Abstract
The plant hormone auxin modulates cell proliferation and cell expansion in part by changing gene expression. Among the three primary auxin response gene families, Aux/IAA, GH3 and SAUR, the function of SAUR genes remains unclear. SAUR transcripts were initially identified in epidermal and cortical cells of elongating tissues and thus were supposed to regulate cell expansion downstream of auxin transport and auxin signaling. Recent studies have proposed that SAUR proteins are able to modulate auxin transport and cell expansion by an unknown mechanism. We present our work on the SAUR41 subfamily genes of Arabidopsis (SAUR41, SAUR40, SAUR71 and SAUR72). Similar to the fusion protein between SAUR41 and EGFP, both SAUR40-EGFP and SAUR71-EGFP were identified in the cytoplasm of all types of root tip cells. This result indicated that the subcellular location pattern of SAUR proteins among the members of the same subfamily could be similar to each other, although the overall location pattern of SAUR proteins appeared to be highly diverse. SAUR41 was distinctively expressed in the quiescent center and cortex/endodermis initials of root stem cell niches and in the endodermis of hypocotyls, whereas SAUR71 and SAUR72 were expressed in the steles of young roots and hypocotyls. In addition, SAUR71 was differentially expressed during stomatal formation. The tissue-specific and developmentally regulated expression patterns of the SAUR41 subfamily genes imply that SAUR transcripts or SAUR proteins might serve as signal molecules to ensure the coordination of cell proliferation and cell expansion. Finally, Arabidopsis seedlings expressing TAP (tandem affinity peptide) tagged SAUR41 displayed phenotypes, indicating that it was rational to use the TAP approach for identification of potential binding partners of SAUR41 proteins.
Keywords: SAUR, Arabidopsis, auxin transport, cell expansion, root meristem patterning, stomatal development
The plant hormone auxin modulates many aspects of plant growth and development and it regulates cell proliferation and cell expansion in part by changing gene expression. Primary auxin response genes consist of members of three gene families: Aux/IAA (auxin/indole-3-acetic acid), GH3 (gretchen hagen3) and SAUR (small auxin up RNA).1 Among these genes, the function of SAUR genes remains unclear, most likely due to genetic redundancy within the large SAUR gene family.1,2
Recent studies have begun to address the function of SAUR genes in Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa). Arabidopsis SAUR32 has been proposed to be involved in apical hook development.3 Rice SAUR39 has been identified as a negative regulator of auxin synthesis and transport, since rice plants overexpressing this gene exhibit reduced lateral root development and shoot and root length.4,5 The SAUR gene family of Arabidopsis could be phylogenetically classified into three clades.6 More recent work has suggested that SAUR19 (clade II) and SAUR63 (clade I) promote cell expansion through the modulation of auxin transport.7,8 Most recently, we presented our work on SAUR41, a clade III SAUR gene with a distinctive expression pattern in root meristems.9 Overexpression of SAUR41 from the CaMV 35S promoter led to pleiotropic auxin-related phenotypes, including long hypocotyls, increased vegetative biomass and lateral root development in young seedlings and expanded petals and twisted inflorescence stems in adult plants.9 Tissue-specific expression of SAUR41 from promoters of auxin transporter genes and root meristem patterning genes (PIN1, PIN2, WOX5, PLT2 and ACR4) differentially modulated root meristem development, root cell expansion and root gravitropic growth.9 In the meantime, SAUR36, another clade III SAUR gene of Arabidopsis has been identified as a positive regulator of leaf senescence and a negative regulator of leaf size.10 In an independent study, SAUR36 has been characterized as a regulator of seed germination in response to gibberellins and abscisic acid.11 Ectopic expression of SAUR36 led to the absence of apical hooks in etiolated seedlings and significantly longer hypocotyls in light-grown seedlings. However, mature plants ectopically expressing SAUR36 displayed dramatically wavy growth of inflorescence axes which could be linked to defects in auxin transport.11 The mechanisms of how SAUR proteins modulate auxin transport and why they sometimes promote auxin transport but in other cases inhibit auxin transport remains unclear.
The subcellular location pattern of SAUR proteins appeared to be highly diverse. By examining GFP-tagged SAUR proteins, rice SAUR39 was found exclusively in the cytoplasm.4 In Arabidopsis, SAUR41 localized to the cytoplasm,9 SAUR19 and SAUR63 localized primarily to the plasma membrane,7,8 whereas SAUR36 targeted the nucleus.12
The SAUR41 subfamily contained four members: SAUR41, At1g16510; SAUR40, At1g79130; SAUR71, At1g56150; SAUR72, At3g12830. Their amino acid sequences differed from other SAUR families in the N-terminus. Herein, to investigate the subcellular localization of SAUR40 and SAUR71 proteins, we generated transgenic constructs in which the CaMV (cauliflower mosaic virus) 35S promoter drove C-terminal translational fusions between the full-length SAUR40 or SAUR71 and the EGFP proteins. Location of fusion proteins in stably-transformed Arabidopsis plants was examined by confocal microscopy. Similar to the SAUR41 fusion protein, SAUR40-EGFP and SAUR71-EGFP were identified in the cytoplasm of all types of root tip cells (Fig. 1A–D). Although we have not generated transgenic lines for the SAUR72-EGFP construction, we suggest that the subcellular location pattern of SAUR proteins among the members of the same gene subfamily could be similar to each other. As a support to this view, the SAUR19 subfamily members (SAUR19, 21, 23 and 24) have been reported to exhibit identical subcellular location patterns.7
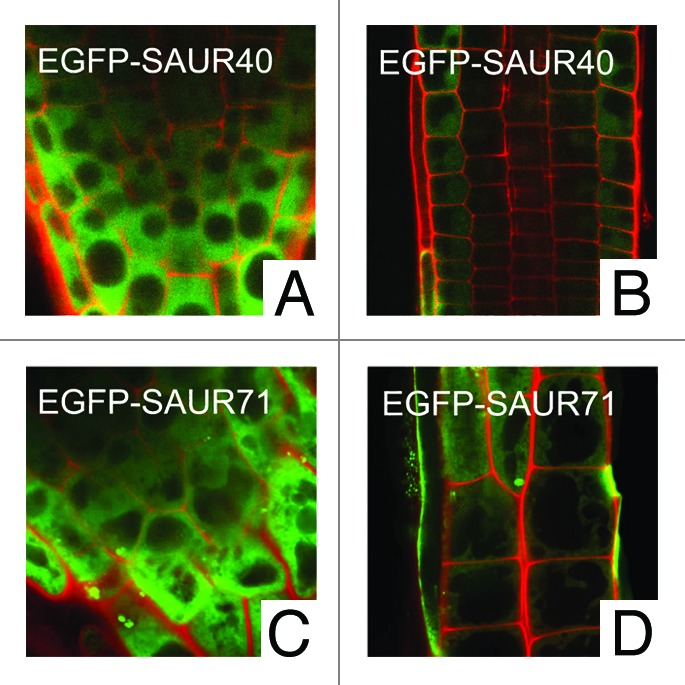
Figure 1. Subcellular location of SAUR40 and SAUR71 fusion proteins. Green EGFP (enhanced green fluorescent protein) signals and red PI (propidium iodide, staining cell wall here) signals were viewed by confocal microscopy.
According to microarray data, the SAUR genes of Arabidopsis in clades I and II had a tendency to display higher expression levels in leaves and lower expressions in roots, while the genes in clade III demonstrated opposite expression patterns.6 The SAUR19 subfamily genes were expressed in growing hypocotyls in response to shade avoidance and in root elongation zones in response to auxin treatment.7 SAUR63 and members of its clade were expressed in growing regions of hypocotyls, cotyledons, petiole, young rosette leaves and inflorescence stems, but not in roots.8 We found that SAUR41 displayed distinctive expression patterns in Arabidopsis roots; it was normally restricted to the quiescent center and cortex/endodermis initials of primary root meristems.9 During lateral root development, SAUR41 was also expressed in prospective stem cell niches of lateral root primordial and in expanding endodermal cells surrounding the lateral root primordia.9
To counteract possible position effects in plant promoter analysis, we used the gypsy-Su(Hw) system of Drosophila in a novel approach that facilitated high and precise expression of reporter genes.13 Using this system, together with the GATEWAY recombination approach, we generated promoter reporter lines for other members of the SAUR41 subfamily genes. We found that the expression pattern of SAUR40 was similar to that of SAUR41, while the expression pattern of SAUR71 was similar to that of SAUR72. Results showed that SAUR72 and SAUR71 were highly expressed in the steles of Arabidopsis roots and hypocotyls (Fig. 2A–C). Interestingly, SAUR71 was extensively expressed in guard mother cells and young guard cells of Arabidopsis cotyledons and leaves during stomatal formation (Fig. 2D).
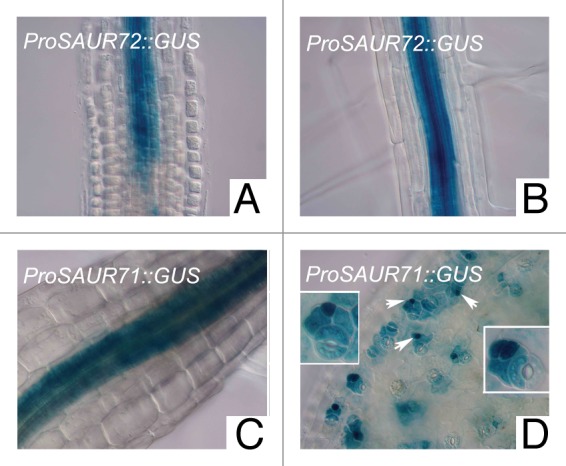
Figure 2. Expression patterns of SAUR71 and SAUR72. GUS (β-glucuronidase) activity stained in blue was viewed by DIC (differential-interference-contrast) microscopy. Arrows indicate guard mother cells.
SAUR transcripts were originally identified in tissues undergoing differential cell expansion during tropic growth and in the epidermal and cortical cells of elongating hypocotyls.14-16 Thus, they were supposed to regulate cell expansion downstream of auxin transport and auxin signaling. Recent work has suggested that they are able to modulate auxin transport and cell expansion by an unknown mechanism.4,7-9 Our work revealed the tissue-specific and developmentally regulated expression patterns of the SAUR41 subfamily genes. SAUR41 was distinctively expressed in the quiescent center and cortex/endodermis initials of root stem cell niches and in the endodermis of hypocotyls. SAUR71 and SAUR72 were expressed in the steles of young roots and hypocotyls (Fig. 2A–C). In addition, SAUR71 was differentially expressed during stomatal formation (Fig. 2D). It has also been reported that SAUR36 was expressed in the regions of shoot apical meristems and in the distinct regions related to the formation of lateral roots.11 Taken together, SAUR transcripts could be identified in stem cells and stele cells of roots and shoots. These results imply that SAUR transcripts or SAUR proteins may serve as signal molecules to ensure the coordination of cell proliferation and cell expansion by unknown pathway(s), because auxin plays a critical role in the integration of cell division, differentiation and expansion. Further investigations to better understand the molecular mechanism of SAUR proteins and to characterize more loss-of-function SAUR mutants are of great interest for expanding our knowledge of auxin biology.
Co-immunoprecipitation (CoIP) is a popular technique to identify physiologically relevant protein-protein interactions. SAUR19 and SAUR63 proteins are highly unstable in Arabidopsis and the addition of epitope tags to these proteins increases the stability of SAUR proteins.7,8 Arabidopsis plants displayed phenotypes only when they expressed stabilized SAUR19 or SAUR63 proteins.7,8 By contrast, Arabidopsis overexpressing untagged SAUR41 had strong phenotypes, similar to that of MYC-tagged SAUR41.9 Here, we found that Arabidopsis seedlings expressing TAP (tandem affinity peptide) tagged SAUR41 displayed phenotypes (Fig. 3), indicating that it was rational to use the TAP approach for identification of potential binding partners of SAUR41 proteins. Taken together, it could be helpful using different SAUR proteins to explore the mechanisms of SAUR proteins and these studies could complement and reinforce each other.
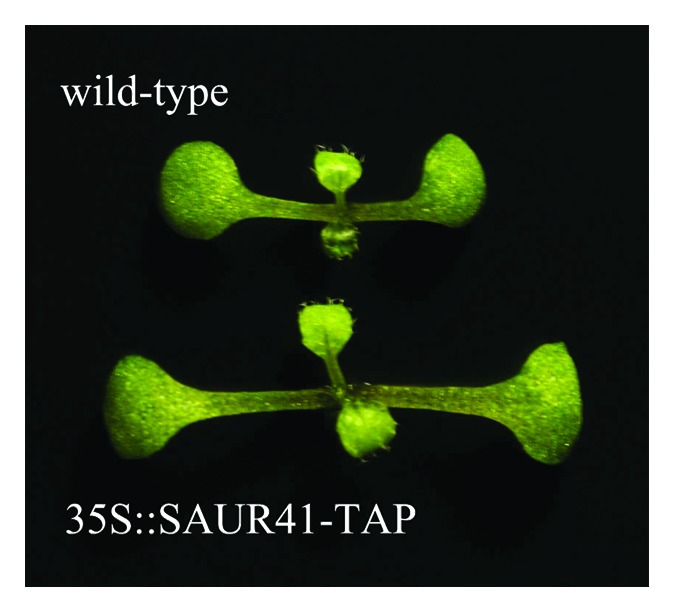
Figure 3.Arabidopsis seedlings expressing TAP (tandem affinity peptide) tagged SAUR41 had longer cotyledon petioles than the wild-type. Seedlings were 7-d-old.
Acknowledgments
This work was supported by funding from the National Natural Science Foundation of China (grant no. 31170211 to J.W.). We are grateful to ABRC and RIKEN-BRC for the distribution of Arabidopsis materials.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/25283
References
- 1. Hagen G, Guilfoyle T. Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol Biol. 2002;49:373–85. doi: 10.1023/A:1015207114117. [DOI] [PubMed] [Google Scholar]
- 2.Jain M, Tyagi AK, Khurana JP. Genome-wide analysis, evolutionary expansion, and expression of early auxin-responsive SAUR gene family in rice (Oryza sativa) Genomics. 2006;88:360–71. doi: 10.1016/j.ygeno.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 3.Park JE, Kim YS, Yoon HK, Park CM. Functional characterization of a small auxin-up RNA gene in apical hook development in Arabidopsis. Plant Sci. 2007;172:150–7. doi: 10.1016/j.plantsci.2006.08.005. [DOI] [Google Scholar]
- 4.Kant S, Bi YM, Zhu T, Rothstein SJ. SAUR39, a small auxin-up RNA gene, acts as a negative regulator of auxin synthesis and transport in rice. Plant Physiol. 2009;151:691–701. doi: 10.1104/pp.109.143875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kant S, Rothstein S. Auxin-responsive SAUR39 gene modulates auxin level in rice. Plant Signal Behav. 2009;4:1174–5. doi: 10.4161/psb.4.12.10043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kodaira KS, Qin F, Tran L-SP, Maruyama K, Kidokoro S, Fujita Y, et al. Arabidopsis Cys2/His2 zinc-finger proteins AZF1 and AZF2 negatively regulate abscisic acid-repressive and auxin-inducible genes under abiotic stress conditions. Plant Physiol. 2011;157:742–56. doi: 10.1104/pp.111.182683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spartz AK, Lee SH, Wenger JP, Gonzalez N, Itoh H, Inzé D, et al. The SAUR19 subfamily of SMALL AUXIN UP RNA genes promote cell expansion. Plant J. 2012;70:978–90. doi: 10.1111/j.1365-313X.2012.04946.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chae K, Isaacs CG, Reeves PH, Maloney GS, Muday GK, Nagpal P, et al. Arabidopsis SMALL AUXIN UP RNA63 promotes hypocotyl and stamen filament elongation. Plant J. 2012;71:684–97. doi: 10.1111/j.1365-313X.2012.05024.x. [DOI] [PubMed] [Google Scholar]
- 9.Kong Y, Zhu Y, Gao C, She W, Lin W, Chen Y, et al. Tissue-specific expression of SMALL AUXIN UP RNA41 differentially regulates cell expansion and root meristem patterning in Arabidopsis. Plant Cell Physiol. 2013;54:609–21. doi: 10.1093/pcp/pct028. [DOI] [PubMed] [Google Scholar]
- 10.Hou K, Wu W, Gan SS. SAUR36, a small auxin up RNA gene, is involved in the promotion of leaf senescence in Arabidopsis. Plant Physiol. 2013;161:1002–9. doi: 10.1104/pp.112.212787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stamm P, Kumar PP. Auxin and gibberellin responsive Arabidopsis SMALL AUXIN UP RNA36 regulates hypocotyl elongation in the light. Plant Cell Rep. 2013;32:759–69. doi: 10.1007/s00299-013-1406-5. [DOI] [PubMed] [Google Scholar]
- 12.Narsai R, Law SR, Carrie C, Xu L, Whelan J. In-depth temporal transcriptome profiling reveals a crucial developmental swith germination in Arabidopsis. Plant Physiol. 2011;157:1342–62. doi: 10.1104/pp.111.183129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.She W, Lin W, Zhu Y, Chen Y, Jin W, Yang Y, et al. The gypsy insulator of Drosophila melanogaster, together with its binding protein suppressor of Hairy-wing, facilitate high and precise expression of transgenes in Arabidopsis thaliana. Genetics. 2010;185:1141–50. doi: 10.1534/genetics.110.117960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McClure BA, Guifoyle TJ. Characterization of a class of small auxin-inducible soybean polyadenylated RNAs. Plant Mol Biol. 1987;9:611–23. doi: 10.1007/BF00020537. [DOI] [PubMed] [Google Scholar]
- 15.McClure BA, Guilfoyle T. Rapid redistribution of auxin-regulated RNAs during gravitropism. Science. 1989;243:91–3. doi: 10.1126/science.11540631. [DOI] [PubMed] [Google Scholar]
- 16.Gee MA, Hagen G, Guilfoyle TJ. Tissue-specific and organ-specific expression of soybean auxin-responsive transcripts GH3 and SAURs. Plant Cell. 1991;3:419–30. doi: 10.1105/tpc.3.4.419. [DOI] [PMC free article] [PubMed] [Google Scholar]


