Abstract
Auxins are crucial for plant growth and development. Auxin signalling primarily depends on four partially redundant F-box proteins of the TIR1/AFB2 Auxin Receptor (TAAR) clade to trigger the degradation of AUX/IAA transcriptional repressors. Auxin signalling is a balanced system which involves complex feedback regulations. miR393 regulation of TAAR genes is important for different developmental programs and for responses to environment. However, so far, the relevance of the two MIR393 genes for Arabidopsis leaf development and their significance for auxin signalling homeostasis have not been evaluated. First, our analyses of mir393a-1 and mir393b-1 mutants and of mir393ab double mutant show that the two genes have only partially redundant functions for leaf development. Expression analyses of typical auxin-induced reporter genes have shown that the loss of miR393 lead to several unanticipated changes in auxin signalling. The expression of DR5pro:GUS is decreased, the expression of primary AUX/IAA auxin-responsive genes is slightly increased and the degradation of the AXR3-NT:GUS reporter protein is delayed in mir393ab mutants. Additional analyses using synthetic auxin and auxin antagonists indicated that miR393 deficient mutants have higher levels of endogenous AUX/IAA proteins, which in turn create a competition for degradation. We propose that the counter-intuitive changes in the expression of AUX/IAA genes and in the accumulation of AUX/IAA proteins are explained by the intrinsic nature of AUX/IAA genes which are feedback regulated by the AUX/IAA proteins which they produce. Altogether our experiments provide an additional highlight of the complexity of auxin signaling homeostasis and show that miR393 is an important component of this homeostasis.
Introduction
Auxins are phytohormones important for plant growth, organogenesis and various responses to environmental changes [1],[2]. Auxin signalling primarily depends on perception by four partially-redundant auxin receptors of the TRANSPORT INHIBITOR RESPONSE 1 (TIR1)/AUXIN SIGNALLING F-BOX PROTEIN 2 (AFB2) clade [3]–[5]. Upon binding auxins, these TAAR proteins, which are the specificity-components of SKIP/CULLIN/F-BOX (SCF)-ubiquitin ligase complexes, form a co-receptor complex with AUXIN/INDOLE-3-ACETIC ACID (AUX/IAA) transcriptional repressors [6]–[8]. AUX/IAA proteins are then ubiquitinated and degraded by the 26S proteasome [7]. This leads to the release of AUXIN RESPONSE FACTOR (ARF) transcription factors to which AUX/IAA were bound and allows ARFs to activate or repress the transcription of primary auxin-responsive genes [9]. The homeostasis of auxin signaling involves feedback-regulation of AUX/IAA genes' expression by the AUX/IAA proteins which they generate. The homeostasis of TAAR genes expression was shown to involve the microRNA miR393. This regulation is important for innate immunity [10], [11], for root response to nitrate, salinity and drought resistance [12]–[15] and for several aspects of rice and Arabidopsis development [15]–[19]. Our own work has shown that this regulation additionally involves the function of secondary siRNAs, the siTAARs, which are generated from TAAR transcripts downstream of the miR393 cleavage site [16], [17]. MiR393 is generated from two distinct genes, MIR393A and MIR393B [20]. AtMIR393A, but not AtMIR393B, was shown to be induced by stress while MIR393B, but not AtMIR393A, was shown to be induced by auxin [10], [19]. Moreover, our work has suggested that miR393 is primarily produced from AtMIR393A in the roots and primarily from AtMIR393B, in the aerial parts of plants grown in normal growth conditions [17]. Thus, these observations suggested that MIR393 genes have major distinct functions: AtMI393A being primarily involved in the plant responses to environment, and, AtMIR393B being primarily involved in the regulation of auxin homeostasis and of auxin-dependent plant development [10]–[19]. Intriguingly, mir393b-1 mutants which accumulate only trace amounts of miR393 in aerial parts of plants exhibit rather mild phenotypes in normal growth conditions essentially characterized by a pronounced leaf epinasty [17]. Thus, this raised the hypothesis that AtMIR393A or other pathways compensate for the loss of AtMIR393B.
MiR393 is important for the regulation of TAAR genes and is part of a complex homeostatic process which involves feedback transcriptional regulations [19]. However, the significance of miR393 for auxin signalling homeostasis has not been evaluated directly in mutants lacking miR393. To gain insights into these important aspects we have obtained single and double mutants of MIR393 genes and we have analyzed the impact of these mutations on plant development, on physiological response to auxin and at the molecular level of auxin signalling. The data which we obtained show that the two MIR393 genes have partially redundant functions for leaf polarity with a primarily role for AtMIR393B. Moreover, we have observed that the expression level of the artificial reporter gene DR5pro:GUS is slightly decreased in these mutants compared to wt plants while the expression of primary auxin-induced genes is increased. Moreover, experiments using synthetic auxin, auxin antagonists and the HSpro:AXR3-NT:GUS reporter gene to monitor the degradation of AUX/IAA proteins showed that the degradation rate of the AXR3-NT:GUS protein is longer in the mutants than in wt plants. These unanticipated results demonstrate that the loss of miR393 leads to complex changes and to simultaneously increase the basal expression level of AUX/IAA genes and the basal level of AUX/IAA protein accumulation. Together our data show that miR393 is an important component of the auxin signalling homeostasis required for the establishment of proper and timely auxin signalling outputs.
Results
MIR393 Genes Have Distinct, but Partially Overlapping Expression Patterns
The microRNA miR393 is encoded by two genes, AtMIR393A (At2g39885) and AtMIR393B (At3g55734) (Fig.1A) [21]. Our earlier studies using mir393b-1 mutants showed that miR393, which primarily arises from AtMIR393B in aerial organs, is important for leaf morphology [17]. However, mir393b-1 mutants exhibit only mild developmental phenotypes. To test whether the weak amounts of miR393 generated from AtMIR393A in miR393b-1 are responsible for this observation, we identified a mutant in the SALK collection which has a T-DNA insertion located in the proximal region of the AtMIR393A promoter. Sequencing of the PCR fragments showed that the insertion is located 74-nt upstream of the transcription start which we have identified by Rapid Amplification of cDNA Ends (RACE) experiments (Fig. 1A and S1). In normal growth conditions, this mutant, which we named mir393a-1, accumulated 30% lower levels of miR393 in the roots than the wt plants and accumulated normal levels of miR393 in the leaves (Fig.1B). The double mutant mir393a-1 mir393b-1 (hereafter noted mir393ab) accumulated 50% lower levels of miR393 than the wt plant in the roots and only 1% of the wt level in the leaves (Fig. 1B). These experiments showed that mir393a-1 is not a null-mutant and that the double mutant accumulates lower levels of miR393 than mir393a-1 or mir393b-1 alone. These experiments also showed that the two MIR393 genes have distinct, but partially overlapping expression profiles.
Figure 1. Identification and characterization of MIR393A T-DNA insertion mutants.
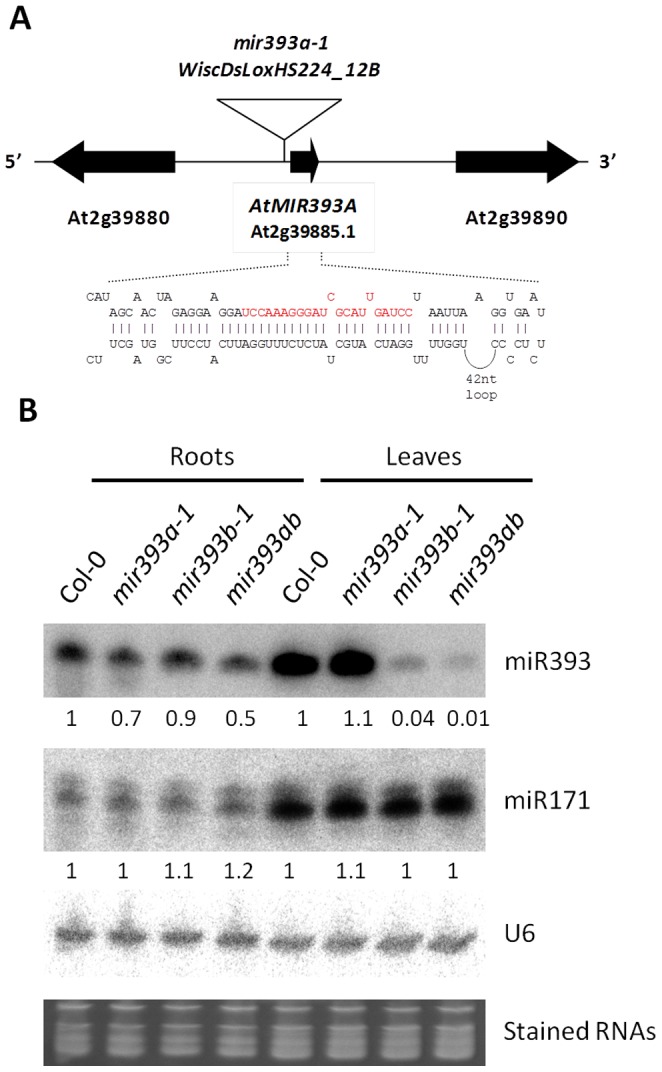
(A) Schematic map showing the 5′-3′ orientation of genes (large arrows and gene names are indicated) flanking AtMIR393A on chromosome II. The arrow representing AtMIR393A indicates the pri-miRNAs which full-length sequence determined by RACE experiments (given in Fig. S1). The expanded region represents the folded pre-miR393 nucleotide sequence with the miR393 sequence indicated in red. The open triangle represents the T-DNA insertion. The scheme is not drawn to scale. (B) RNA-blot hybridization of RNA prepared from roots and leaves of 34d-old plants. Probed RNAs are indicated on the right. The signal detected for mutants relative to wild-type Col-0 are normalized relative to the Midori Green stained RNA signals. Similar results were obtained in three independent experiments.
MIR393 Genes Have Overlapping, Partially Redundant Functions in Leaf Development
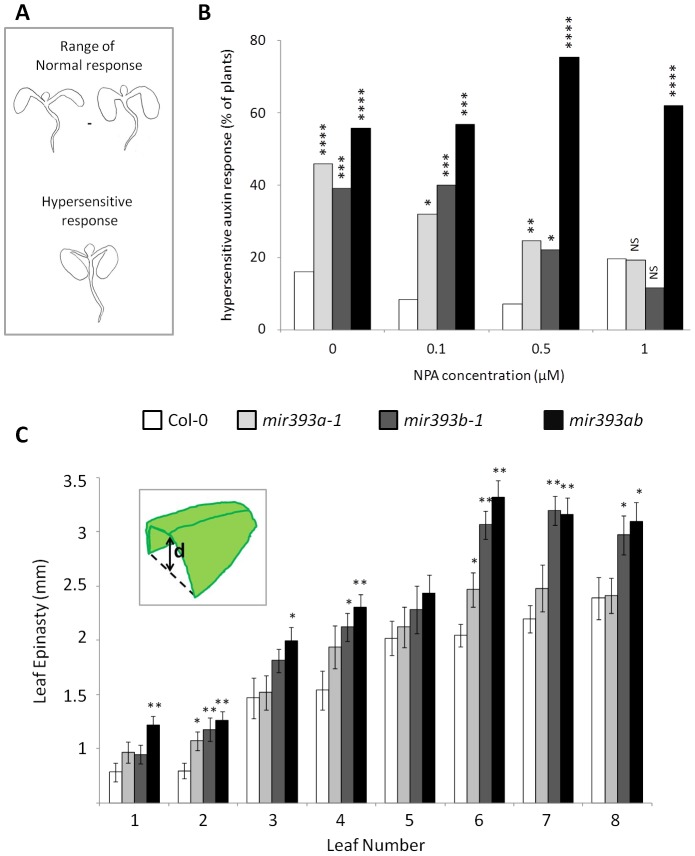
A first inspection of the different mutant plants showed that neither mir393a-1 nor mir393ab mutants exhibit a more drastic developmental defects than mir393b-1. We analyzed whether the two MIR393 genes have redundant functions in leaf development by, first, recording the incidence of cotyledon epinasty (ICE) which is a typical auxin hypersensitive response regulated by miR393 [17], [22]. When grown on standard medium, a high and significantly greater fraction of mir393a-1 (46%), of mir393b-1 (39%) and of mir393ab mutants (56%) than wild-type plants (16%) exhibited the extreme cotyledon epinasty phenotype (Fig. 2A). In the cases of mir393a-1 and miR393b-1, this ICE was similarly decreased by increasing the concentration of the auxin transport inhibitor NPA (1-N-naphthylphthalamic acid) to 0.1 or 0.5 µM and this ICE was similar to that of wt at a concentration of 1 µM (Fig. 2B) [23]. In mir393ab however the incidence of cotyledon epinasty remained significantly higher even at high NPA concentrations. These observations established that the cotyledon epinasty response to auxin depends on the overlapping function of both MIR393 genes.
Figure 2. AtMIR393A and AtMIR393B are partially redundant for proper leaf morphogenesis.
(A) Schematic representation of the normal range of cotyledon epinasty (top) and the extreme cotyledon epinasty (bottom) typical of the auxin-hypersensitive response. (B) The incidence of cotyledon auxin-hypersensitive response in populations of Col-0 (open bars), mir393a-1 (light grey bars), mir393b-1 (dark grey bars), and mir393ab double mutants (dark bars). Seedlings (n>40 for each condition and genotype) were grown on media containing the concentration of NPA indicated and harvested 4 d after germination. P values (two-tailed Fisher's exact test) for significant differences towards Col-0 are indicated; NS for P>0.05, * for P≤0.05, ** for P≤0.01, *** for P≤0.001, **** for P≤0.0001. P values for significant differences between mutants are given in Fig. S2. (C) Epinasty of leaf number 1 to 8 for Col-0, mir393a-1, mir393b-1 and mir393ab was measured by the vertical distance between the adaxial leaf side and the leaf margin (in mm ± SEM) (see drawing in the insert). Significant difference towards Col-0 is indicated (two-tailed student t-test). * for P≤0.05, ** for P≤0.01. N = 10. P values for significant differences between mutants are given in Fig. S2.
Next, we measured the degree of leaf epinasty for the first 8 leaves of the plants. For mir393a-1 mutants, most of the leaves were more epinastic than wt plants but the difference to wt was significant only for leaves #2 and #6 (Fig. 2C). For mir393b-1, the epinasty was greater than that recorded for mir393a-1 and was significantly different from wt for more than half of the leaves. For mir393ab mutants, the leaf epinasty was again greater than that of mir393b-1 and the difference to wt was highly significant for all leaves except for leaf #5. These observations showed that AtMIR393B has a main role in the regulation of leaf epinasty and that AtMIR393A contributes in a slightly redundant manner to regulate the underlying developmental process.
MiR393-Deficient Mutants Exhibit Decreased, rather than increased, Expression of DR5pro:GUS
The loss of miR393 and the consequent increase in TAAR genes' expression [10], [17] is expected to increase the degradation of AUX/IAA proteins and to increase the expression of AUX/IAA genes. However, because the expression of AUX/IAA genes is feedback-regulated by the AUX/IAA which they generate, it is uncertain at which level the homeostasis of AUX/IAA genes will then be maintained in miR393-deficient mutants. To gain insights into the role of miR393 in this homeostasis, we first analyzed the expression of the artificial auxin-induced gene DR5pro:GUS, which contains seven soybean auxin response elements (ARE) and serves to report the state of the auxin signalling output [24].
Without treatments, the expression of DR5pro:GUS was similar in the distal tips of mir393a-1, mir393b-1 and mir393ab and wt leaves while it was slightly lower in the margins of mir393 mutants' leaves than in those of wt leaves (Fig. 3A). Treatment with 2,4-D, a synthetic diffusible auxin, induced the expression of DR5pro:GUS in all plant genotypes. However, its expression was lower in the blades of mir393 mutants leaves, in the order mir393a-1>mir393b-1 = mir393ab, than in those of wt leaves (Fig. 3A). Thus, these results showed that the expression of DR5pro:GUS is changed in a counter-intuitive manner by the loss of miR393. DR5pro:GUS is more stably repressed in mir393 mutants than in wt plants. Thus, these results suggest that the loss of miR393 leads to more complex changes than initially anticipated, especially at the level of AUX/IAA proteins.
Figure 3. Basal expression level of primary auxin-inducible AUX/IAA genes in mir393 mutants compared to wt plants.
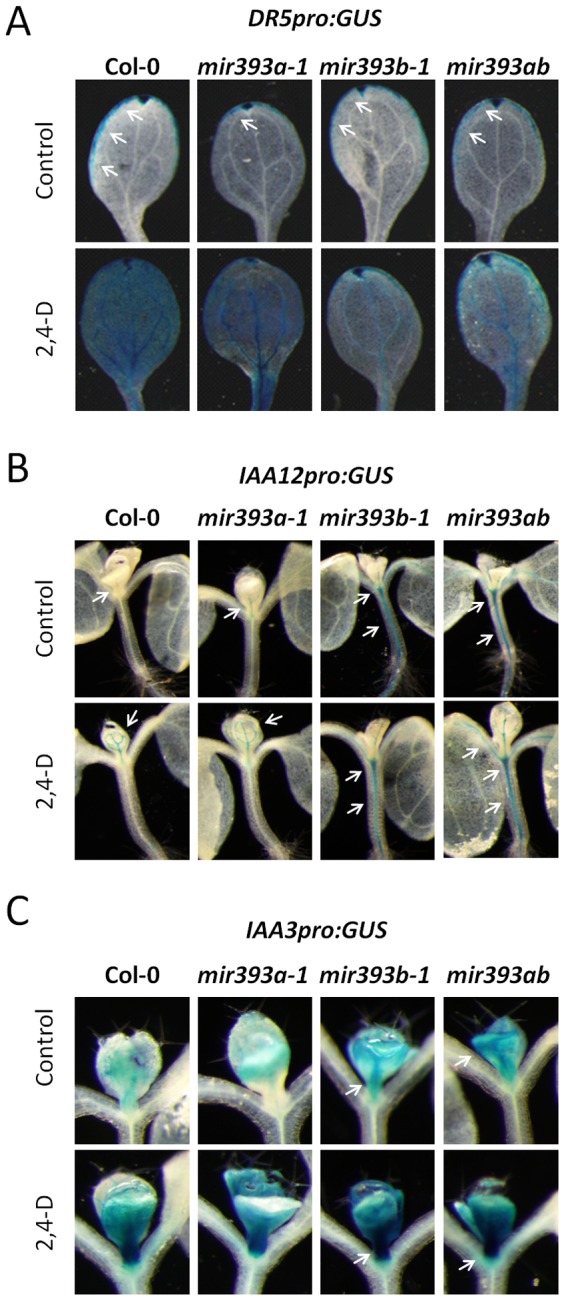
Pictures of representative wt and mir393 mutant plants expressing the DR5pro:GUS (A), the IAA12pro:GUS (B), or the IAA3pro:GUS gene (C) upon treatment or not with 10 µM 2,4-D for 8 h. Arrows highlight the GUS staining detected.
MiR393-Deficient Mutants Exhibit Increased Basal Expression of AUX/IAA Genes
To better understand the changes triggered by the loss of miR393, we analyzed the expression of artificial genes reporting the activity of the primary auxin-induced promoters of IAA12/BDL and IAA3/SHY2 genes in wt and in miR393-deficient mutants (Fig. 3B–C). Without treatment, the expression level of IAA12pro:GUS was undetectable in wt plants, was weakly detectable in the shoot tips and roots of mir393a-1 mutants, and was ectopically expressed at high levels in the shoot tips and hypocotyls of mir393b-1 mutants (Fig. 3B). Interestingly, IAA12pro:GUS was also ectopically expressed in the shoot tips, in the hypocotyls and in the leaf veins of mir393ab mutants but at even higher levels. For IAA3pro:GUS we did not observe ectopic expression in the mutants but it was detected at high and increasing levels in the order mir393b-1<mir393ab while it was only weakly expressed in emerging leaves of wt and mir393a-1 plants (Fig. 3C). Together these observations indicated that the two primary auxin-induced genes IAA3/SHY2 and IAA12/BDL have higher basal expression levels in miR393-deficient mutants than in wt plants. Moreover, the mutation in AtMIR393B has a greater effect on AUX/IAA genes expression than the mutation in AtMIR393A.
Importantly, treatments of plantlets with 2,4-D for 8h induced the expression of IAA12pro:GUS and IAA3pro:GUS genes in all plant genotypes (Fig. 3B–C). In these conditions, the two reporter genes were also more expressed in mir393a-1, mir393b-1 and mir393ab mutants than in wt plants. These data showed that miR393 is important to maintain the proper basal expression level of these two AUX/IAA genes and for their proper induction.
MiR393-Deficient Mutants Exhibit Complex Changes in the steady state of AUX/IAA and GH3 mRNAs
To determine whether the observations made with the two reporter genes also apply to endogenous genes, we analyzed the mRNA steady state of few auxin-induced genes (Fig. 4). We selected 5 AUX/IAA and 3 GH3 genes based on their high expression level reported in the Arabidopsis eFP browser hormone series (www.bar.utoronto.ca) [25]. Without auxin treatment, IAA17 and IAA19 mRNAs accumulated to higher levels in mir393ab mutants than in wt plants, and, the expression of the other 6 genes was not detected. The expression of all genes tested was induced by treatment with 2,4-D. In these conditions, GH3.5 mRNAs accumulated to similar levels in wt and mir393ab mutants, GH3.2, IAA19, IAA29, IAA5 and IAA2 accumulated to higher levels in mir393ab mutants than in wt plants, while GH3.6 and IAA17 accumulated to lower levels in mir393ab mutants than in wt plants. Thus, although all genes tested were induced by auxin treatments in the mir393ab mutants, their accumulation was substantially different from that observed in wt plants. Together with the reporter genes analyses, these data showed that miR393 plays an important role to maintain the proper basal expression and for the accurate induction of AUX/IAA and GH3 genes.
Figure 4. miR393 is required for proper induction of AUX/IAA and GH3 genes.
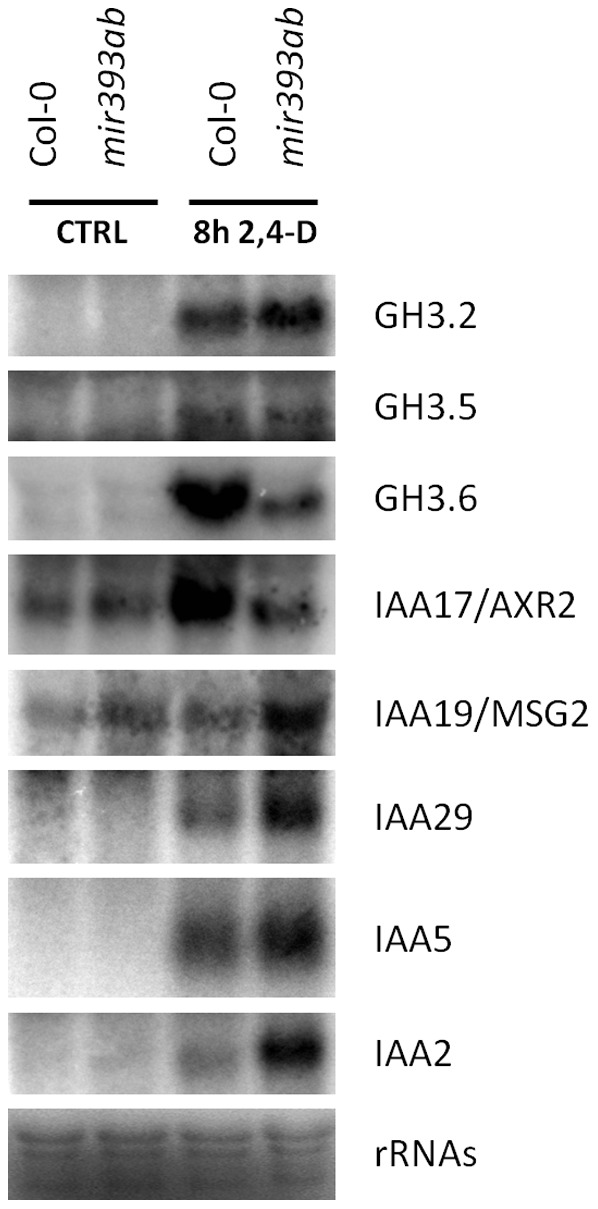
Northern blots of RNAs from wt and mir393ab mutant plantlets treated or not with 10 µM 2,4-D for 8 h. Without treatments, IAA17 and IAA19 have higher steady state levels in mir393ab mutants than in wt plants. Treatment with 2,4-D for 8 h induces the expression of all genes tested. GH3.5 accumulates to similar levels in wt and mutants, GH3.2, IAA19, IAA29, IAA5 and IAA2 accumulate to higher levels in mir393ab mutants than in wt plants while GH3.6 and IAA17 accumulate to lower levels in mir393ab mutants than in wt plants.
MIR393 is Necessary for Proper Degradation of AXR3NT:GUS Proteins
Next, we used the heat shock inducible HSpro:AXR3-NT:GUS line to assay the level of accumulation of AUX/IAA proteins and the efficiency of their degradation [7]. We anticipated that an increase in AUX/IAA protein steady-state levels should create a competition for ubiquitination and/or for degradation between AUX/IAAs and might therefore affect the degradation rate of AXR3-NT:GUS. After induction of the HSpro:AXR3-NT:GUS gene and without other treatments (Fig. 5; control) the level of AXR3-NT:GUS proteins was consistently higher in the roots of mir393a-1, mir393b-1 and mir393ab mutants than in wt plants (first row). This result showed that in miR393-deficient mutants the AXR3-NT:GUS proteins are not degraded as efficiently as in wt plants. Brief treatment with 2,4-D during induction to enhance AUX/IAA protein degradation lead to a marked decrease in the accumulation of AXR3-NT:GUS proteins in mir393 mutants compared to the control condition; Although the level was still higher in mir393 mutants than in wt plants (second row). Similarly, brief treatment with auxinole (α-[2,4-dimethylphenylethyl-2-oxo]-IAA) which prevents the degradation of AUX/IAA proteins by binding to TIR1/AFBs [26] lead to increase the accumulation of AXR3-NT:GUS proteins (Fig. 5; third row). Brief treatment with MG132 which specifically inhibits the function of the proteasome didn't lead to substantial changes in the accumulation of AXR3-NT:GUS proteins (Fig. 5; fourth row). Thus, these experiments showed that the SCFTIR/AFB complexes and the proteasome are functional, although less efficient, in mir393 mutants than in wt plants. These experiments suggest that the level of auxins is the limiting factor and that the higher steady state level of endogenous AUX/IAA proteins in mir393 mutants cannot be degraded as fast as in the wt. We further challenged this possibility by treating the plants with 2,4-D four hours before induction of the HSpro:AXR3-NT:GUS gene. As we anticipated, chasing the higher levels of AUX/IAA proteins by inducing their degradation with high quantities of auxin lead to restore the proper degradation of the AXR3-NT:GUS protein (Fig. 5; fifth row). Together these experiments showed that miR393 is required to ensure the proper degradation of the AXR3-NT:GUS fusion proteins and of other AUX/IAA proteins. Moreover, miR393-deficient mutants seem to indeed accumulate higher steady state levels of endogenous AUX/IAA proteins.
Figure 5. miR393 is required for proper degradation of AXR3-NT:GUS proteins.
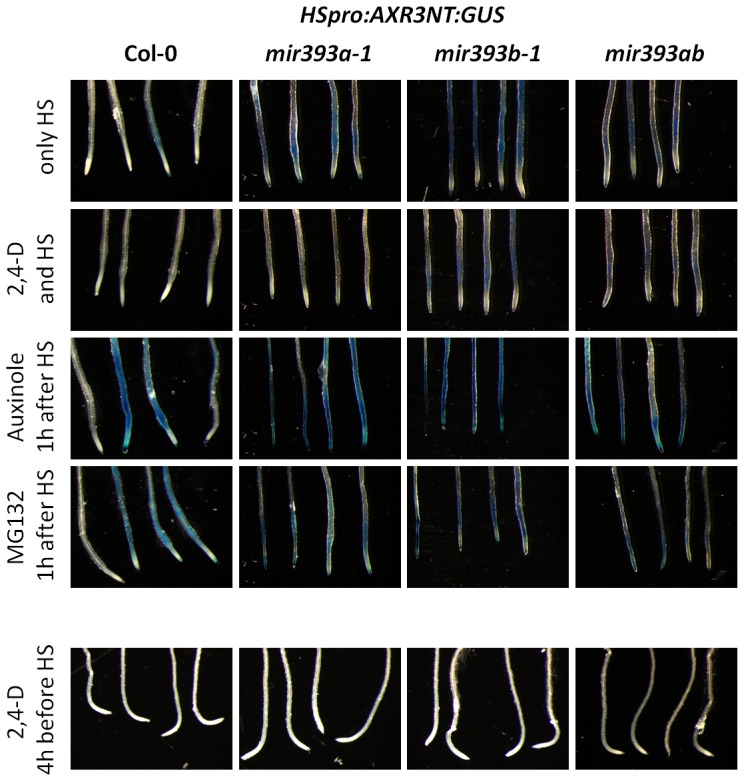
Representative pictures of wt and mir393 mutant roots expressing the HSpro:AXR3-NT:GUS gene. Without treatments, the AXR3-NT:GUS fusion protein is more stable in mir393a-1, mir393b-1 and mir393ab mutants compared to wt plants. Short treatments with 10 µM auxin induces the degradation of AXR3-NT:GUS, while treatment with 20 µM auxinole or MG132 blocks the degradation of AXR3-NT:GUS. This shows that the SCF complexes and proteasome are functional in the mutants as well. Clearance of presumably high levels of endogenous AUX/IAA proteins by treatment with 2,4-D for 4 h restores the proper degradation of AXR3-NT:GUS.
Discussion
Auxins are major plant hormones with crucial functions in almost every aspect of plant life [1], [2]. Identification of mutants affected in synthesis, transport and perception of auxins have demonstrated the importance of auxins in developmental and morphogenetic programs [5], [27]. MiR393 is encoded by two distinct genes and regulates the expression of TIR1/AFB2 auxin receptor genes. According to the crucial role of auxins, we anticipated that miR393-deficient mutants, which we showed in mir393b-1 mutants, fail to regulate TAAR mRNA levels, should exhibit pleiotropic developmental defects. However, no obvious developmental defects other than mild leaf polarity defects were observed in the single mutants mir393a-1, mir393b-1 or in the double mutants mir393ab grown in standard laboratory conditions. We suspect that mir393a-1 is not a null mutant since this mutation leads to a 30% decrease in the level of mature miR393 in the roots. Another possibility is that a third unidentified MIR393 gene produces the remaining miR393 in the roots. Chen and colleagues have shown that the expression of MIR393 and TAAR genes are under transcriptional feedback regulations [19]. Thus, it is possible that these feedbacks can compensate for the loss of miR393. However, we do not favor this possibility since we have observed clear effects on expression of primary auxin-induced genes and on degradation of AUX/IAA proteins in miR393-deficient mutants. Thus, we suspect that other unrelated pathways, rather than the above mentioned feedback mechanisms solely, act redundantly with miR393 and somehow compensate for its loss.
Although mir393a-1 is unlikely a null mutant, our studies with mir393a-1, mir393b-1 and mir393ab has shown that both MIR393 genes contribute in a partially redundant fashion to the establishment of leaf polarity in arabidopsis; with AtMIR393B gene playing the predominant role. We found that all miR393-deficient mutants exhibit a hypersensitive auxin response characterized by an extreme epinasty of the cotyledons. For mir393a-1 and mir393b-1, the hypersensitivity phenotype was prevented when the plants were grown on increasing concentrations of the auxin transport inhibitor NPA. For mir393ab double mutants the extreme auxin response was not prevented by the highest concentrations of NPA. When we increased the concentration of NPA to 2 µM or higher, the cotyledon epinasty phenotype could not be analyzed appropriately since the plants did not germinate or grow correctly (not shown).
Auxin homeostasis relies on intricate feedback regulations at several levels including synthesis, transport and signaling via AUX/IAA proteins. The data which we report here add to the current picture and show that the regulation of TAAR genes homeostasis by miR393 plays a significant role for the homeostasis of auxin signaling. We found that the basal steady-state expression level of IAA12pro:GUS, of IAA3pro:GUS and of several endogenous AUX/IAA genes is higher in miR393-deficient mutants than in wt plants. Importantly, this and additional experiments suggest that the increase in AUX/IAA gene expression leads to a concomitant increase in the basal steady state level of AUX/IAA proteins. These observations are counter-intuitive on first sight since an increase in the expression level of AUX/IAA genes leading to an increase in the level of AUX/IAA proteins should result in a decrease in the expression of AUX/IAA genes. Conversely, a decrease in the expression level of AUX/IAA genes leading to a decrease in the level of AUX/IAA proteins should result in an increase in the expression of AUX/IAA genes. We believe that the counter-intuitive observations are due to the intrinsic homeostatic nature of AUX/IAA genes which are feedback regulated by the AUX/IAA proteins which they generate. Indeed, such feedback-regulated systems lead to meta-stable steady state levels. Thus, we believe that the AUX/IAA genes, which exhibit a concomitant increase in gene expression and in the level of the corresponding AUX/IAA proteins, have actually reach a higher meta-stable steady-state level in miR393-deficient mutants than in wt plants. We have not been able to challenge this hypothesis by straightforward measurements of endogenous AUX/IAA protein levels because antibodies for AUX/IAA proteins or arabidopsis lines expressing tagged versions of AUX/IAA proteins under their native promoters are not available. We speculate that the expression of a given AUX/IAA gene should be even higher in a mir393ab mutant expressing non functional AUX/IAA proteins. However, this approach would require the production of mutants with several orders of magnitude. However, we have been able to provide indirect evidences supporting our hypothesis. Indeed, experiments using HSpro:AXR3-NT:GUS showed that the AXR3-NT:GUS fusion proteins are not properly degraded in miR393-deficient mutants. This indicated that the proper degradation of the AXR3-NT:GUS proteins in auxin-limiting conditions is prevented by the high levels of AUX/IAA proteins. This conclusion was also supported by the observations that treatment of plants with 2,4-D for 4 h before inducing the HSpro:AXR3-NT:GUS gene, which induces the degradation of endogenous AUX/IAA proteins, could restore the proper degradation of AXR3-NT:GUS. Finally, expression analysis of DR5pro:GUS, the universal auxin signalling output marker gene, also support our hypothesis since its expression level was slightly lower in miR393-deficient mutants than in wild type plants, and thus indicate that it is slightly more repressed in mR393-deficient mutants than in wt plants.
The delay of AXR3-NT:GUS degradation is more pronounced in the roots of mir393b-1 than in those of mir393a-1. However, our sRNA blot experiments showed than miR393 accumulates to lower levels in the roots of mir393a-1 than in those of mir393b-1. Thus, this discrepancy suggests that the loss of miR393 observed in the shoots of mir393b-1 leads to increase the level of competing AUX/IAA proteins in the roots. We speculate that this is achieved either by an increased synthesis or by an increased transport.
Earlier studies had use miR393-resistant target genes and miR393 overexpressers to unravel the function and roles of miR393 [10]-[12], [14], [15], [19]. However, although miR393 had been shown to regulate the expression of TAAR genes and of auxin-responsive genes, and moreover to be involved in important biological processes, its functional significance for auxin signalling and its homeostasis had not really been evaluated directly. Our experiments now clarify the picture and demonstrate that miR393 is necessary to maintain low basal expression levels of AUX/IAA genes, low basal levels of AUX/IAA proteins and that these features are important for the proper degradation of AUX/IAA proteins. Thus, miR393 is not only important for TAAR genes homeostasis but also for the establishment of proper auxin signalling outputs and for auxin signalling homeostasis per se.
Materials and Methods
Plant Material
miR393a-1 mutant was obtained by PCR-based genotyping of plants from the seed batch WiscDsLoxHS224_12B obtained from the NASC stock center. The mir393ab double mutant was obtained by PCR-based genotyping of F2 seedlings obtained from the cross of mir393a-1 and mir393b-1 [17]. The DR5pro:GUS, IAA12pro:GUS, IAA3pro:GUS and HSpro:AXR3-NT:GUS reporters were introgressed in mir393a-1, mir393b-1 and mir393ab mutants by crossing and homozygous plants were found by PCR-based genotyping in the F2 population. In all cases, double homozygosis was ascertained on at least 9 plants of the F3 population.
Growth Conditions & Treatments
Arabidopsis thaliana plants used to prepare RNA blots and to measure leaf epinasty were grown on soil in the greenhouse as described previously [28].
Studies of cotyledon epinasty were done as we previously described [17] on Murashige and Skoog (MS) solid medium supplemented as indicated with NPA (1-N-naphthylphthalamic acid, Fluka) in DMSO or DMSO alone as a control.
Measurements of leaf epinasty were made on plants grown for 45 days in short-day (SD) conditions (8 h light/16 h dark). Root measurements were made on plantlets germinated on MS medium and transferred for 8 days in vertically oriented square-plates on solid MS medium supplemented as indicated with 0.1 µM 2,4-D or 20 µM auxinole in SD conditions. For studies of IAA3pro:GUS, IAA12pro:GUS and DR5pro:GUS expression, 7 days old seedlings were incubated in MS medium containing 10 µM 2,4-D in ethanol or ethanol alone as a control for the time indicated before proceeding to staining. For studies of AXR3-NT:GUS fusion protein stability, 7 days old seedlings were placed in water at 37°C for 2 h to induce the expression of the HSpro:AXR3-NT:GUS gene and incubated for 30 minutes at room temperature before proceeding to GUS staining. For the treatments, either 10 µM 2,4-D were added to the solution just before the heat shock or 20 µM auxinole were added after 1 h of heat-shock as it was described [7], [26].
RNA preparation and RNA Analysis
Extraction of total RNAs, preparation of RNA blots and qRT-PCRs were done as previously described [28].
Histochemical GUS Assays
Arabidopsis seedlings were incubated into staining solution containing 1 mM X-Gluc in 100 mM Na3PO4 (pH 7.2), 0.1% Triton X-100, 5 mM K3Fe(CN)6 and 5 mM K4Fe(CN)6 for 24 h at 37°C in the dark [29]. Seedlings were then cleared in 70% ethanol for 2 days and mounted in 50% v/v glycerol before observations.
Supporting Information
Position of miR393a-1 T-DNA insertion in AtMIR393A (At2g39885). The pri-miRNA sequence (546-nt) which we identified by RACE experiments is indicated in orange color. The pre-miRNA sequence (133-nt) is in typed in capital letters and the miR393 sequence (22-nt) is underlined. The T-DNA insertion is located between the two nucleotides highlighted in red.
(DOCX)
AtMIR393A and AtMIR393B are partially redundant for proper leaf morphogenesis. (A–B) The incidence of cotyledon auxin-hypersensitive response in populations of Col-0 (open bars), mir393a-1 (light grey bars), mir393b-1 (dark grey bars), and mir393ab double mutants (dark bars). Seedlings (n>40 for each condition and genotype) were grown on media containing the concentration of NPA indicated and harvested 4 d after germination. P values (two-tailed Fisher's exact test) for significant differences towards mir393a-1 (A) or mir393b-1 (B) are indicated; NS for P>0.05, * for P≤0.05, ** for P≤0.01, *** for P≤0.001, **** for P≤0.0001. (C–D) Epinasty of leaf number 1 to 8 for Col-0, mir393a-1, mir393b-1 and mir393ab was measured by the vertical distance between the adaxial leaf side and the leaf margin (in mm ± SEM). Significant difference towards mir393a-1 (C) or mir393b-1 (D) are indicated (two-tailed student t-test). * for P≤0.05, ** for P≤0.01. N = 10.
(TIF)
Acknowledgments
We thank Thomas Boller for providing laboratory space at the Botanical Institute of the University of Basel. We thank Frederick Meins Jr. and Mark Estelle for helpful comments and discussions, Ken-Ichiro Hayashi for samples of auxinole, Miltos Tsiantis for seeds of DR5pro:GUS and Gerd Juergens for seeds of IAA12pro:GUS and IAA3pro:GUS.
Funding Statement
This work was supported by the Swiss National Science Foundation (Ambizione grant PZ00P3_126329 and PZ00P3_142106 to FV) and by the Rector's Conference of the Swiss Universities (SCIEX-NMS fellowship 11.115 to F.V., Z.S.K. and D.B). The Ph.D. fellowship of DB is part of the International Ph.D. Program “From genome to phenotype: A multidisciplinary approach to functional genomics” funded by the Foundation for Polish Science (FNP). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Mockaitis K, Estelle M (2008) Auxin receptors and plant development: a new signaling paradigm. Annu Rev Cell Dev Biol 24: 55–80. [DOI] [PubMed] [Google Scholar]
- 2. Vanneste S, Friml J (2009) Auxin: a trigger for change in plant development. Cell 136: 1005–1016. [DOI] [PubMed] [Google Scholar]
- 3. Dharmasiri N, Dharmasiri S, Estelle M (2005) The F-box protein TIR1 is an auxin receptor. Nature 435: 441–445. [DOI] [PubMed] [Google Scholar]
- 4. Kepinski S, Leyser O (2005) The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435: 446–451. [DOI] [PubMed] [Google Scholar]
- 5. Dharmasiri N, Dharmasiri S, Weijers D, Lechner E, Yamada M, et al. (2005) Plant development is regulated by a family of auxin receptor F box proteins. Dev Cell 9: 109–119. [DOI] [PubMed] [Google Scholar]
- 6. Calderon Villalobos LI, Lee S, De Oliveira C, Ivetac A, Brandt W, et al. (2012) A combinatorial TIR1/AFB-Aux/IAA co-receptor system for differential sensing of auxin. Nat Chem Biol 8: 477–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M (2001) Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature 414: 271–276. [DOI] [PubMed] [Google Scholar]
- 8. Lechner E, Achard P, Vansiri A, Potuschak T, Genschik P (2006) F-box proteins everywhere. Curr Opin Plant Biol 9: 631–638. [DOI] [PubMed] [Google Scholar]
- 9. Ulmasov T, Hagen G, Guilfoyle TJ (1999) Activation and repression of transcription by auxin-response factors. Proc Natl Acad Sci U S A 96: 5844–5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Robert-Seilaniantz A, MacLean D, Jikumaru Y, Hill L, Yamaguchi S, et al. (2011) The microRNA miR393 re-directs secondary metabolite biosynthesis away from camalexin and towards glucosinolates. Plant J 67: 218–231. [DOI] [PubMed] [Google Scholar]
- 12. Vidal EA, Araus V, Lu C, Parry G, Green PJ, et al. (2010) Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana. Proc Natl Acad Sci U S A 107: 4477–4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen H, Li Z, Xiong L (2012) A plant microRNA regulates the adaptation of roots to drought stress. FEBS Lett 586: 1742–1747. [DOI] [PubMed] [Google Scholar]
- 14. Gao P, Bai X, Yang L, Lv D, Pan X, et al. (2011) osa-MIR393: a salinity- and alkaline stress-related microRNA gene. Mol Biol Rep 38: 237–242. [DOI] [PubMed] [Google Scholar]
- 15. Xia K, Wang R, Ou X, Fang Z, Tian C, et al. (2012) OsTIR1 and OsAFB2 downregulation via OsmiR393 overexpression leads to more tillers, early flowering and less tolerance to salt and drought in rice. PLoS One 7: e30039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Windels D, Vazquez F (2011) miR393: Integrator of environmental cues in auxin signaling? Plant Signal Behav 6: 1672–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Si-Ammour A, Windels D, Arn-Bouldoires E, Kutter C, Ailhas J, et al. (2011) miR393 and Secondary siRNAs Regulate Expression of the TIR1/AFB2 Auxin Receptor Clade and Auxin-Related Development of Arabidopsis Leaves. Plant Physiol 157: 683–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bian H, Xie Y, Guo F, Han N, Ma S, et al. (2012) Distinctive expression patterns and roles of the miRNA393/TIR1 homolog module in regulating flag leaf inclination and primary and crown root growth in rice (Oryza sativa). New Phytol 196: 149–161. [DOI] [PubMed] [Google Scholar]
- 19. Chen ZH, Bao ML, Sun YZ, Yang YJ, Xu XH, et al. (2011) Regulation of auxin response by miR393-targeted transport inhibitor response protein 1 is involved in normal development in Arabidopsis. Plant Mol Biol 77: 619–629. [DOI] [PubMed] [Google Scholar]
- 20. Jones-Rhoades MW, Bartel DP (2004) Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell 14: 787–799. [DOI] [PubMed] [Google Scholar]
- 21. Sunkar R, Zhu JK (2004) Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 16: 2001–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hayashi K, Kamio S, Oono Y, Townsend LB, Nozaki H (2009) Toyocamycin specifically inhibits auxin signaling mediated by SCFTIR1 pathway. Phytochemistry 70: 190–197. [DOI] [PubMed] [Google Scholar]
- 23. Scanlon MJ (2003) The polar auxin transport inhibitor N-1-naphthylphthalamic acid disrupts leaf initiation, KNOX protein regulation, and formation of leaf margins in maize. Plant Physiol 133: 597–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ (1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, et al. (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hayashi KI, Neve J, Hirose M, Kuboki A, Shimada Y, et al. (2012) Rational Design of an Auxin Antagonist of the SCF(TIR1) Auxin Receptor Complex. ACS Chem Biol 7: 590–598. [DOI] [PubMed] [Google Scholar]
- 27. Friml J (2003) Auxin transport - shaping the plant. Curr Opin Plant Biol 6: 7–12. [DOI] [PubMed] [Google Scholar]
- 28. Vazquez F, Blevins T, Ailhas J, Boller T, Meins F Jr (2008) Evolution of Arabidopsis MIR genes generates novel microRNA classes. Nucleic Acids Res 36: 6429–6438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. Embo J 6: 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Position of miR393a-1 T-DNA insertion in AtMIR393A (At2g39885). The pri-miRNA sequence (546-nt) which we identified by RACE experiments is indicated in orange color. The pre-miRNA sequence (133-nt) is in typed in capital letters and the miR393 sequence (22-nt) is underlined. The T-DNA insertion is located between the two nucleotides highlighted in red.
(DOCX)
AtMIR393A and AtMIR393B are partially redundant for proper leaf morphogenesis. (A–B) The incidence of cotyledon auxin-hypersensitive response in populations of Col-0 (open bars), mir393a-1 (light grey bars), mir393b-1 (dark grey bars), and mir393ab double mutants (dark bars). Seedlings (n>40 for each condition and genotype) were grown on media containing the concentration of NPA indicated and harvested 4 d after germination. P values (two-tailed Fisher's exact test) for significant differences towards mir393a-1 (A) or mir393b-1 (B) are indicated; NS for P>0.05, * for P≤0.05, ** for P≤0.01, *** for P≤0.001, **** for P≤0.0001. (C–D) Epinasty of leaf number 1 to 8 for Col-0, mir393a-1, mir393b-1 and mir393ab was measured by the vertical distance between the adaxial leaf side and the leaf margin (in mm ± SEM). Significant difference towards mir393a-1 (C) or mir393b-1 (D) are indicated (two-tailed student t-test). * for P≤0.05, ** for P≤0.01. N = 10.
(TIF)