Abstract
CD1 proteins are antigen-presenting molecules that evolved to present lipids rather than peptides to T cells. However, unlike major histocompatibility complex genes, CD1 genes show low rates of polymorphism and have not been clearly associated with human disease. We report that an intronic polymorphism in CD1A (rs411089) is associated with susceptibility to tuberculosis in two cohorts of Vietnamese adults (combined cohort odds ratio 1.78; 95%CI: 1.24-2.57; p=0.001). These data strengthen the hypothesis that CD1A-mediated lipid antigen presentation is important for controlling tuberculosis in humans.
Keywords: Human, Tuberculosis, Host Response, CD1A, Genetic Association, Single Nucleotide Polymorphism
Introduction
Mycobacterium tuberculosis is a pathogen of worldwide importance, but its mechanisms of virulence are only partially understood. Infection with M. tuberculosis may result in several outcomes including latent infection and active disease, which can be confined to the lungs or manifest at distant sites. The most common manifestation in adults is pulmonary tuberculosis, while dissemination to the central nervous system is less common but results in greater morbidity and mortality. These myriad outcomes have been difficult to model in animal systems, and human studies are limited1.
Several lines of evidence indicate a role for T-cell mediated immunity in controlling the clinical course of tuberculosis in humans. First, CD4+ T-cell depletion as a result of HIV co-infection has been associated with increased rates of pulmonary (primary progressive and reactivation disease) and extrapulmonary tuberculosis2. Second, candidate gene association studies have revealed links between MHC Class II alleles and susceptibility to tuberculosis3. Finally, Mendelian studies indicate that the inability to respond to IFN-γ, a cytokine produced by T-cells, is associated with susceptibility to disseminated infection after vaccination with BCG4. The identification of the antigens mediating protective T cell immunity is critical for the rational design of novel vaccine candidates. These studies reveal that immunogenetics can illuminate the mechanisms by which protective T cell immunity is generated.
Classically, T cells recognize foreign peptide antigens in the context of polymorphic MHC Class I or Class II molecules. However, T cells can also recognize bacterial lipids in the context of non-classical CD1 molecules5. Thus, the potential catalog of protective antigens extends beyond the proteome of M. tuberculosis. A number of CD1-restricted mycobacterial lipid antigens have been characterized, and T-cell responses to these antigens are detectable in the blood of M. tuberculosis infected humans6,7. Whether CD1-restricted T-cell responses provide protective immunity to tuberculosis remains to be determined.
CD1 proteins are evolutionarily conserved; however, unlike MHC genes, CD1 genes show low rates of genetic polymorphism and have not been clearly associated with human disease8. The human CD1 locus contains five genes (CD1A, CD1B, CD1C, CD1D, and CD1E) clustered on chromosome 1. Notably, the mouse genome contains only an ortholog of human CD1D, thus limiting the utility of mouse models for studying CD1 biology9. Human CD1A-restricted T cells recognize M. tuberculosis infected dendritic cells in-vitro and respond to the mycobacterial cell wall lipopeptide antigen, dideoxymycobactin10,11. Therefore, we hypothesized that CD1A might play a protective role in human tuberculosis.
We recently demonstrated that a polymorphism in the 5′ UTR of CD1A (rs366316) is common and associated with functional CD1A-deficiency in dendritic cells12. The identification of genetic markers for variation in CD1A expression and function provided us with a tool to probe for human disease association. Here, we report that an intronic CD1A polymorphism (rs411089) is reproducibly associated with susceptibility to tuberculosis in Vietnam. These data demonstrate the utility of population genetics studies in elucidating the role of CD1 in human disease.
Results and Discussion
We used a case-population study design to evaluate whether haplotype-tagging SNPs in CD1A were associated with the development of active tuberculosis disease in Vietnamese adults. We genotyped rs366316, rs2269714, rs411089, and rs389293 in a discovery cohort of 352 cases and 382 controls (Table 1). Polymorphisms rs366316 and rs411089 were significantly associated with the development of tuberculosis with a genotypic model (p=0.003 and p=0.036, respectively). We next performed a recessive model analysis since we previously found that the minor homozygous genotypes of both of these SNPs were associated with functional CD1A-deficiency. The minor homozygous genotype of rs411089 was associated with an increased risk of tuberculosis with an odds ratio (OR) of 1.94 (95%CI: 1.13–3.4; p=0.011). The association between the minor homozygous genotype of rs366316 and risk of tuberculosis showed a trend toward significance (OR=1.73; 95%CI: 0.96-3.15; p=0.051). Notably, rs2269714, which codes for a non-synonymous mutation, and rs389293 were not associated with disease. Thus, CD1A SNPs in non-coding or regulatory regions were associated with tuberculosis in this cohort.
Table 1. Candidate gene association study of CD1A and tuberculosis in Vietnam.
Association between SNPs in CD1A and pulmonary (PTB), meningeal (TBM), or pulmonary + meningeal (TB) tuberculosis in Vietnam. SNP genotypes are shown as n(frequency) of total for common homozygous (AA), heterozygous (Aa), and minor homozygous (aa) genotypes. The pvalue for deviation from Hardy-Weinberg equilibrium among cord blood controls was calculated using a χ2 goodness-of-fit test and is indicated under “HWE.” Genotypic analysis compares frequencies in a 2×3 contingency table. For the recessive model analysis, AA + Aa was compared to aa in a 2×2 contingency table. Two-sided hypothesis testing was used for all comparisons. Unadjusted p-values of significance are noted in bold italic.
| Discovery Cohort | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| SNP | Group | HWE | Genotypic Analysis | Recessive Analysis | ||||||||
| AA | Aa | aa | χ 2 | p-value | AA+Aa | aa | χ 2 | p-value | OR 95% CI | |||
|
| ||||||||||||
| rs366316 T → C |
CTRL ALL |
0.20 | 192 (0.5) 161 (0.58) |
164 (0.43) 86 (0.31) |
25 (0.07) 30 (0.11) |
11.36 | 0.003 | 356 (0.93) 247 (0.89) |
25 (0.07) 30 (0.11) |
3.82 | 0.051 | 1.73 (0.96-3.15) |
|
| ||||||||||||
| rs411089 T → C |
CTRL ALL |
0.18 | 192 (0.5) 171 (0.49) |
165 (0.43) 138 (0.39) |
25 (0.07) 42 (0.12) |
6.64 | 0.036 | 357 (0.93) 309 (0.88) |
25 (0.07) 42 (0.12) |
6.47 | 0.011 | 1.94 (1.13-3.4) |
|
| ||||||||||||
| rs2269714 C → T |
CTRL ALL |
0.56 | 120 (0.32) 116 (0.33) |
192 (0.51) 168 (0.48) |
68 (0.18) 68 (0.19) |
0.60 | 0.742 | 312 (0.82) 284 (0.81) |
68 (0.18) 68 (0.19) |
0.24 | 0.621 | 1.10 (0.74-1.62) |
|
| ||||||||||||
| rs389293 G → A |
CTRL ALL |
0.80 | 159 (0.42) 161 (0.47) |
176 (0.46) 143 (0.42) |
46 (0.12) 38 (0.11) |
2.09 | 0.352 | 335 (0.88) 304 (0.89) |
46 (0.12) 38 (0.11) |
0.16 | 0.687 | 0.91 (0.56-1.47) |
|
| ||||||||||||
| Validation Cohort | ||||||||||||
|
| ||||||||||||
| rs366316 T → C |
CTRL ALL |
0.91 | 214 (0.58) 156 (0.59) |
133 (0.36) 93 (0.35) |
20 (0.05) 17 (0.06) |
0.31 | 0.858 | 347 (0.95) 249 (0.94) |
20 (0.05) 17 (0.06) |
0.25 | 0.618 | 1.18 (0.57-2.43) |
|
| ||||||||||||
| rs411089 T → C |
CTRL ALL |
0.56 | 171 (0.47) 161 (0.51) |
159 (0.44) 110 (0.35) |
32 (0.09) 46 (0.15) |
8.80 | 0.012 | 330 (0.91) 271 (0.85) |
32 (0.09) 46 (0.15) |
5.35 | 0.021 | 1.75 (1.06-2.92) |
|
| ||||||||||||
| Combined Cohort | ||||||||||||
|
| ||||||||||||
| rs366316 T → C |
CTRL ALL |
0.33 | 406 (0.54) 317 (0.58) |
297 (0.4) 179 (0.33) |
45 (0.06) 47 (0.09) |
7.90 | 0.019 | 703 (0.94) 496 (0.91) |
45 (0.06) 47 (0.09) |
3.31 | 0.069 | 1.48 (0.95-2.32) |
|
| ||||||||||||
| PTB TBM |
217 (0.57) 100 (0.61) |
126 (0.33) 53 (0.32) |
35 (0.09) 12 (0.07) |
6.88 3.35 |
0.032 0.188 |
343 (0.91) 153 (0.93) |
35 (0.09) 12 (0.07) |
4.00 0.36 |
0.045 0.546 |
1.59 (0.97-2.59) 1.23 (0.58-2.42) |
||
|
| ||||||||||||
| rs411089 T → C |
CTRL ALL |
0.27 | 367 (0.49) 327 (0.5) |
325 (0.43) 245 (0.37) |
59 (0.08) 87 (0.13) |
12.96 | 0.002 | 692 (0.92) 572 (0.87) |
59 (0.08) 87 (0.13) |
10.81 | 0.001 | 1.78 (1.24-2.57) |
|
| ||||||||||||
| PTB TBM |
182 (0.49) 145 (0.5) |
137 (0.37) 108 (0.38) |
52 (0.14) 35 (0.12) |
11.96 6.01 |
0.003
0.050 |
319 (0.86) 253 (0.88) |
52 (0.14) 35 (0.12) |
10.57 4.67 |
0.001
0.031 |
1.91 (1.26-2.89) 1.62 (1.01-2.57) |
||
Next, we genotyped rs366316 and rs411089 in a validation cohort of 339 cases and 376 controls. Again, we found a significant association between rs411089 and tuberculosis (p=0.012) in a genotypic analysis; however, there was no association with rs366316 (p=0.858) (Table 1). We confirmed the association of the minor homozygous genotype of rs411089 in a recessive model analysis (OR=1.75, 95%CI: 1.06–2.92; p=0.021). In a combined cohort analysis, we found a highly significant association between rs411089 and tuberculosis (OR=1.78, 95%CI: 1.24–2.57; p=0.001). Because of its previously documented functional importance, we also examined rs366316 in a combined cohort even though the initial association did not validate. We found a statistically significant association between rs366316 and the risk of tuberculosis (p=0.019). Because rs366316 and rs411089 are in low linkage disequilibrium in Vietnam (R2=0.15; Figure 1), we tested for evidence of genetic interaction with a haplotype analysis but found no evidence for this (data not shown). These data identify rs411089 as a second CD1A locus of potential functional importance that is reproducibly associated with susceptibility to tuberculosis.
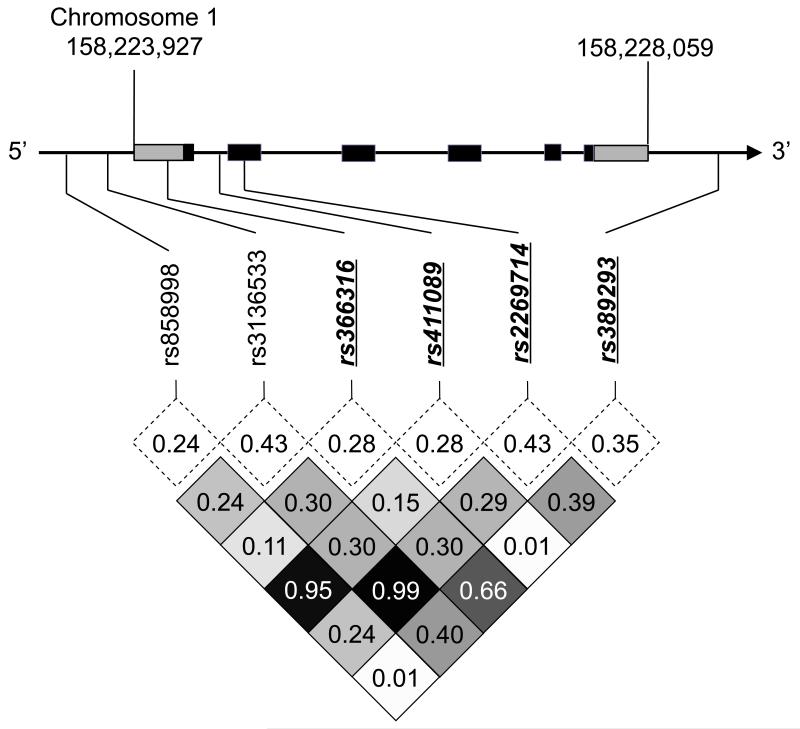
Figure 1. CD1A single nucleotide polymorphisms (SNPs) in Vietnam.
(a) Linkage disequilibrium plot of SNPs in CD1A coding region ± 10kB flanking regions among cord blood controls in Vietnam. CD1A spans 4132 bases on chromosome 1 and consists of six exons (black) and two untranslated regions (gray). Minor allele frequencies (dotted box) and linkage disequilibrium as measured by R2 values (shaded box) are indicated. Four haplotype-tagging SNPs selected for genotyping are emphasized in bold italic underline.
Finally, we evaluated whether rs366316 and rs411089 were associated with clinical subtypes of tuberculosis, either pulmonary or meningeal disease. Stratification by site of disease showed that rs366316 was modestly associated with pulmonary disease (p=0.032) and was not associated with meningeal tuberculosis (p=0.188). In a recessive model analysis, rs366316 showed a borderline association with pulmonary disease (OR=1.59, 95%CI 0.97–2.59; p=0.045). By contrast, rs411089 was associated with both pulmonary and meningeal tuberculosis (p=0.003 and p=0.050 respectively) in a genotypic model. In a recessive model analysis, rs411089 was also associated with both pulmonary (OR=1.91, 95%CI 1.26–2.89; p=0.001) and meningeal (OR=1.62, 95%CI 1.01–2.57; p=0.031) tuberculosis (Table 1). These data are consistent with our findings in the unstratified analysis, and demonstrate that CD1A SNPs are associated with both pulmonary and meningeal tuberculosis in Vietnamese adults.
The capacity for T cells to respond to mycobacterial lipid antigens in the context of CD1 molecules was described almost twenty years ago, yet the importance of this observation for human disease pathogenesis is still not clear. We found that rs411089, an intronic SNP within CD1A, was associated with susceptibility to tuberculosis in two cohorts of Vietnamese adults. We also found that rs366316, which is associated with functional CD1A-deficiency in dendritic cells, was not reproducibly associated with tuberculosis in this study. These data demonstrate the utility of a population genetics approach to understanding the role of CD1-mediated lipid antigen presentation in human disease. The information gained from studies like this one could be important in the design of novel vaccines. For example, T cell immunity against lipids might be expected after administration of a whole cell vaccine but not with a protein subunit vaccine.
The findings reported here are subject to at least two limitations. First, our study design included cord blood controls, and some of these individuals will surely develop tuberculosis as adults. However, misclassification of controls would indicate that the associations reported here underestimate the true genetic risk. Second, we did not validate an association between a previously known functional variant (rs366316) and tuberculosis. Rather than indicating that there is no association between CD1A-deficiency and tuberculosis, these data could be the result of a lack of statistical power, a possibility underscored by the marginal association still present in the combined cohort analysis. The other possibility is that the immunologic mechanisms involved in pulmonary and extrapulmonary infection are very different from what we have described when studying in-vitro derived dendritic cells. Thus, the reproducible association between rs411089 and tuberculosis could be the consequence of an additional unknown genetic regulatory mechanism for CD1A. We do not know if rs411089 modulates transcription of CD1A independent of rs366316, but the positive findings reported here and low linkage disequilibrium between the two SNPs suggest this is possible. It is notable that rs411089 is strongly linked to rs858998, which is located in the CD1A promoter region and might regulate transcription independently of rs366316 (Supplementary Figure 1). If confirmed through mechanistic studies, impaired lipid antigen presentation would be a plausible biological mechanism for the association between SNPs in CD1A and susceptibility to tuberculosis.
Materials and Methods
Human Subjects
Details of clinical characteristics and enrollment criteria for Vietnam study subjects have been previously published13. Population controls were enrolled at Hung Vuong Hospital where umbilical cord blood was collected from newborns. Subjects with pulmonary tuberculosis were recruited from a network of district tuberculosis control units and defined by typical clinical symptoms in addition to sputum smear positive for acid-fast bacilli and/or culture positive for M. tuberculosis. Subjects with meningeal tuberculosis (TBM) were classified as definite or probable. All cases in the discovery cohort had definite TBM with clinical meningitis and either positive Ziehl-Neelsen stain for acid-fast bacilli or positive M. tuberculosis culture from CSF. The validation cohort included subjects with both definite (N=54) and probable (N=73) meningitis defined as clinical meningitis plus one or more of the following: chest radiograph consistent with active tuberculosis, acid-fast bacilli found in any specimen other than CSF, and clinical evidence of extrapulmonary tuberculosis at an additional site. All case and control participants were unrelated and greater than 95% were of the Vietnamese Kinh ethnicity.
Ethics
Written, informed consent was obtained from patients or their relatives if the patient could not provide consent. Parents provided consent for cord-blood controls. All protocols were approved by human subject review committees at Pham Ngoc Thach Hospital for Tuberculosis, the Hospital for Tropical Diseases, Health Services of Ho Chi Minh City, and Hung Vuong Hosptial in Vietnam as well as the Oxford Tropical Research Ethics Committee and the University of Washington.
Selection of Single Nucleotide Polymorphisms (SNPs) for Genotyping
We used data from the International Haplotype Mapping Project (www.hapmap.org, version 3, release 2) to select SNPs within ten kilobases of CD1A from the Han Chinese in Beijing, China (CHB) or Japanese in Tokyo, Japan (JPT). We previously demonstrated similar genome-wide haplotype frequencies between Vietnam and CHB/JPT populations14. There are seven SNPs with a minor allele frequency greater than 4% within the CD1A gene region, and all but one of these SNPs is located within regulatory or non-coding regions of the gene (Supplementary Figure 1). We genotyped six of these SNPs in 382 control subjects from Vietnam, and confirmed a similar minor allele frequency and linkage pattern when compared to the CHB/JPT HapMap populations. Haplotype-tagging SNPs with a R2 cutoff of 0.80 for linkage disequilibrium were identified using Haploview v4.2 (www.broad.mit.edu/haploview). Thus, we were able to select only four tagged SNPs as markers for a candidate gene association study (Figure 1).
Genomic techniques
Genomic DNA was prepared from peripheral blood (Qiagen). Because of limiting quantities, genomic DNA from cases in Vietnam was amplified using Repli-G (Qiagen). Multiplex genotyping of rs411089, rs2269714, and rs389293 in the discovery cohort was performed using allele-specific primer extension on the MassARRAY (Sequenom) platform. Genotyping for rs366316 in the discovery cohort and rs366316 and rs411089 in the validation cohort was performed using TaqMan SNP Genotyping Assay (Applied Biosystems, Catalog# 4351379). In all cohorts and platforms, automatic call rates exceeded 95% and less than 3% of calls were assigned manually. Genotypes were confirmed in a subset of individuals by DNA sequencing or by genotyping on an alternate platform. All SNPs were in Hardy-Weinberg equilibrium among control subjects, effectively ruling out errors in genotyping (Table 1). We have previously demonstrated a lack of population stratification in this cohort13,14.
Statistics
Statistical analyses were performed using Stata Statistical Software: Release 11 (StataCorp LP, College Station, TX). Function ‘pwld’ was used to calculate R2 measurements of linkage disequilibrium between polymorphisms. SNPs were assessed for association with tuberculosis using “genassoc”15
Supplementary Material
Acknowledgements
We would like to acknowledge the work of the clinical staff from the Hospital of Tropical Diseases and Pham Ngoc Thach Hospital who initially diagnosed and studied the patients with TBM and PTB. We would like to thank Dr. Nguyen Thi Hieu from Hung Vuong Obstetric Hospital Vietnam, Dr Tran Tinh Hien from the Hospital for Tropical Diseases Vietnam, and all the Vietnamese Doctors and patients who participated in this study. We thank Drs. Alan Aderem (Seattle BioMed), Marta Janer and Sarah Li (Institute for Systems Biology) for advice and technical assistance.
Funding
This study was funded in part by the NIH (K24AI089794 to TRH and K08AI089938 to CS), the Burroughs Wellcome Foundation (TRH), and the Irvington Institute Fellowship Program (CS). The clinical component of this study was funded through the Wellcome Trust Major Overseas Program in Vietnam (089276/Z/09/Z).
Footnotes
Conflict of Interest Statement
All authors declare that they have no competing financial interests with this research.
Conference Presentations
The findings described here are unpublished but were presented as an oral abstract at the 6th International Symposium on CD1 and NKT Cells (Chicago, USA September 23-27, 2011) and a poster at the Host Response in Tuberculosis Keystone Symposium (Whistler, CA March 13-18, 2013)
References
- 1.Russell DG, Barry CE, 3rd, Flynn JL. Tuberculosis: what we don’t know can, and does, hurt us. Science. 2010;328(5980):852–6. doi: 10.1126/science.1184784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zumla A, Raviglione M, Hafner R, von Reyn CF. Tuberculosis. N Engl J Med. 2013;368(8):745–55. doi: 10.1056/NEJMra1200894. [DOI] [PubMed] [Google Scholar]
- 3.Remus N, Alcais A, Abel L. Human genetics of common mycobacterial infections. Immunol Res. 2003;28(2):109–29. doi: 10.1385/IR:28:2:109. [DOI] [PubMed] [Google Scholar]
- 4.Casanova JL, Abel L. Genetic dissection of immunity to mycobacteria: the human model. Annu Rev Immunol. 2002;20:581–620. doi: 10.1146/annurev.immunol.20.081501.125851. [DOI] [PubMed] [Google Scholar]
- 5.Beckman EM, Porcelli SA, Morita CT, Behar SM, Furlong ST, Brenner MB. Recognition of a lipid antigen by CD1-restricted alpha beta+ T cells. Nature. 1994;372(6507):691–4. doi: 10.1038/372691a0. [DOI] [PubMed] [Google Scholar]
- 6.Montamat-Sicotte DJ, Millington KA, Willcox CR, Hingley-Wilson S, Hackforth S, Innes J, et al. A mycolic acid-specific CD1-restricted T cell population contributes to acute and memory immune responses in human tuberculosis infection. J Clin Invest. 2011;121(6):2493–503. doi: 10.1172/JCI46216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van Rhijn I, Ly D, Moody DB. CD1a, CD1b, and CD1c in immunity against mycobacteria. Adv Exp Med Biol. 2013;783:181–97. doi: 10.1007/978-1-4614-6111-1_10. [DOI] [PubMed] [Google Scholar]
- 8.Dascher CC. Evolutionary biology of CD1. Curr Top Microbiol Immunol. 2007;314:3–26. doi: 10.1007/978-3-540-69511-0_1. [DOI] [PubMed] [Google Scholar]
- 9.Balk SP, Bleicher PA, Terhorst C. Isolation and expression of cDNA encoding the murine homologues of CD1. J Immunol. 1991;146(2):768–74. [PubMed] [Google Scholar]
- 10.Moody DB, Young DC, Cheng TY, Rosat JP, Roura-Mir C, O’Connor PB, et al. T cell activation by lipopeptide antigens. Science. 2004;303(5657):527–31. doi: 10.1126/science.1089353. [DOI] [PubMed] [Google Scholar]
- 11.Rosat JP, Grant EP, Beckman EM, Dascher CC, Sieling PA, Frederique D, et al. CD1-restricted microbial lipid antigen-specific recognition found in the CD8+ alpha beta T cell pool. J Immunol. 1999;162(1):366–71. [PubMed] [Google Scholar]
- 12.Seshadri C, Shenoy M, Wells RD, Hensley-McBain T, Andersen-Nissen E, McElrath MJ, et al. Human CD1a Deficiency Is Common and Genetically Regulated. J Immunol. 2013 doi: 10.4049/jimmunol.1300575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horne DJ, Randhawa AK, Chau TT, Bang ND, Yen NT, Farrar JJ, et al. Common Polymorphisms in the PKP3-SIGIRR-TMEM16J Gene Region Are Associated With Susceptibility to Tuberculosis. J Infect Dis. 2012 doi: 10.1093/infdis/jir785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khor CC, Chau TN, Pang J, Davila S, Long HT, Ong RT, et al. Genome-wide association study identifies susceptibility loci for dengue shock syndrome at MICB and PLCE1. Nat Genet. 2011;43(11):1139–41. doi: 10.1038/ng.960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shephard N. GENASS: Stata module to perform Genetic Case-control Association tests. Boston College Department of Economics; 2005. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.