Abstract
miR820 is a small RNA species (22 and 24 nucleotides), produced from transcripts originated from a region inside CACTA DNA transposons in rice. Because MIR820 is a transposon gene, its expression may depend on the transposon copy number. Here, we investigated the copy number of MIR820 and its expression levels in various cultivars and wild species of rice. We found no correlation between copy number and expression level, suggesting that MIR820 transcription is regulated not by the copy dosage but by the epigenetic state of each copy.
Keywords: transposon, Oryza sativa, DNA methyltransferase, miR820, OsDRM2
Transposable elements (TEs) and their remnants are the major components of eukaryotic genomes.1 TEs have increased their copy number in the host genome because they replicate faster than the host genome. Thus, TEs are referred to as ultimate parasites that proliferate selfishly in the genome.2,3 TE transposition induces insertion mutations and chromosome aberrations, causing instability of the host genome. Therefore, most TEs are kept silent by their host. Small RNA-mediated RNA silencing participates in suppression of TEs; this mechanism has a similar role in plants and animals.4-6
The prominent presence of TEs in the host genome suggests the existence of a running battle between the host defense machinery suppressing transposition of TEs and TEs’ countermeasures against host-mediated silencing. However, little is known about the strategies that TEs have escaped the host silencing. We have previously shown that members of the microRNA820 (miR820) family negatively regulate OsDRM2 (a de novo DNA methyltransferase gene), which allows transposons to escape silencing by the host. We also found a dramatic proliferation of CACTA transposons carrying MIR820 in some wild rice accessions, such as BB- and BBCC-genome species.7 Therefore, we assumed that MIR820 expression would be extremely high in these species.
To test this, we conducted northern blot analysis to detect miR820 in three different cultivated rice accessions (Oryza sativa) and one wild rice species (Oryza punctata, accession W1514) (Fig. 1A). Surprisingly, no expression was detected in W1514 despite the presence of more than 18 copies of MIR820.7 Accordingly, no cleavage of OsDRM2 by miR820 was detected in W1514 (Fig. 1B). Next, we determined the levels of pre-miR820 by quantitative RT-PCR (qRT-PCR) in two O. sativa cultivars (AA) and four wild rice accessions belonging to two species containing the BB or BBCC genome, in which CACTA carrying MIR820 is highly amplified (Fig. 1C).7 The qRT-PCR revealed that the absence of miR820 in W1514 is due to a reduced level of the pre-miR820 transcript, rather than to its reduced processing. This implies that the high copy number of MIR820 in wild rice accession with BB or BBCC genome may cause a stronger MIR820 silencing in these species.
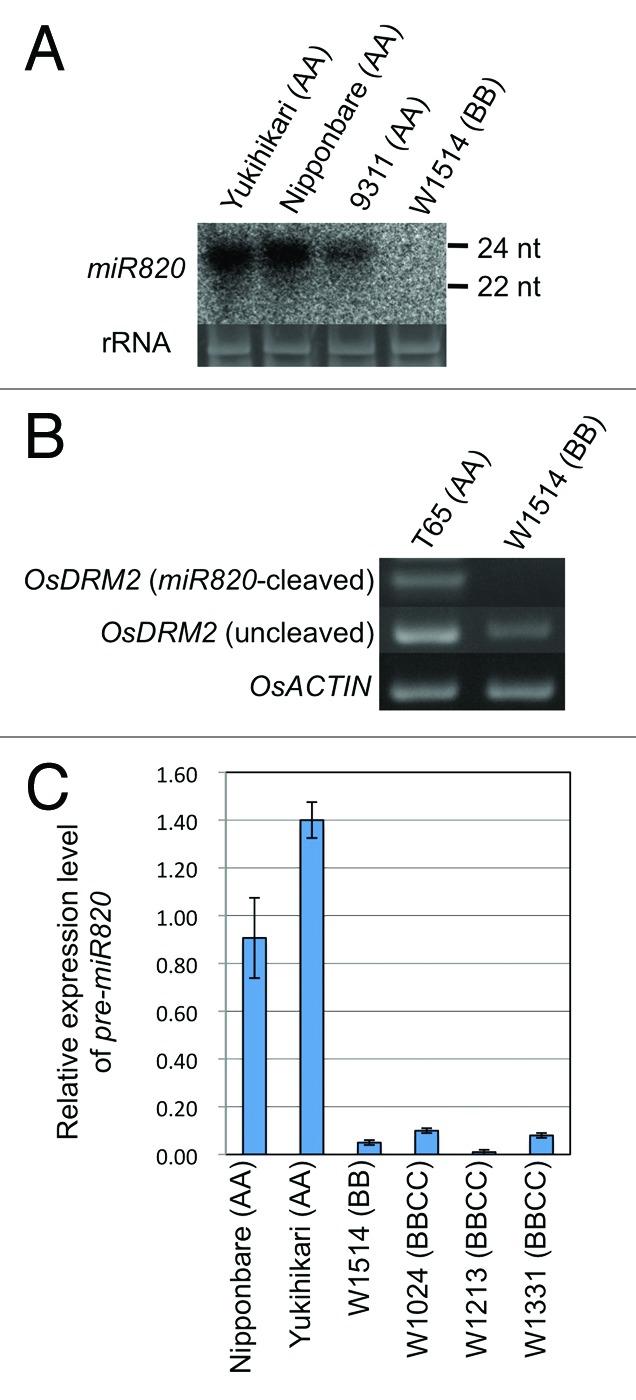
Figure 1. Expression and functional analysis of miR820 in cultivated rice and wild rice species. (A) Northern blot analysis of miR820 expression in AA and BB Oryza species. (B) Detection of miR820-cleaved OsDRM2 mRNA by RNA ligation-mediated 5′-RACE (upper panel) in AA and BB species. The same cDNA templates were used for PCR to amplify uncleaved OsDRM2 (middle panel) and OsACTIN (bottom panel) as controls. (C) The relative expression levels of pre-MIR820 measured by qRT-PCR in AA, BB and BBCC Oryza species and calculated by subtracting the values with RT reaction by the values without RT reaction, then normalized by OsACTIN. Values are means of triplicate experiments, with bars showing standard errors. Probes and primers are described in our previous work.7
To explore whether the expression level of MIR820 depends on its copy number, we also analyzed rice accessions with low or moderate copy numbers, as determined by Southern hybridization. We used 45 cultivars from the Japanese Rice Core Collection (JRC) and 56 cultivars from the World Rice Core Collection (WRC) (Tables 1 and 2).8,9 Among the JRC cultivars, the average copy number of MIR820 was 4.5 (minimum: 2; maximum: 6). Among the WRC cultivars, the average copy number was 6.3 (minimum: 3; maximum: 11) (Fig. 2A). Then, we determined the levels of pre-miR820 by qRT-PCR in the same accessions and found no correlation between the MIR820 copy number and pre-miR820 levels (Fig. 2B).
Table 1. Copy numbers of MIR820 in the Japanese Rice Core Collection (JRC).
| JRC No. | Name | Origin | Copy number |
|---|---|---|---|
| JRC01 | Gaisen Mochi | Japan (unknown) | 4 |
| JRC03 | Hinode | Kinki | 5 |
| JRC04 | Senshou | Tokyo | 4 |
| JRC05 | Yamada Bake | Kagoshima | 4 |
| JRC06 | Kaneko B | Kantou Touzan | 5 |
| JRC07 | Iruma Nishiki | Saitama | 5 |
| JRC08 | Okka Modoshi | Japan (unknown) | 4 |
| JRC10 | Hirayama | Tokyo | 4 |
| JRC11 | Kahei | Kagoshima | 4 |
| JRC12 | Oiran | Kumamoto | 5 |
| JRC13 | Bouzu Mochi | Ooita | 5 |
| JRC17 | Akage | Akita | 4 |
| JRC19 | Wataribune | Shiga | 3 |
| JRC20 | Hosogara | Aomori | 4 |
| JRC21 | Akamai | Kouchi | 5 |
| JRC22 | Mansaku | Nagano | 5 |
| JRC23 | Ishijiro | Toyama | 5 |
| JRC24 | Joushuu | Yamagata | 5 |
| JRC25 | Dango | Japan (unknown) | 5 |
| JRC26 | Aikoku | Fukui | 5 |
| JRC27 | Ginbouzu | Ishikawa | 5 |
| JRC28 | Shinriki Mochi | Kumamoto | 5 |
| JRC29 | Shichimenchou Mochi | Japan (unknown) | 5 |
| JRC30 | Morita Wase | Yamagata | 5 |
| JRC31 | Kameji | Shimane | 6 |
| JRC32 | Omachi | Okayama | 5 |
| JRC33 | Shinriki | Hyougo | 5 |
| JRC34 | Kyoutoasahi | Kyoto | 5 |
| JRC35 | Kabashiko | Miyazaki | 5 |
| JRC37 | Shinyamadaho 2 | Hyougo | 5 |
| JRC38 | Nagoya Shiro | Akita | 2 |
| JRC39 | Shiroine | Tokushima | 4 |
| JRC40 | Akamai | Nagasaki | 5 |
| JRC41 | Akamai | Tokushima | 3 |
| JRC42 | Touboshi | Kagoshima | 5 |
| JRC43 | Akamai | Kantou Touzan | 3 |
| JRC44 | Karahoushi | Kagoshima | 3 |
| JRC46 | Fukoku | Hokkaido | 5 |
| JRC47 | Okabo | Japan (unknown) | 4 |
| JRC48 | Hakamuri | Kagoshima | 4 |
| JRC49 | Rikutou Rikuu 2 | Japan (unknown) | 5 |
| JRC51 | Shinshuu | Nagano | 5 |
| JRC52 | Aichiasahi | Aichi | 3 |
| JRC53 | Raiden | Kantou Touzan | 5 |
| JRC54 | Houmanshinden Ine | Kagoshima | 4 |
Table 2. Copy numbers of MIR820 in the World Rice Core Collection (WRC).
| WRC No. | Name | Origin | Copy number |
|---|---|---|---|
| WRC01 | Nipponbare | Japan | 5 |
| WRC02 | Kasalath | India | 6 |
| WRC03 | Bei Khe | Cambodia | 6 |
| WRC04 | Jena 035 | Nepal | 5 |
| WRC05 | Naba | India | 8 |
| WRC06 | Puluik Arang | Indonesia | 3 |
| WRC07 | Davao 1 | Philippines | 3 |
| WRC09 | Ryou Suisan Koumai | China | 5 |
| WRC10 | Shuusoushu | China | 9 |
| WRC11 | Jinguoyin | China | 5 |
| WRC12 | Dahonggu | China | 7 |
| WRC13 | Asu | Bhutan | 6 |
| WRC14 | IR 58 | Philippines | 4 |
| WRC15 | Co 13 | India | 7 |
| WRC16 | Vary Futsi | Madagascar | 7 |
| WRC17 | Keiboba | China | 11 |
| WRC18 | Qingyu (Seiyu) | China | 8 |
| WRC19 | Deng Pao Zhai | China | 4 |
| WRC20 | Tadukan | Philippines | 6 |
| WRC21 | Shwe Nang Gyi | Myanmar | 6 |
| WRC22 | Calotoc | Philippines | 8 |
| WRC23 | Lebed | Philippines | 5 |
| WRC24 | Pinulupot 1 | Philippines | 5 |
| WRC25 | Muha | Indonesia | 10 |
| WRC26 | Jhona 2 | India | 10 |
| WRC27 | Nepal 8 | Nepal | 9 |
| WRC28 | Jarjan | Bhutan | 7 |
| WRC29 | Kalo Dhan | Nepal | 7 |
| WRC30 | Anjana Dhan | Nepal | 5 |
| WRC31 | Shoni | Bangladesh | 6 |
| WRC32 | Tupa 121–3 | Bangladesh | 6 |
| WRC33 | Surjamukhi | India | 9 |
| WRC34 | ARC 7291 | India | 9 |
| WRC35 | ARC 5955 | India | 9 |
| WRC36 | Ratul | India | 7 |
| WRC37 | ARC 7047 | India | 8 |
| WRC39 | Badari Dhan | Nepal | 8 |
| WRC40 | Nepal 555 | India | 6 |
| WRC41 | Kaluheenati | Sri Lanka | 5 |
| WRC42 | Local Basmati | India | 5 |
| WRC43 | Dianyu 1 | China | 4 |
| WRC44 | Basilanon | Philippines | 6 |
| WRC45 | Ma sho | Myanmar | 6 |
| WRC46 | Khao Nok | Laos | 5 |
| WRC47 | Jaguary | Brazil | 8 |
| WRC48 | Khau Mac Kho | Vietnam | 8 |
| WRC49 | Padi Perak | Indonesia | 8 |
| WRC50 | Rexmont | USA | 5 |
| WRC51 | Urasan 1 | Japan | 7 |
| WRC52 | Khau Tan Chiem | Vietnam | 3 |
| WRC53 | Tima | Bhutan | 5 |
| WRC55 | Tupa729 | Bangladesh | 7 |
| WRC57 | Milyang 23 | Korea | 4 |
| WRC98 | Deejiaohualuo | China | 4 |
| WRC99 | Hong Cheuh Zai | China | 5 |
| WRC100 | Vandaran | Sri Lanka | 5 |
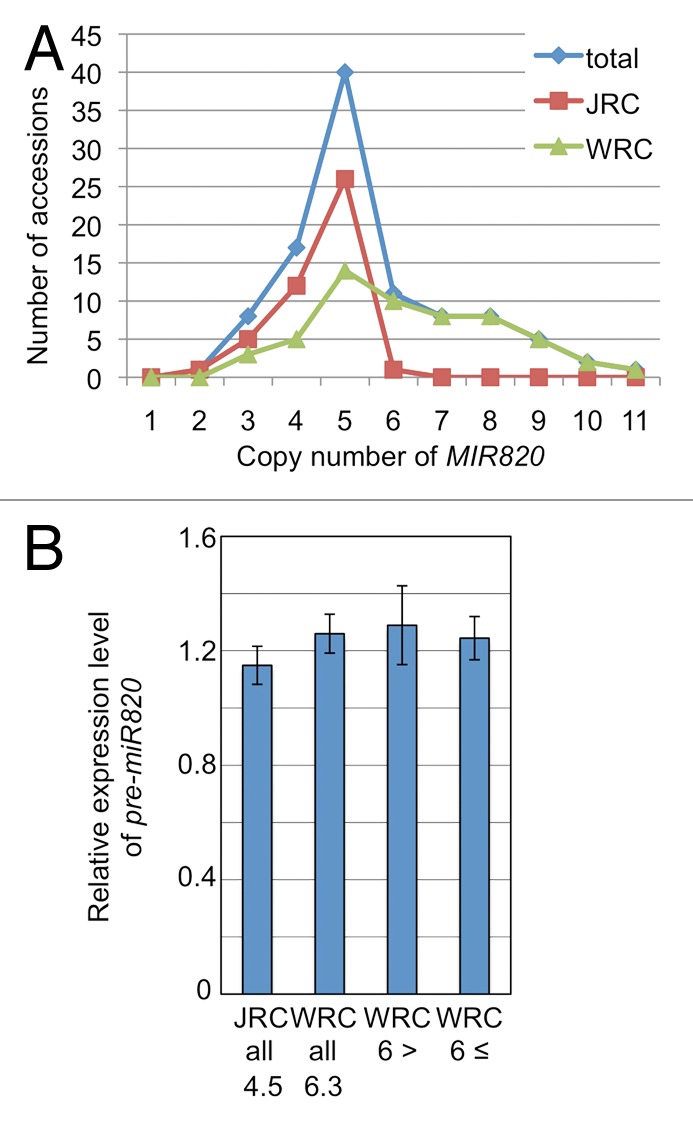
Figure 2. Copy numbers and expression levels of MIR820 in the Japanese (JRC) and World (WRC) rice collections. (A) Distribution of the copy number of CACTA transposons carrying MIR820 determined by Southern blot analysis. Genomic DNAs from the NIAS JRC and WRC rice core collections were digested with EcoRI and EcoRV and probed with the pre-miR820 DNA fragment. “Total” is the combination of both collections. (B) The relative expression levels of MIR820 measured by qRT-PCR as the same method mentioned before. The expression levels in all JRC and WRC accessions, in WRC accessions with less than six copies and those with six or more copies of CACTA are indicated. Numbers below JRC all or WRC all indicate the average copy numbers. Values are means of triplicate experiments, with bars showing standard errors. The numbers below JRC or WRC indicate the copy number of MIR820. Probes and primers are described in our previous work.7
Previously, we have shown that miR820 downregulates the expression of de novo DNA methyltransferase, responsible for transposon inactivation.7 Therefore, one would assume that once miR820 effectively suppresses OsDRM2, silencing of transposons (including CACTA carrying MIR820) would be released, resulting in an increased transcription of MIR820. This would lead to a feed-forward loop, reinforcing the function of miR820. However, in this study, we found that the expression of MIR820 is relatively constant in many cultivated rice cultivars, despite its copy number varying from 2 to 11. This suggests that MIR820 transcription is regulated not by copy dosage, but rather by the epigenetic state of each locus. This mechanism may have evolved during the “arms race” between the host and the parasite and may allow the host to inhibit the feed-forward loop triggered by miR820 and thus to prevent the overwhelming victory of the parasites.
Acknowledgments
We thank Ms Tomoko Atsumi for technical assistance. The wild rice accessions and Core Collections of rice cultivars used in this study were obtained from the National Institute of Genetics, supported by the National Bioresource Project, MEXT and the Genebank at the National Institute of Agrobiological Sciences (NIAS), Japan. This work was supported by JSPS KAKENHI Grant 23658006 to Y.S.
Disclosure of Potential Conflict of Interest
No potential conflicts of interest were disclosed
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/25169
References
- 1.Kidwell MG. Transposable elements and the evolution of genome size in eukaryotes. Genetica. 2002;115:49–63. doi: 10.1023/A:1016072014259. [DOI] [PubMed] [Google Scholar]
- 2.Doolittle WF, Sapienza C. Selfish genes, the phenotype paradigm and genome evolution. Nature. 1980;284:601–3. doi: 10.1038/284601a0. [DOI] [PubMed] [Google Scholar]
- 3.Orgel LE, Crick FHC. Selfish DNA: the ultimate parasite. Nature. 1980;284:604–7. doi: 10.1038/284604a0. [DOI] [PubMed] [Google Scholar]
- 4.Zilberman D, Henikoff S. Silencing of transposons in plant genomes: kick them when they’re down. Genome Biol. 2004;5:249. doi: 10.1186/gb-2004-5-12-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 6.Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, Ikawa M, et al. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 2008;22:908–17. doi: 10.1101/gad.1640708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nosaka M, Itoh JI, Nagato Y, Ono A, Ishiwata A, Sato Y. Role of transposon-derived small RNAs in the interplay between genomes and parasitic DNA in rice. PLoS Genet. 2012;8:e1002953. doi: 10.1371/journal.pgen.1002953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kojima Y, Ebana K, Fukuoka S, Nagamine T, Kawase M. Development of an RFLP-based rice diversity research set of germplasm. Breed Sci. 2005;55:431–40. doi: 10.1270/jsbbs.55.431. [DOI] [Google Scholar]
- 9.Ebana K, Kojima Y, Fukuoka S, Nagamine T, Kawase M. Development of mini core collection of Japanese rice landrace. Breed Sci. 2008;58:281–91. doi: 10.1270/jsbbs.58.281. [DOI] [Google Scholar]


