homoeosis in Drosophila: a new enhancer of polycomb and related homoeotic mutations. Genetics. 1983;105(2):357–70. doi: 10.1093/genetics/105.2.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stankunas K, Berger J, Ruse C, Sinclair DA, Randazzo F, Brock HW. The enhancer of polycomb gene of Drosophila encodes a chromatin protein conserved in yeast and mammals. Development. 1998;125(20):4055–66. doi: 10.1242/dev.125.20.4055. [DOI] [PubMed] [Google Scholar]
- 23.Boudreault AA, Cronier D, Selleck W, Lacoste N, Utley RT, Allard S, et al. Yeast enhancer of polycomb defines global Esa1-dependent acetylation of chromatin. Genes Dev. 2003;17(11):1415–28. doi: 10.1101/gad.1056603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doyon Y, Selleck W, Lane WS, Tan S, Cote J. Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Mol Cell Biol. 2004;24(5):1884–96. doi: 10.1128/MCB.24.5.1884-1896.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim JR, Kee HJ, Kim JY, Joung H, Nam KI, Eom GH, et al. Enhancer of polycomb1 acts on serum response factor to regulate skeletal muscle differentiation. J Biol Chem. 2009;284(24):16308–16. doi: 10.1074/jbc.M807725200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fuchs M, Gerber J, Drapkin R, Sif S, Ikura T, Ogryzko V, et al. The p400 complex is an essential E1A transformation target. Cell. 2001;106(3):297–307. doi: 10.1016/s0092-8674(01)00450-0. [DOI] [PubMed] [Google Scholar]
- 27.Kim J, Woo AJ, Chu J, Snow JW, Fujiwara Y, Kim CG, et al. A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell. 2010;143(2):313–24. doi: 10.1016/j.cell.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin CY, Loven J, Rahl PB, Paranal RM, Burge CB, Bradner JE, et al. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151(1):56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nie Z, Hu G, Wei G, Cui K, Yamane A, Resch W, et al. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell. 2012;151(1):68–79. doi: 10.1016/j.cell.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sears R, Leone G, DeGregori J, Nevins JR. Ras enhances Myc protein stability. Mol Cell. 1999;3(2):169–79. doi: 10.1016/s1097-2765(00)80308-1. [DOI] [PubMed] [Google Scholar]
- 31.Yin X, Giap C, Lazo JS, Prochownik EV. Low molecular weight inhibitors of Myc-Max interaction and function. Oncogene. 2003;22(40):6151–9. doi: 10.1038/sj.onc.1206641. [DOI] [PubMed] [Google Scholar]
- 32.Huang MJ, Cheng YC, Liu CR, Lin S, Liu HE. A small-molecule c-Myc inhibitor, 10058-F4, induces cell-cycle arrest, apoptosis, and myeloid differentiation of human acute myeloid leukemia. Exp Hematol. 2006;34(11):1480–9. doi: 10.1016/j.exphem.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 33.Chittuluru JR, Chaban Y, Monnet-Saksouk J, Carrozza MJ, Sapountzi V, Selleck W, et al. Structure and nucleosome interaction of the yeast NuA4 and Piccolo-NuA4 histone acetyltransferase complexes. Nat Struct Mol Biol. 2011;18(11):1196–203. doi: 10.1038/nsmb.2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sinclair DA, Clegg NJ, Antonchuk J, Milne TA, Stankunas K, Ruse C, et al. Enhancer of Polycomb is a suppressor of position-effect variegation in Drosophila melanogaster. Genetics. 1998;148(1):211–20. doi: 10.1093/genetics/148.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Micci F, Panagopoulos I, Bjerkehagen B, Heim S. Consistent rearrangement of chromosomal band 6p21 with generation of fusion genes JAZF1/PHF1 and EPC1/PHF1 in endometrial stromal sarcoma. Cancer Res. 2006;66(1):107–12. doi: 10.1158/0008-5472.CAN-05-2485. [DOI] [PubMed] [Google Scholar]
- 36.Nakahata S, Saito Y, Hamasaki M, Hidaka T, Arai Y, Taki T, et al. Alteration of enhancer of polycomb 1 at 10p11.2 is one of the genetic events leading to development of adult T-cell leukemia/lymphoma. Genes Chromosomes Cancer. 2009;48(9):768–76. doi: 10.1002/gcc.20681. [DOI] [PubMed] [Google Scholar]
- 37.Squatrito M, Gorrini C, Amati B. Tip60 in DNA damage response and growth control: many tricks in one HAT. Trends Cell Biol. 2006;16(9):433–42. doi: 10.1016/j.tcb.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 38.McMahon SB, Van Buskirk HA, Dugan KA, Copeland TD, Cole MD. The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell. 1998;94(3):363–74. doi: 10.1016/s0092-8674(00)81479-8. [DOI] [PubMed] [Google Scholar]
- 39.Wood MA, McMahon SB, Cole MD. An ATPase/helicase complex is an essential cofactor for oncogenic transformation by c-Myc. Mol Cell. 2000;5(2):321–30. doi: 10.1016/s1097-2765(00)80427-x. [DOI] [PubMed] [Google Scholar]
- 40.Attwooll C, Oddi S, Cartwright P, Prosperini E, Agger K, Steensgaard P, et al. A novel repressive E2F6 complex containing the polycomb group protein, EPC1, that interacts with EZH2 in a proliferation-specific manner. J Biol Chem. 2005;280(2):1199–208. doi: 10.1074/jbc.M412509200. [DOI] [PubMed] [Google Scholar]
- 41.Osaki H, Walf-Vorderwulbecke V, Mangolini M, Zhao L, Horton SJ, Morrone G, et al. The AAA ATPase RUVBL2 is a critical mediator of MLL-AF9 oncogenesis. Leukemia. 2013 doi: 10.1038/leu.2013.42. [DOI] [PubMed] [Google Scholar]
- 42.Soucek L, Evan GI. The ups and downs of Myc biology. Curr Opin Genet Dev. 2010 Feb;20(1):91–95. doi: 10.1016/j.gde.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murphy DJ, Junttila MR, Pouyet L, Karnezis A, Shchors K, Bui DA, et al. Distinct thresholds govern Myc’s biological output in vivo. Cancer Cell. 2008 Dec 9;14(6):447–457. doi: 10.1016/j.ccr.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pelengaris S, Khan M, Evan GI. Suppression of Myc-induced apoptosis in beta cells exposes multiple oncogenic properties of Myc and triggers carcinogenic progression. Cell. 2002 May 3;109(3):321–334. doi: 10.1016/s0092-8674(02)00738-9. [DOI] [PubMed] [Google Scholar]
- 45.Evan GI, Wyllie AH, Gilbert CS, Littlewood TD, Land H, Brooks M, et al. Induction of apoptosis in fibroblasts by c-myc protein. Cell. 1992 Apr 3;69(1):119–128. doi: 10.1016/0092-8674(92)90123-t. [DOI] [PubMed] [Google Scholar]
- 46.Thomas LR, Tansey WP. Proteolytic control of the oncoprotein transcription factor Myc. Adv Cancer Res. 2011;110:77–106. doi: 10.1016/B978-0-12-386469-7.00004-9. [DOI] [PubMed] [Google Scholar]
- 47.Luscher B, Vervoorts J. Regulation of gene transcription by the oncoprotein MYC. Gene. 2012;494(2):145–60. doi: 10.1016/j.gene.2011.12.027. [DOI] [PubMed] [Google Scholar]
- 48.Malempati S, Tibbitts D, Cunningham M, Akkari Y, Olson S, Fan G, et al. Aberrant stabilization of c-Myc protein in some lymphoblastic leukemias. Leukemia. 2006 Sep;20(9):1572–1581. doi: 10.1038/sj.leu.2404317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hemann MT, Bric A, Teruya-Feldstein J, Herbst A, Nilsson JA, Cordon-Cardo C, et al. Evasion of the p53 tumour surveillance network by tumour-derived MYC mutants. Nature. 2005;436(7052):807–811. doi: 10.1038/nature03845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.King B, Trimarchi T, Reavie L, Xu L, Mullenders J, Ntziachristos P, et al. The ubiquitin ligase FBXW7 modulates leukemia-initiating cell activity by regulating MYC stability. Cell. 2013 Jun 20;153(7):1552–1566. doi: 10.1016/j.cell.2013.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vervoorts J, Luscher-Firzlaff JM, Rottmann S, Lilischkis R, Walsemann G, Dohmann K, et al. Stimulation of c-MYC transcriptional activity and acetylation by recruitment of the cofactor CBP. EMBO Rep. 2003;4(5):484–90. doi: 10.1038/sj.embor.embor821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Faiola F, Liu X, Lo S, Pan S, Zhang K, Lymar E, et al. Dual regulation of c-Myc by p300 via acetylation-dependent control of Myc protein turnover and coactivation of Myc-induced transcription. Mol Cell Biol. 2005;25(23):10220–34. doi: 10.1128/MCB.25.23.10220-10234.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Patel JH, Du Y, Ard PG, Phillips C, Carella B, Chen CJ, et al. The c-MYC oncoprotein is a substrate of the acetyltransferases hGCN5/PCAF and TIP60. Mol Cell Biol. 2004;24(24):10826–34. doi: 10.1128/MCB.24.24.10826-10834.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Luscher B, Vervoorts J. Regulation of gene transcription by the oncoprotein MYC. Gene. 2012;494(2):145–60. doi: 10.1016/j.gene.2011.12.027. [DOI] [PubMed] [Google Scholar]
- 55.Lynch JT, Somerville TD, Spencer GJ, Huang X, Somervaille TC. TTC5 is required to prevent apoptosis of acute myeloid leukemia stem cells. Cell Death Dis. 2013;4:e573. doi: 10.1038/cddis.2013.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39(Database issue):D945–50. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.