Abstract
Cerebral cavernous malformations (CCM) are neurovascular dysplasias that result in mulberry-shaped lesions predominantly located in brain and spinal tissues. Mutations in three genes are associated with CCM. These genes encode for the proteins KRIT1/CCM1 (krev interaction trapped 1/cerebral cavernous malformations 1), cerebral cavernous malformations 2, osmosensing scaffold for MEKK3 (CCM2/malcavernin/OSM), and cerebral cavernous malformations 3/programmed cell death 10 (CCM3/PDCD10). There have been many significant recent advances in our understanding of the structure and function of these proteins, as well as in their roles in cellular signaling. Here, we provide an update on the current knowledge of the structure of the CCM proteins and their functions within cellular signaling, particularly in cellular adhesion complexes and signaling cascades. We go on to discuss subcellular localization of the CCM proteins, the formation and regulation of the CCM complex signaling platform, and current progress towards targeted therapy for CCM disease. Recent structural studies have begun to shed new light on CCM protein function, and we focus here on how these studies have helped inform the current understanding of these roles and how they may aid future studies into both CCM-related biology and disease mechanisms.
Keywords: Cerebral cavernous malformations, CCM complex signaling platform, Neurovasculature, Integrin signaling, Kinases
Introduction
Cerebral cavernous malformations (CCM) are dysplasias that occur predominantly in the neurovasculature, but can also occur in the eye or skin [1–3]. The lesions caused by the disease, also known as cavernoma, can lead to stroke, intracranial hemorrhage, and focal neurological outcomes [4]. Occurrence of CCM is relatively common, with up to 0.5 % of the population harboring at least one cavernoma in postmortem studies [5]. The cause of CCM can be either sporadic or genetic. Sporadic cases account for approximately 80 % of CCM [6] and are mostly associated with formation of a single lesion [6, 7]. In contrast, the heritable form is thought to occur following a ‘second-hit’, or Knudsonian, mutation [8–10], although there remains the possibility that additional factors which are potentially specific to the neurovascular environment could be necessary for acquisition of lesions [11, 12]. Nonetheless, the genetic form of the disease is usually more severe because these patients tend to have multiple lesions [13], with the number and size of lesions increasing as the patient ages [14].
Identification of the genes responsible for CCM disease began with the discovery of a chromosome 7q founder mutation in a Hispanic population [15–17] that genetic sequencing identified as the KRIT1/CCM1 gene [18, 19]. Subsequently, two other genes were also identified to be associated with CCM acquisition: CCM2/MGC4607/OSM/Malcavernin [20] and CCM3/PDCD10/TFAR15 [21, 22]. Following the identification of these genes, several transgenic mouse and zebrafish models [23–33] validated the correlation of these genes to disease [11, 23–30]. Cerebral cavernous malformations are associated with heterozygous loss of one allele for KRIT1, CCM2, or CCM3, followed by a second-hit mutation that usually results in the complete loss of one of their protein products [8–10]. Mutations in CCM3 tend to result in a more aggressive form of the disease than those in KRIT1 or CCM2 [14], suggesting potential differences in the signaling pathways in which CCM3 is involved.
KRIT1, CCM2, and CCM3 encode for the KRIT1, CCM2, and CCM3 proteins, respectively. Given that the architectural features of the proteins are distinct from one another and that they may play roles in different signaling pathways, it has become important to understand how KRIT1, CCM2, and CCM3 function, what roles they play in signaling transduction, and where their signaling pathways cross. In the past several years, structural biology has begun to shed light on the domain architecture of KRIT1, CCM2, and CCM3. These studies have both uncovered unpredicted domains within each of the proteins and elucidated novel modes of binding with some of their interaction partners. Although there is much yet to be learned about CCM protein structure and function, we are now significantly closer to understanding what these proteins look like, and, by extension, are in an optimal position to use this new information to more deeply and comprehensively probe their cellular functions. Understanding where the key nodal points reside that allow cross-talk between the signaling pathways could potentially facilitate a therapeutically useful strategy for all CCM patients. The recent discoveries of structures of all three CCM proteins, including some of complexes with binding partners, will be invaluable towards this understanding, and will help to guide future studies probing the biological roles of these proteins.
Architecture of the CCM proteins
Recent studies have significantly improved the understanding of the molecular architecture of the CCM proteins (KRIT1, CCM2, CCM3), having implications for understanding how these proteins function in their respective signaling pathways.
KRIT1 (CCM1)
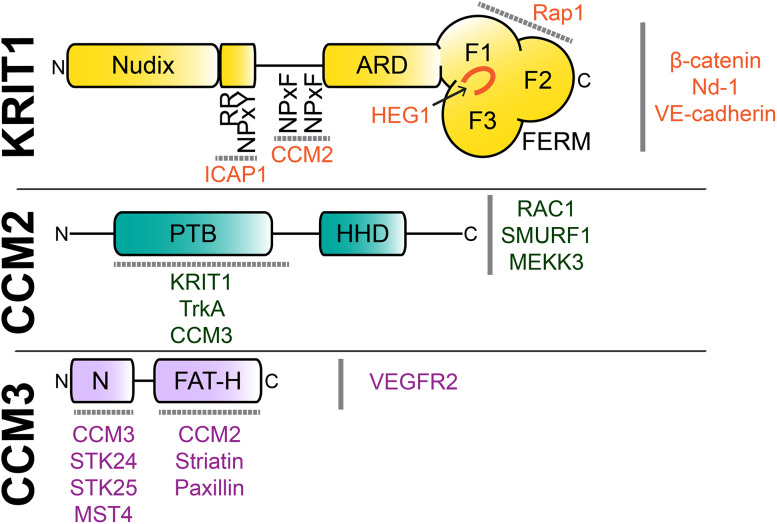
KRIT1 is a 736 amino acid protein that was originally described to contain a C-terminal FERM (band 4.1, ezrin, radixin, moesin) domain that interacts with the small GTPase Krev-1 (Rap1) and an ankyrin repeat domain N-terminal to the FERM domain consisting of 4 ankyrin repeats [34]. KRIT1 was later discovered to contain three canonical motifs for direct binding to PTB (phosphotyrosine binding) domains [35]. These NPxY/F motifs (192NPAY, 231NPLF, 250NPYF) are important for the protein–protein interactions of KRIT1 and have also been suggested to play a role in regulating intra-molecular KRIT1 conformational changes and their functional outputs [36, 37].
Until recently, the region of KRIT1 consisting of the 170 residues at its N-terminus that precede its first NPxY/F motif had been thought to be disordered [38–40]. This N-terminal region contains a Nuclear Localization Sequence [39], a putative Nuclear Export Sequence [41], and a tubulin binding sequence [37], but very little functional work has been conducted to investigate its role. Crystallographic studies have now discovered that this region encompasses a nucleotide diphosphate linked to an X moiety (Nudix) domain [42]. This fold is adopted by an extremely diverse superfamily of hydrolases [43] that have a large scope of substrates, but most frequently hydrolyze diphosphate linkages. Based on structural analysis, the KRIT1 Nudix domain cannot be classified into any of the known Nudix domain sub-families and, furthermore, it lacks conserved residues required for enzymatic activity [42]. Although the function of the KRIT1 Nudix domain has yet to be elucidated, these unusual attributes resemble those of the pseudokinase class of protein kinases, which maintain a protein kinase fold, but not enzymatic activity [44]. It is therefore possible that KRIT1 is a ‘pseudonudix’ domain protein, but further work will be required to determine its precise function.
FERM domains are archetypal modular domains that contain three lobes (F1, F2, and F3), each of which fold similarly to other known protein domains. The FERM domain F1 lobe adopts a ubiquitin-like fold, the F2 lobe adopts an acyl-CoA binding protein fold, and the F3 lobe adopts a PH/PTB domain-like fold. These domains are often used as sites of intermolecular interaction, as is also observed for KRIT1 in the context of its associations with both Rap1 [45, 46] and HEG1 [46, 47], discussed in detail below.
It has also been suggested that KRIT1 can engage in a head-to-tail interaction with itself. Studies by multiple groups have suggested that the KRIT1 FERM domain interacts with N-terminal portions of the protein. As the NPxY/F motifs of KRIT1 are within the N-terminus, and NPxY/F motifs are known to serve as PTB-domain interaction partners, the PTB-like F3 lobe of the FERM domain is believed to engage with one or more of the NPxY/F motifs at the N-terminus [36, 37]. There is controversy, however, over which of the NPxY/F motifs is the critical factor in this putative ‘head–tail’ KRIT1 interaction [36, 37], and there remains the potential that the Nudix domain might play some role in the interaction [36]. Nonetheless, interruption of this interaction by binding partners (e.g., ICAP1, Rap1) is suggested to be important for altering the subcellular localization of KRIT1. Conformational lability of KRIT1 is therefore thought to be important for its sub-cellular localization and, consequently, the localization of the CCM complex signaling platform [36, 37]. Further studies are required to better understand how its binding partners regulate the conformational rearrangement of KRIT1 and what impact this has on signal transduction.
CCM2 (malcavernin/OSM)
CCM2 is a 444 amino acid protein originally predicted to contain a folded domain only at its N-terminus thought to belong to the PH/PTB superfamily [20, 48]. This predicted CCM2 PTB domain has been studied to investigate whether it could mediate interactions with KRIT1. Yeast two-hybrid studies suggest that it binds both the second and third of the three conserved NPxY/F motifs in the KRIT1 N-terminus [41], although other studies have failed to show a CCM2 PTB interaction with the KRIT1 NPxY/F motifs [49]. The CCM2 PTB domain also seems to function in several other signaling pathways. For example, it has interactions with the juxtamembrane region of the TrkA receptor tyrosine kinase [50] and with the E3 ubiquitin ligase SMURF1 (SMAD ubiquitination regulatory factor 1) [51], although it is not clear how these interactions may occur. Recent work has also discovered that there is a folded domain at the C-terminus of CCM2 [52] with strong structural homology to a protein interaction domain in the cochlear hair cell scaffolding protein harmonin [52, 53]. This fold has only been identified in CCM2, in harmonin itself, and in RTEL1, a DNA helicase of unknown function [54]. While the role of this domain in RTEL1 has not been determined, harmonin uses its equivalent domain as a scaffold that mediates interactions with binding partners such as cadherin-23 [53]. Therefore, it is likely that CCM2 also uses its harmonin-homology domain (HHD) to engage in interactions with binding partners. It will be intriguing to discover with which of CCM2’s binding partners the HHD interacts, and how these interactions might help to regulate specific signaling events.
Recently, a CCM2 paralog termed CCM2L was identified [55, 56]. This protein has high sequence similarity to CCM2 but is significantly larger (613 amino acids) due to inclusion of large loops within the PTB domain and between the PTB and HHD domains. CCM2L can bind to KRIT1 in competition with CCM2, but does not bind CCM3. The role of CCM2L seems to be to compete with the vascular stabilizing effects of the CCM complex to allow cell growth and proliferation [56].
CCM3 (PDCD10)
CCM3, a 212 amino acid protein, initially evaded structural characterization [57], but crystallographic studies of CCM3 surprisingly revealed it to fold as a two-domain protein [58]. The N-terminal domain is a unique fold consisting of four helices that interlock with a second CCM3 molecule to form a tight homodimer. The C-terminus of CCM3 is a core four-helix bundle that displays striking similarities to the focal adhesion targeting (FAT) domain found in non-receptor tyrosine kinases Pyk2 and FAK. A flexible hinge region links CCM3’s N-terminal dimerization and C-terminal FAT-homology (FAT-H) domains [58]. Structural studies have suggested that this region is important for the interaction of CCM3 with inositol-(1,3,4,5)-tetrakisphosphate [59] and with paxillin [60]. Additionally, a significant conformational change occurs upon CCM3 heterodimerization with members of the germinal center kinase III (GCKIII) sub-family of the Sterile-20 group of serine/threonine kinases [61, 62]. These conformational movements, combined with the discovery of specific domains within CCM3, have allowed some of the roles of CCM3 in specific signaling pathways to be parsed out, the details of which will be discussed below.
Formation of a heterotrimeric KRIT1–CCM2–CCM3 ‘CCM complex signaling platform’
Early work suggested the direct interaction of the CCM proteins with one another to form a signaling platform around a ternary KRIT1–CCM2–CCM3 complex [27, 41, 49, 63, 64]. In this platform, CCM2 acts as the central hub, using two independent binding sites to simultaneously interact with KRIT1 and CCM3 [27, 65]. CCM2 is thought to use its PTB domain to bind KRIT1, potentially via the second and third KRIT1 NPxY/F motifs [41]. In contrast, CCM2 is predicted to use a conserved motif slightly C-terminal to its PTB domain to interact with the CCM3 FAT-H domain [66], but this has yet to be demonstrated experimentally. Although the three CCM proteins have been found to be capable of existing within the same complex [27, 41, 49, 63, 64], there is also a great deal of evidence suggesting that this complex has a dynamic nature. For example, loss of CCM3 results in a significantly more aggressive disease phenotype than that observed upon loss of either KRIT1 or CCM2 (which are clinically indistinguishable from one another) [14]. Furthermore, the interactions of CCM3 with members of the GCKIII family are observed more readily than CCM3 interactions with CCM2 using proteomics techniques [63]. Additionally, CCM3 is more evolutionarily conserved than either KRIT1 or CCM2. CCM3 is conserved from humans to insects [58], while KRIT1 and CCM2 are strongly conserved from humans to fish [42, 45, 52], with no invertebrate orthologs of CCM2 and only the kri-1 ortholog of KRIT1 in Caenorhabditis elegans [67]. The direct interaction of these proteins may therefore describe only one of several multi-protein complexes involving the CCM proteins and could potentially imply a cell-type-specific function of the CCM proteins. The interplay between association with the CCM complex and with association with other proteins that may or may not involve all three CCM proteins will need to be addressed in the future. However, with the initial groundwork laid out by the structural studies of recent years that have explored the architecture of the three CCM proteins at the atomic level, the framework is now in place for future studies to look more deeply at not only the roles of CCM proteins in various pathways but also which of their domains are involved (Figs. 1, 2).
Fig. 1.
Domain diagrams and interaction partners of the CCM complex signaling platform. Current knowledge of the domains of the CCM proteins is shown. Binding partners are shown, with the binding location indicated where known. Four previously unpredicted domains within the CCM proteins have been discovered by X-ray crystallography and have opened up important new avenues to better understand CCM complex signal transduction. These previously unpredicted domains are the KRIT1 N-terminal Nudix domain [42], the CCM2 C-terminal harmonin-homology domain [52] and the CCM3 N-terminal dimerization domain and C-terminal FAT-homology domain [58]. Nudix nucleotide diphosphate linked to an X moiety, RR arginine–arginine motif, NPxY Asn-Pro-X-Tyr motif, NPxF Asn-Pro-X-Phe motif, ARD ankyrin repeat domain, FERM band 4.1, ezrin, radixin, moesin, PTB phosphotyrosine binding domain, HHD harmonin-homology domain, FAT-H focal adhesion targeting-homology
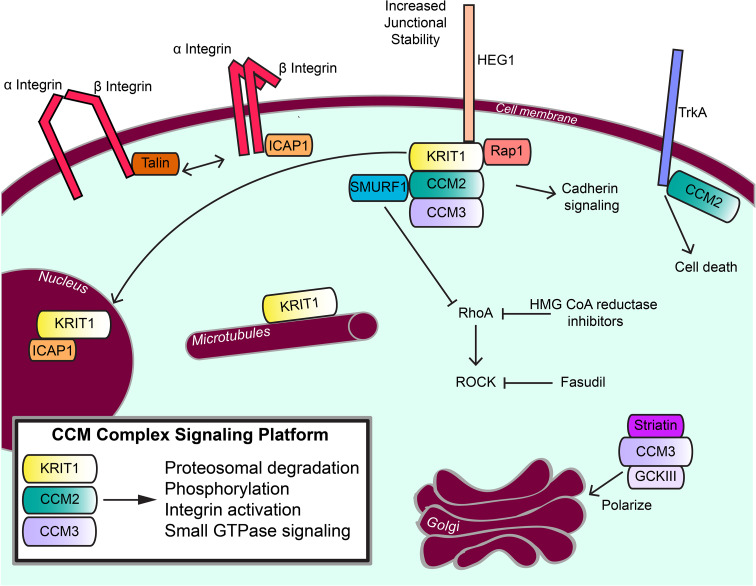
Fig. 2.
CCM proteins in signal transduction. Signal transduction from the CCM complex signaling platform is shown and the roles of CCM proteins outside of the CCM complex signaling platform are also indicated. The CCM proteins play roles in cell adhesion complexes and integrin signaling, in kinase signaling cascades, and in degradation/regulation of the Rho family of small GTPases
CCM proteins and their involvement in cellular pathways
The CCM proteins and adhesion complexes
In addition to their interactions with one another, the CCM proteins have also been implicated in directly binding protein partners that affect cell adhesion. These partners include the small GTPase Rap1, integrin cytoplasmic-associated protein 1 (ICAP1), the orphan cell adhesion receptor heart-of-glass 1 (HEG1), vascular endothelial cadherin, and β-catenin. Recent structural studies have led to a much better understanding of some of these critical CCM interactions.
Rap1 (Krev-1)-related signaling pathways
KRIT1 was initially discovered as a binding partner for Ras-related protein Rap-1 (Rap1; Ras-related protein Krev-1; KREV1) [34]. Rap1 is a small GTPase that functions to control cell polarity, to regulate cell–cell contacts, and to control integrin-mediated cell adhesion via inside-out integrin activation [68]. Like most small GTPases, Rap1 cycles between inactive and active states, a process facilitated by the actions of GTPase activating proteins (GAPs) and guanine nucleotide exchange factors (GEFs) [69]. KRIT1 is an effector of Rap1 and binds specifically to the GTP-loaded (active) form [37, 70] with an affinity of ~2 μM [71]. Because both FERM domain F1 lobes and small GTPase RA (Ras-association) domains adopt a ubiquitin-like fold, it was predicted that the KRIT1 FERM domain would bind to Rap1 using its F1 lobe [71]. Recent crystallographic analyses of KRIT1, however, have shown that the KRIT1 binding interface with Rap1 is significantly larger than those usually found in small GTPase interactions with RA domains [45]. The KRIT1–Rap1 binding surface encompasses regions from both the F1 and F2 lobes of the KRIT1 FERM domain and is completely conserved over evolution [45]. This new structural data explains the specificity that KRIT1 has for Rap1 over H-Ras [45, 46] and places KRIT1 relocalization to cell membranes firmly under the control of Rap1 [37, 70, 71]. The interaction of these proteins affects KRIT1 localization in the cell by targeting KRIT1 to cell–cell junctions [37, 70], and consequently affects KRIT1 interactions with other binding partners. The molecular basis for the specificity of KRIT1 for Rap1 over other Ras-family small GTPases was recently discovered by structural analysis [45, 46], and will help guide further studies into how Rap1 regulates subcellular localization of KRIT1.
KRIT1 has also been found to co-localize with β-catenin, VE-cadherin, and other junctional proteins in a Rap1-dependent manner to stabilize cell–cell junctions [70]. Indeed, loss of KRIT1 seems to increase nuclear localization of β-catenin, implicating a junctional stabilization role for KRIT1 in both endothelial and non-endothelial tissues [72] that may represent a critical node in cell–cell and cell–ECM contacts. Whether the interaction of KRIT1 with β-catenin or VE-cadherin is direct has not yet been determined, so intermediary proteins may act to stabilize this complex. There is also some evidence emerging for cross-talk between integrin-mediated and cadherin-mediated signaling via the CCM complex [73], although how this occurs is still to be defined.
ICAP1
KRIT1 has also been shown to interact with ICAP1, a small 200 amino acid protein that encodes a short N-terminal region that can be phosphorylated by CaMKII [74] and a C-terminal PTB domain [75]. The PTB domain of ICAP1 has long been thought to directly interact with integrin cytoplasmic tails [75–78]. Because it was believed to compete with talin, the master regulator of integrin activation, ICAP1 was thought to be an integrin suppressor [79–81], and this was recently clearly demonstrated [42]. ICAP1 is also a known binder of KRIT1 [35, 82], and was thought to sequester ICAP1 away from integrin tails, allowing talin- and kindlin-mediated activation [74, 79, 80, 83]. Recent structural work has revealed that KRIT1 and integrin β1 directly compete for binding to ICAP1 by its PTB domain, and that both the first KRIT1 NPxY/F motif and the integrin β1 NPxY motif occupy the same binding site on ICAP1 [42]. Interestingly, the KRIT1 binding site for ICAP1 encompasses the first of the three KRIT1 NPxY/F motifs plus a region N-terminal of this motif (residues 170–198). These interfaces make a bidentate KRIT1–ICAP1 binding surface, with a canonical PTB–NPxY interaction and a previously unpredicted interface that is unique among PTB–fold interactions [42]. Functionally, these recent studies clearly showed that the role of the KRIT1–ICAP1 interaction is to modulate the integrin activation state by sequestration of ICAP1 away from the membrane, allowing consequent talin-mediated integrin activation. However, ICAP1 may also have a role in stabilization of KRIT1, an effect that seems to be cell-type-specific [73]. CCM2 may also play a role in stabilization of KRIT1 and ICAP1 [73], but how this may occur given the lack of direct interaction between CCM2 and ICAP1 is not yet understood. ICAP1 may also play a role in regulation of junctional stability by having an impact on ECM remodeling and integrin activation [73]. KRIT1 is also thought to sequester ICAP1 to the nucleus [84], although the nuclear roles of KRIT1 and ICAP1 are still to be discerned. In addition, loss of KRIT1, ICAP1, and CCM3 downregulate DELTA-NOTCH signaling, leading to increased angiogenesis [85–87]. The mechanism by which this occurs is not yet known, and currently there is no evidence to support direct interactions between the CCM complex and NOTCH.
HEG1
The heart-of-glass receptor (HEG1) is an orphan transmembrane receptor whose only known binding partner is KRIT1. The role of HEG1 is still being determined, but multiple studies now suggest that it is an important binding partner of KRIT1, suggesting a role for it within the CCM complex and in control of junctional stability [25, 47]. Because HEG1 contains an NPxY motif in its cytoplasmic region, the expectation was that the KRIT1 FERM domain would interact with this motif via a canonical FERM domain F3 lobe–NPxY motif interaction. Unexpectedly, however, point mutagenesis and determination of the co-crystal structure of the KRIT1 FERM domain in complex with the HEG1 cytoplasmic tail showed that the extreme C-terminus of HEG1, a sequence encoding residues Asp-Tyr-Phe, inserts into a hydrophobic pocket located between the F1 and F3 lobes. The location of this binding site closely correlates to the inositol phosphate binding site seen in other FERM domains [88]. Although the HEG1 binding site is adjacent to the canonical F3 lobe binding site for NPxY motifs, these two sites to do not overlap with one another, implying that KRIT1 may have the capability to bind HEG1 receptor and an NPxY motif protein simultaneously. Rap1 binding to KRIT1 also does not alter HEG1 binding [46], so the KRIT1 FERM domain may be able to simultaneously bind Rap1, HEG1, and an NPxY motif protein. The structural data therefore suggest that the KRIT1 FERM domain may act as a node, or hub, that functions by scaffolding multiple proteins together. Supporting this idea, loss of heg1 in zebrafish results in a similar phenotype to loss of the fish homologues of KRIT1 and CCM2 (santa and valentine), potentially linking them to a common pathway [25, 28]. This is further confirmed by the finding that loss of heg1 binding by mutated krit1 expressed in zebrafish results in similar cardiovascular development phenotypes to krit1 mutants that cannot bind rap1 or ccm2 [47].
Paxillin
The C-terminal domain of CCM3 folds extremely similarly to the Focal Adhesion Targeting domains of the FAK and Pyk2 non-receptor tyrosine kinases. This observation led to the hypothesis that CCM3 might interact with a known binding partner of FAK and Pyk2: the scaffolding protein paxillin [58]. This was confirmed by biochemical and crystallographic experiments, and CCM3 and paxillin were observed to co-localize in leading edges in pericytes [60]. Although the full functional significance of this interaction is not yet understood, there is some evidence for a role of the GCKIII kinase MST3 in cell migration by regulation of paxillin phosphorylation [89].
CCM proteins and signaling cascades
The CCM proteins directly interact with multiple protein kinases, but it is not known if the CCM proteins act as direct modifiers of protein kinase enzymatic activity for any of their binding partners. Although our understanding of CCM proteins in the context of kinase signaling is limited at the moment, recent structural work has helped to shed more light on some of these pathways.
CCM3, Germinal Center Kinases, and the STRIPAK complex
CCM3 directly interacts with each of the three germinal center kinase III (GCKIII) serine/threonine kinases: STK24 (MST3), STK25 (Ysk1; Sok1), and MST4 (MASK) [57, 61, 90, 91]. These three proteins are members of the Sterile 20-like group of human kinases and play roles in cell polarization and migration [89, 92, 93]. Recent crystal structures reveal that CCM3 and MST4, and CCM3 and STK25 both form heterodimers through an interaction between the dimerization domain of CCM3 and the C-terminal domain of the kinase [61, 62]. CCM3 binds with nanomolar affinity to GCKIII kinases and this interaction increases cell migration and proliferation [61]. The CCM3–GCKIII interactions are likely important for targeting STK24, STK25, and MST4 to specific locations in the cell, including the cell membrane, where they can phosphorylate the ERM proteins (ezrin, radixin, and moesin) that link the actin cytoskeleton to the membrane. Knockdown of the ERM proteins activates RhoA [94], potentially tying a CCM3-mediated targeting of GCKIII kinases to Rho signaling [95]. The GCKIII kinases have also been shown to phosphorylate PTP-PEST to regulate paxillin phosphorylation [89], further linking the CCM3–GCKIII complex with paxillin. Additionally, the GCKIII kinases and CCM3 are also part of the STRIPAK (striatin-interacting phosphatase and kinase) complex, a multiprotein complex implicated in regulation of Golgi polarization [66, 96]. CCM3 and striatin have opposite roles in Golgi polarization, as CCM3 promotes correct Golgi positioning while striatin reduces it [66]. CCM3 is thought to bind CCM2, striatin and paxillin using the same interface, a hydrophobic cleft between two helices in the CCM3 FAT-homology domain [58, 60, 66], necessitating the exquisite evolutionary conservation observed for this surface [58]. Based on these structural findings, it seems likely that CCM3 can interact with both GCKIII kinases (through the dimerization domain) and with striatin, a regulatory subunit of the PP2A phosphatase holoenzyme (through the FAT-homology domain). Therefore, CCM3 likely functions as a bridge within the complex, bringing the GCKIII proteins to the STRIPAK phosphatase, which is thought to play a role in cell polarity [66, 96].
Signaling to cell death pathways
One interaction partner of CCM2 is the TrkA receptor tyrosine kinase. The CCM2 PTB domain was shown to bind to a portion of the juxtamembrane region of TrkA [50], while its C-terminus then signals to cell death pathways by an unknown mechanism. Interestingly, CCM3 and the GCKIII kinases seem also to be recruited to TrkA [64], but this may happen through an indirect CCM2–CCM3–GCKIII interaction. Given the newly identified CCM2 HHD, it will be interesting to determine if the mechanism for cell death is mediated through an interaction between the HHD and another yet-to-be-determined protein.
RhoA, ROCK and SMURF1
Knockdown of the CCM proteins in endothelial cell culture results in increased activity of the small GTPase RhoA and consequent increased stress fiber formation [24, 70, 95, 97, 98]. Small molecule inhibition of the RhoA-activated kinase ROCK (Rho-associated coiled coil-forming kinase) rescues the CCM phenotype in cells that do not express CCM1, CCM2, or CCM3 [98], correlating well with CCM complex regulation of RhoA activity. The CCM complex seems to regulate RhoA activity by using the CCM2 PTB domain to recruit SMURF1, a RhoA-specific E3 ubiquitin ligase [51]. Although the mechanism for this is still not defined at the molecular level, it appears that CCM2 uses its PTB domain to interact with the HECT domain of SMURF1 [51]. Recent work further points out that integrin β1 can also control ROCK, potentially suggesting an alternative mechanism for signaling [73]. Nonetheless, the CCM complex can regulate localized RhoA activity at the cell membrane, with loss of the CCM complex resulting in the phenotypes that correlate well with increased RhoA activity.
CCM2 and MAP kinase signaling
CCM2 was initially named osmosensing scaffold for MEKK3 [99], as it was discovered to play a scaffolding role between RAC1 and MEKK3 in the p38 MAP kinase cascade response to osmotic stress [99]. This work revealed that CCM2 binds to MEKK3 in yeast two-hybrid assays, and recruits MEKK3 to membrane ruffles [99]. Recent work confirms that RAC1 and CCM2 signal in the same pathway in response to osmotic stress, but suggests that the signaling occurs through PLC-γ1 [100]. Nonetheless, further studies are still required to understand the role of the CCM complex in this pathway, the requirements for the RAC1 interaction with the CCM complex, and whether this is a GTP-dependent mechanism.
DELTA-NOTCH
KRIT1 seems also to be important as an upstream regulator of DELTA-NOTCH signaling, playing a role in regulation of the NOTCH ligand DLL4, and the NOTCH target genes HEY1 and HEY2, although the molecular basis for how regulation of DELTA-NOTCH signaling by KRIT1 occurs is not yet clearly defined [85].
ROS Signaling
Loss of KRIT1 is also associated with increased intracellular reactive oxygen species (ROS) by regulating expression of the transcription factor FoxO1 and the ROS scavenger SOD2 [101], possibly by cooperation with Nd1-L [102]. This may be important in the context of CCM disease because altered dosage of KRIT1 could result in localized responses to oxidative stress events, resulting in differential levels of endothelial dysfunction and vascular permeability [101–103].
SMAD regulation
Very recent work has shown that one of the roles of the CCM complex is to regulate TGF-β (transforming growth factor-β) and BMP6 (bone morphogenic protein 6) signaling by modulating SMAD (mothers against decapentaplegic homolog) transcription factors. Loss of KRIT1 or CCM3 increases SMAD activation, and, consequently, signals from TGF-β and BMP6 to induce an endothelial-to-mesenchymal transition (EndMT), a critical step in CCM lesion formation [104]. The molecular basis for how this occurs is not yet understood, but it seems likely that the CCM complex plays an important role in direct regulation of SMAD activation.
Exocytosis
Two very recent studies have also discovered a role for CCM3 in exocytosis [105, 106]. The first showed that loss of CCM3 or GCKIII kinase in Drosophila results in dilated tracheal tubes that can be suppressed by reduced expression of NSF2, a protein required for vesicle-mediated transport [106]. The second study showed that CCM3–STK24 is a regulator of neutrophil degranulation by interacting with the fusion regulator UNC13D in a calcium-dependent manner. The CCM3–STK24 complex inhibits UNC13D interaction with the plasma membrane, preventing vesicle fusion by an STK24–UNC13D interaction. Upon increases in calcium levels, CCM3 binds UNC13D reducing STK24 binding to UNC13D and allowing vesicle fusion to occur. Loss of CCM3 or STK24 results in increased exocytosis. This regulation of exocytosis is independent of STK24 kinase activity and represents a novel function of the CCM proteins that may be important for the CCM disease phenotype [105].
Subcellular localization of the CCM proteins
The sub-cellular localization of the CCM proteins seems to be controlled by multiple factors. Localization of KRIT1 has been studied more extensively than that of CCM2 and CCM3, and KRIT1 has been found in multiple sub-cellular compartments.
KRIT1 localization, tubulin, and the nuclear question
KRIT1 subcellular localization seems to be controlled by its interactions with partner proteins. It is observed to be a microtubule-binding protein [107] whose interactions with microtubules can be disrupted by mutation of a basic residue patch (residues K47KRK) [37] within a flexible loop of the Nudix domain [42]. Its interaction with microtubules can also be reduced by co-expression with either Rap1 or ICAP1 [37], although how interactions with these proteins might regulate microtubule binding is an open question. It may be that KRIT1 exists in ‘open’ and ‘closed’ states where binding partners can regulate conformational rearrangements by interrupting potential interactions between the KRIT1 FERM domain and its N-terminal NPxY/F motifs [37]. It will be interesting to investigate whether CCM2 or CCM3 can also localize to microtubules, as this has not yet been shown. Following release from microtubules, KRIT1 seems to localize to cell membranes driven predominantly by its interaction with Rap1 [37, 71]. Once at the membrane, KRIT1 plays a role in endothelial cell–cell junctions, cell polarity, and correct lumen formation [108]. Other interactions may also play a role in retention of KRIT1 at the membrane, particularly that with HEG1 [47]. In addition, CCM2 may also bind to membrane-targeted proteins including small GTPases [99, 100], and CCM3 can bind paxillin [58, 60], so there may be multi-dentate forces retaining the CCM complex membrane localization. A driving force for KRIT1 release from the membrane seems to be its direct interaction with ICAP1 [49]. Once these proteins interact with one another, the KRIT1–ICAP1 complex is targeted to the cell nucleus (thus allowing integrin activation). Interestingly, ICAP1-driven relocalization of KRIT1 to the nucleus may release the KRIT1–CCM2 interaction [49]. CCM2 is a cytoplasmic protein, but a detailed description of its subcellular localization is not yet available. There are currently very little data that describe the role of KRIT1 or ICAP1 in the nucleus, but ICAP1 may play a role in c-Myc promotor activation [109]. KRIT1 may also interact with the sorting nexin family member SNX17 at intracellular vesicles, potentially affecting sorting nexin-driven integrin recycling and degradation [110–112].
CCM3 localization
CCM3 subcellular localization is different than that observed for either KRIT1 or CCM2. It seems to promote Golgi assembly and polarization, and its interactions with the GCKIII kinases and striatin occur in the Golgi [66, 91]. CCM3 is also recruited to cell membranes upon VEGF stimulation where it protects VEGFR2 from endocytosis [26], and interaction with PtdIns(3,4,5) may also play a role in CCM3 localization to the plasma membrane [113]. Recent work has also shown that both CCM3 and GCKIII are important for tubulogenesis in the Drosophila tracheal system [106]. CCM3 therefore seems to maintain functions outside the CCM complex. Given the close correlation between acquisition of CCM disease and improper formation of the CCM complex, it is interesting to speculate whether accurate CCM complex formation is critical for the normal functions of the CCM proteins.
Progress towards targeted therapy
CCM disease is still predominantly treated by neurosurgery to remove CCM cavernomas. As neurosurgery is not optimal in all cases (for example, for cavernomas that occur in regions that are surgically inaccessible), a long-term goal in the field is to discover a non-invasive treatment for the disease. Current studies for treatment strategies have focused on reducing RhoA signaling, which is increased upon loss of the CCM proteins. The two complementary strategies are to inhibit kinase activity of ROCK, currently by using a selective small molecule, Fasudil, which is approved for clinical use in Japan [97, 114], and to reduce or remove RhoA from the cell membrane by using HMG-CoA reductase inhibitors that block the pathway that gernanylgeranylates RhoA [24, 115]. Although these therapeutic targets have clinical potential, concern remains that chronic targeted inhibition of one of the key enzymes for actin remodeling (ROCK) or the high-doses of HMG-CoA reductase inhibitors required to affect geranylgeranylation of RhoA in the clinical setting could represent high hurdles to overcome. Another interesting insight is the recent work into the link between CCM and the TGF-β and BMP6 pathways and its implication for potential therapeutics [104]. Whether RhoA/ROCK signaling is a unique target for CCM disease, or whether there are also other targets, remains an open question. Further work is therefore still necessary to discover novel mechanisms that can inhibit the dysregulated signaling that occurs on loss of the CCM proteins.
Conclusions and outstanding questions
In the past few years, many studies have shed new light on the structure and function of KRIT1, CCM2, and CCM3, as well as begun to address how these three proteins interact with their many binding partners. Until recent years, with minimal information concerning the domain architecture of these proteins, it has been difficult to establish the intricacies of their roles within the cell. We now have a basic understanding of the structure of these proteins, allowing for future studies to more readily determine how they interact with each other and with non-CCM binding partners, and how these interactions can be formed and broken, which will provide better tools for understanding CCM biology. This has already proven useful, for example, in probing the balance between the KRIT1 and integrin β1 interactions with ICAP [42]. These studies highlight the many roles the CCM proteins play in the cell, and will serve to guide future work into the biological roles of these proteins, why their absence or mutation leads to disease, and therapeutic options for cerebral cavernous malformations. However, many outstanding questions concerning the CCM proteins remain. These include: What is the function of the newly discovered KRIT1 Nudix domain? What is the role of the newly discovered CCM2 HHD? How do the CCM proteins interact with β-catenin or VE-cadherin and RAC1, and what is the functional role of these interactions? How is recruitment and detachment of CCM complex binding partners controlled, how does it affect signal transduction, and is this spatially and/or temporally controlled? Do proposed conformational rearrangements of KRIT1 associate with specific binding partners and functional roles for the CCM complex? What is the role of KRIT1 at microtubules and in the nucleus? And how is SMAD signaling controlled by the CCM complex? Answers to these and other questions over the coming years, aided by past and future structural studies, will provide significant improvements in our understanding of CCM disease and increase the potential of treating this disease with targeted therapies.
Acknowledgments
O.S.F. is funded by a National Science Foundation Graduate Research Fellowship. T.J.B. is funded by National Institutes of Health grants R01GM102262, R01NS085078, and R01GM100411.
References
- 1.Labauge P, Denier C, Bergametti F, Tournier-Lasserve E. Genetics of cavernous angiomas. Lancet Neurol. 2007;6(3):237–244. doi: 10.1016/S1474-4422(07)70053-4. [DOI] [PubMed] [Google Scholar]
- 2.Cavalcanti DD, Kalani MY, Martirosyan NL, Eales J, Spetzler RF, Preul MC. Cerebral cavernous malformations: from genes to proteins to disease. J Neurosurg. 2012;116(1):122–132. doi: 10.3171/2011.8.JNS101241. [DOI] [PubMed] [Google Scholar]
- 3.Revencu N, Vikkula M. Cerebral cavernous malformation: new molecular and clinical insights. J Med Genet. 2006;43(9):716–721. doi: 10.1136/jmg.2006.041079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yadla S, Jabbour PM, Shenkar R, Shi C, Campbell PG, Awad IA. Cerebral cavernous malformations as a disease of vascular permeability: from bench to bedside with caution. Neurosurg Focus. 2010;29(3):E4. doi: 10.3171/2010.5.FOCUS10121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Otten P, Pizzolato GP, Rilliet B, Berney J. [131 cases of cavernous angioma (cavernomas) of the CNS, discovered by retrospective analysis of 24,535 autopsies] Neurochirurgie. 1989;35(2):82–83. [PubMed] [Google Scholar]
- 6.Riant F, Bergametti F, Ayrignac X, Boulday G, Tournier-Lasserve E. Recent insights into cerebral cavernous malformations: the molecular genetics of CCM. FEBS J. 2010;277(5):1070–1075. doi: 10.1111/j.1742-4658.2009.07535.x. [DOI] [PubMed] [Google Scholar]
- 7.Krisht KM, Whitehead KJ, Niazi T, Couldwell WT. The pathogenetic features of cerebral cavernous malformations: a comprehensive review with therapeutic implications. Neurosurg Focus. 2010;29(3):E2. doi: 10.3171/2010.6.FOCUS10135. [DOI] [PubMed] [Google Scholar]
- 8.Akers AL, Johnson E, Steinberg GK, Zabramski JM, Marchuk DA. Biallelic somatic and germline mutations in cerebral cavernous malformations (CCMs): evidence for a two-hit mechanism of CCM pathogenesis. Hum Mol Genet. 2009;18(5):919–930. doi: 10.1093/hmg/ddn430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pagenstecher A, Stahl S, Sure U, Felbor U. A two-hit mechanism causes cerebral cavernous malformations: complete inactivation of CCM1, CCM2 or CCM3 in affected endothelial cells. Hum Mol Genet. 2009;18(5):911–918. doi: 10.1093/hmg/ddn420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gault J, Shenkar R, Recksiek P, Awad IA. Biallelic somatic and germ line CCM1 truncating mutations in a cerebral cavernous malformation lesion. Stroke. 2005;36(4):872–874. doi: 10.1161/01.STR.0000157586.20479.fd. [DOI] [PubMed] [Google Scholar]
- 11.Boulday G, Rudini N, Maddaluno L, Blecon A, Arnould M, Gaudric A, Chapon F, Adams RH, Dejana E, Tournier-Lasserve E. Developmental timing of CCM2 loss influences cerebral cavernous malformations in mice. J Exp Med. 2011;208(9):1835–1847. doi: 10.1084/jem.20110571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dammann P, Hehr U, Weidensee S, Zhu Y, Gerlach R, Sure U. Two-hit mechanism in cerebral cavernous malformation? A case of monozygotic twins with a CCM1/KRIT1 germline mutation. Neurosurg Rev. 2013;36(3):483–486. doi: 10.1007/s10143-013-0456-z. [DOI] [PubMed] [Google Scholar]
- 13.Plummer NW, Zawistowski JS, Marchuk DA. Genetics of cerebral cavernous malformations. Curr Neurol Neurosci Rep. 2005;5(5):391–396. doi: 10.1007/s11910-005-0063-7. [DOI] [PubMed] [Google Scholar]
- 14.Denier C, Labauge P, Bergametti F, Marchelli F, Riant F, Arnoult M, Maciazek J, Vicaut E, Brunereau L, Tournier-Lasserve E. Genotype-phenotype correlations in cerebral cavernous malformations patients. Ann Neurol. 2006;60(5):550–556. doi: 10.1002/ana.20947. [DOI] [PubMed] [Google Scholar]
- 15.Dubovsky J, Zabramski JM, Kurth J, Spetzler RF, Rich SS, Orr HT, Weber JL. A gene responsible for cavernous malformations of the brain maps to chromosome 7q. Hum Mol Genet. 1995;4(3):453–458. doi: 10.1093/hmg/4.3.453. [DOI] [PubMed] [Google Scholar]
- 16.Gunel M, Awad IA, Anson J, Lifton RP. Mapping a gene causing cerebral cavernous malformation to 7q11.2-q21. Proc Natl Acad Sci USA. 1995;92(14):6620–6624. doi: 10.1073/pnas.92.14.6620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gunel M, Awad IA, Finberg K, Anson JA, Steinberg GK, Batjer HH, Kopitnik TA, Morrison L, Giannotta SL, Nelson-Williams C, Lifton RP. A founder mutation as a cause of cerebral cavernous malformation in Hispanic Americans. N Engl J Med. 1996;334(15):946–951. doi: 10.1056/NEJM199604113341503. [DOI] [PubMed] [Google Scholar]
- 18.Sahoo T, Johnson EW, Thomas JW, Kuehl PM, Jones TL, Dokken CG, Touchman JW, Gallione CJ, Lee-Lin SQ, Kosofsky B, Kurth JH, Louis DN, Mettler G, Morrison L, Gil-Nagel A, Rich SS, Zabramski JM, Boguski MS, Green ED, Marchuk DA. Mutations in the gene encoding KRIT1, a Krev-1/rap1a binding protein, cause cerebral cavernous malformations (CCM1) Hum Mol Genet. 1999;8(12):2325–2333. doi: 10.1093/hmg/8.12.2325. [DOI] [PubMed] [Google Scholar]
- 19.Laberge-le Couteulx S, Jung HH, Labauge P, Houtteville JP, Lescoat C, Cecillon M, Marechal E, Joutel A, Bach JF, Tournier-Lasserve E. Truncating mutations in CCM1, encoding KRIT1, cause hereditary cavernous angiomas. Nat Genet. 1999;23(2):189–193. doi: 10.1038/13815. [DOI] [PubMed] [Google Scholar]
- 20.Liquori CL, Berg MJ, Siegel AM, Huang E, Zawistowski JS, Stoffer T, Verlaan D, Balogun F, Hughes L, Leedom TP, Plummer NW, Cannella M, Maglione V, Squitieri F, Johnson EW, Rouleau GA, Ptacek L, Marchuk DA. Mutations in a gene encoding a novel protein containing a phosphotyrosine-binding domain cause type 2 cerebral cavernous malformations. Am J Hum Genet. 2003;73(6):1459–1464. doi: 10.1086/380314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guclu B, Ozturk AK, Pricola KL, Bilguvar K, Shin D, O’Roak BJ, Gunel M. Mutations in apoptosis-related gene, PDCD10, cause cerebral cavernous malformation 3. Neurosurgery. 2005;57(5):1008–1013. doi: 10.1227/01.neu.0000180811.56157.e1. [DOI] [PubMed] [Google Scholar]
- 22.Bergametti F, Denier C, Labauge P, Arnoult M, Boetto S, Clanet M, Coubes P, Echenne B, Ibrahim R, Irthum B, Jacquet G, Lonjon M, Moreau JJ, Neau JP, Parker F, Tremoulet M, Tournier-Lasserve E. Mutations within the programmed cell death 10 gene cause cerebral cavernous malformations. Am J Hum Genet. 2005;76(1):42–51. doi: 10.1086/426952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boulday G, Blecon A, Petit N, Chareyre F, Garcia LA, Niwa-Kawakita M, Giovannini M, Tournier-Lasserve E. Tissue-specific conditional CCM2 knockout mice establish the essential role of endothelial CCM2 in angiogenesis: implications for human cerebral cavernous malformations. Dis Model Mech. 2009;2(3–4):168–177. doi: 10.1242/dmm.001263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whitehead KJ, Chan AC, Navankasattusas S, Koh W, London NR, Ling J, Mayo AH, Drakos SG, Jones CA, Zhu W, Marchuk DA, Davis GE, Li DY. The cerebral cavernous malformation signaling pathway promotes vascular integrity via Rho GTPases. Nat Med. 2009;15(2):177–184. doi: 10.1038/nm.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kleaveland B, Zheng X, Liu JJ, Blum Y, Tung JJ, Zou Z, Sweeney SM, Chen M, Guo L, Lu MM, Zhou D, Kitajewski J, Affolter M, Ginsberg MH, Kahn ML. Regulation of cardiovascular development and integrity by the heart of glass-cerebral cavernous malformation protein pathway. Nat Med. 2009;15(2):169–176. doi: 10.1038/nm.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He Y, Zhang H, Yu L, Gunel M, Boggon TJ, Chen H, Min W. Stabilization of VEGFR2 signaling by cerebral cavernous malformation 3 is critical for vascular development. Sci Signal. 2010;3(116):ra26. doi: 10.1126/scisignal.2000722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Voss K, Stahl S, Schleider E, Ullrich S, Nickel J, Mueller TD, Felbor U. CCM3 interacts with CCM2 indicating common pathogenesis for cerebral cavernous malformations. Neurogenetics. 2007;8(4):249–256. doi: 10.1007/s10048-007-0098-9. [DOI] [PubMed] [Google Scholar]
- 28.Mably JD, Chuang LP, Serluca FC, Mohideen MA, Chen JN, Fishman MC. Santa and valentine pattern concentric growth of cardiac myocardium in the zebrafish. Development. 2006;133(16):3139–3146. doi: 10.1242/dev.02469. [DOI] [PubMed] [Google Scholar]
- 29.Chan AC, Drakos SG, Ruiz OE, Smith AC, Gibson CC, Ling J, Passi SF, Stratman AN, Sacharidou A, Revelo MP, Grossmann AH, Diakos NA, Davis GE, Metzstein MM, Whitehead KJ, Li DY. Mutations in 2 distinct genetic pathways result in cerebral cavernous malformations in mice. J Clin Invest. 2011;121(5):1871–1881. doi: 10.1172/JCI44393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Louvi A, Chen L, Two AM, Zhang H, Min W, Gunel M. Loss of cerebral cavernous malformation 3 (Ccm3) in neuroglia leads to CCM and vascular pathology. Proc Natl Acad Sci USA. 2011;108(9):3737–3742. doi: 10.1073/pnas.1012617108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cunningham K, Uchida Y, O’Donnell E, Claudio E, Li W, Soneji K, Wang H, Mukouyama YS, Siebenlist U. Conditional deletion of Ccm2 causes hemorrhage in the adult brain: a mouse model of human cerebral cavernous malformations. Hum Mol Genet. 2011;20(16):3198–3206. doi: 10.1093/hmg/ddr225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McDonald DA, Shenkar R, Shi C, Stockton RA, Akers AL, Kucherlapati MH, Kucherlapati R, Brainer J, Ginsberg MH, Awad IA, Marchuk DA. A novel mouse model of cerebral cavernous malformations based on the two-hit mutation hypothesis recapitulates the human disease. Hum Mol Genet. 2011;20(2):211–222. doi: 10.1093/hmg/ddq433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu H, Rigamonti D, Badr A, Zhang J. Ccm1 regulates microvascular morphogenesis during angiogenesis. J Vasc Res. 2011;48(2):130–140. doi: 10.1159/000316851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Serebriiskii I, Estojak J, Sonoda G, Testa JR, Golemis EA. Association of Krev-1/rap1a with Krit1, a novel ankyrin repeat-containing protein encoded by a gene mapping to 7q21-22. Oncogene. 1997;15(9):1043–1049. doi: 10.1038/sj.onc.1201268. [DOI] [PubMed] [Google Scholar]
- 35.Zhang J, Clatterbuck RE, Rigamonti D, Chang DD, Dietz HC. Interaction between krit1 and icap1alpha infers perturbation of integrin beta1-mediated angiogenesis in the pathogenesis of cerebral cavernous malformation. Hum Mol Genet. 2001;10(25):2953–2960. doi: 10.1093/hmg/10.25.2953. [DOI] [PubMed] [Google Scholar]
- 36.Francalanci F, Avolio M, De Luca E, Longo D, Menchise V, Guazzi P, Sgro F, Marino M, Goitre L, Balzac F, Trabalzini L, Retta SF. Structural and functional differences between KRIT1A and KRIT1B isoforms: a framework for understanding CCM pathogenesis. Exp Cell Res. 2009;315(2):285–303. doi: 10.1016/j.yexcr.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 37.Beraud-Dufour S, Gautier R, Albiges-Rizo C, Chardin P, Faurobert E. Krit 1 interactions with microtubules and membranes are regulated by Rap1 and integrin cytoplasmic domain associated protein-1. FEBS J. 2007;274(21):5518–5532. doi: 10.1111/j.1742-4658.2007.06068.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sahoo T, Goenaga-Diaz E, Serebriiskii IG, Thomas JW, Kotova E, Cuellar JG, Peloquin JM, Golemis E, Beitinjaneh F, Green ED, Johnson EW, Marchuk DA. Computational and experimental analyses reveal previously undetected coding exons of the KRIT1 (CCM1) gene. Genomics. 2001;71(1):123–126. doi: 10.1006/geno.2000.6426. [DOI] [PubMed] [Google Scholar]
- 39.Zhang J, Clatterbuck RE, Rigamonti D, Dietz HC. Cloning of the murine Krit1 cDNA reveals novel mammalian 5’ coding exons. Genomics. 2000;70(3):392–395. doi: 10.1006/geno.2000.6410. [DOI] [PubMed] [Google Scholar]
- 40.Eerola I, McIntyre B, Vikkula M. Identification of eight novel 5′-exons in cerebral capillary malformation gene-1 (CCM1) encoding KRIT1. Biochim Biophys Acta. 2001;1517(3):464–467. doi: 10.1016/s0167-4781(00)00303-1. [DOI] [PubMed] [Google Scholar]
- 41.Zhang J, Rigamonti D, Dietz HC, Clatterbuck RE. Interaction between krit1 and malcavernin: implications for the pathogenesis of cerebral cavernous malformations. Neurosurgery. 2007;60(2):353–359. doi: 10.1227/01.NEU.0000249268.11074.83. [DOI] [PubMed] [Google Scholar]
- 42.Liu W, Draheim KM, Zhang R, Calderwood DA, Boggon TJ. Mechanism for KRIT1 release of ICAP1-mediated suppression of integrin activation. Mol Cell. 2013;49(4):719–729. doi: 10.1016/j.molcel.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bessman MJ, Frick DN, O’Handley SF. The MutT proteins or “Nudix” hydrolases, a family of versatile, widely distributed, “housecleaning” enzymes. J Biol Chem. 1996;271(41):25059–25062. doi: 10.1074/jbc.271.41.25059. [DOI] [PubMed] [Google Scholar]
- 44.Boudeau J, Miranda-Saavedra D, Barton GJ, Alessi DR. Emerging roles of pseudokinases. Trends Cell Biol. 2006;16(9):443–452. doi: 10.1016/j.tcb.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 45.Li X, Zhang R, Draheim KM, Liu W, Calderwood DA, Boggon TJ. Structural basis for small G protein effector interaction of Ras-related protein 1 (Rap1) and adaptor protein Krev interaction trapped 1 (KRIT1) J Biol Chem. 2012;287(26):22317–22327. doi: 10.1074/jbc.M112.361295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gingras AR, Puzon-McLaughlin W, Ginsberg MH. The structure of the ternary complex of krev interaction trapped 1 (KRIT1) bound to both the Rap1 GTPase and the heart of glass (HEG1) cytoplasmic tail. J Biol Chem. 2013;288(33):23639–23649. doi: 10.1074/jbc.M113.462911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gingras AR, Liu JJ, Ginsberg MH. Structural basis of the junctional anchorage of the cerebral cavernous malformations complex. J Cell Biol. 2012;199(1):39–48. doi: 10.1083/jcb.201205109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Denier C, Goutagny S, Labauge P, Krivosic V, Arnoult M, Cousin A, Benabid AL, Comoy J, Frerebeau P, Gilbert B, Houtteville JP, Jan M, Lapierre F, Loiseau H, Menei P, Mercier P, Moreau JJ, Nivelon-Chevallier A, Parker F, Redondo AM, Scarabin JM, Tremoulet M, Zerah M, Maciazek J, Tournier-Lasserve E. Mutations within the MGC4607 gene cause cerebral cavernous malformations. Am J Hum Genet. 2004;74(2):326–337. doi: 10.1086/381718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zawistowski JS, Stalheim L, Uhlik MT, Abell AN, Ancrile BB, Johnson GL, Marchuk DA. CCM1 and CCM2 protein interactions in cell signaling: implications for cerebral cavernous malformations pathogenesis. Hum Mol Genet. 2005;14(17):2521–2531. doi: 10.1093/hmg/ddi256. [DOI] [PubMed] [Google Scholar]
- 50.Harel L, Costa B, Tcherpakov M, Zapatka M, Oberthuer A, Hansford LM, Vojvodic M, Levy Z, Chen ZY, Lee FS, Avigad S, Yaniv I, Shi L, Eils R, Fischer M, Brors B, Kaplan DR, Fainzilber M. CCM2 mediates death signaling by the TrkA receptor tyrosine kinase. Neuron. 2009;63(5):585–591. doi: 10.1016/j.neuron.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 51.Crose LE, Hilder TL, Sciaky N, Johnson GL. Cerebral cavernous malformation 2 protein promotes smad ubiquitin regulatory factor 1-mediated RhoA degradation in endothelial cells. J Biol Chem. 2009;284(20):13301–13305. doi: 10.1074/jbc.C900009200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fisher OS, Zhang R, Li X, Murphy JW, Demeler B, Boggon TJ. Structural studies of cerebral cavernous malformations 2 (CCM2) reveal a folded helical domain at its C-terminus. FEBS Lett. 2013;587(3):272–277. doi: 10.1016/j.febslet.2012.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pan L, Yan J, Wu L, Zhang M. Assembling stable hair cell tip link complex via multidentate interactions between harmonin and cadherin 23. Proc Natl Acad Sci USA. 2009;106(14):5575–5580. doi: 10.1073/pnas.0901819106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Faure G, Revy P, Schertzer M, Londono-Vallejo A, Callebaut I (2013) The C-terminal extension of human RTEL1, mutated in Hoyeraal–Hreidarsson syndrome, contains Harmonin-N-like domains. Proteins. doi:10.1002/prot.24438 [DOI] [PubMed]
- 55.Rosen JN, Sogah VM, Ye LY, Mably JD. ccm2-like is required for cardiovascular development as a novel component of the Heg-CCM pathway. Dev Biol. 2013;376(1):74–85. doi: 10.1016/j.ydbio.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zheng X, Xu C, Smith AO, Stratman AN, Zou Z, Kleaveland B, Yuan L, Didiku C, Sen A, Liu X, Skuli N, Zaslavsky A, Chen M, Cheng L, Davis GE, Kahn ML. Dynamic regulation of the cerebral cavernous malformation pathway controls vascular stability and growth. Dev Cell. 2012;23(2):342–355. doi: 10.1016/j.devcel.2012.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Voss K, Stahl S, Hogan BM, Reinders J, Schleider E, Schulte-Merker S, Felbor U. Functional analyses of human and zebrafish 18-amino acid in-frame deletion pave the way for domain mapping of the cerebral cavernous malformation 3 protein. Hum Mutat. 2009;30(6):1003–1011. doi: 10.1002/humu.20996. [DOI] [PubMed] [Google Scholar]
- 58.Li X, Zhang R, Zhang H, He Y, Ji W, Min W, Boggon TJ. Crystal structure of CCM3, a cerebral cavernous malformation protein critical for vascular integrity. J Biol Chem. 2010;285(31):24099–24107. doi: 10.1074/jbc.M110.128470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ding J, Wang X, Li DF, Hu Y, Zhang Y, Wang DC. Crystal structure of human programmed cell death 10 complexed with inositol-(1,3,4,5)-tetrakisphosphate: a novel adaptor protein involved in human cerebral cavernous malformation. Biochem Biophys Res Commun. 2010;399(4):587–592. doi: 10.1016/j.bbrc.2010.07.119. [DOI] [PubMed] [Google Scholar]
- 60.Li X, Ji W, Zhang R, Folta-Stogniew E, Min W, Boggon TJ. Molecular recognition of LD motifs by the FAT-homology domain of cerebral cavernous malformation 3 (CCM3) J Biol Chem. 2011;286(29):26138–26147. doi: 10.1074/jbc.M110.211250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang M, Dong L, Shi Z, Jiao S, Zhang Z, Zhang W, Liu G, Chen C, Feng M, Hao Q, Wang W, Yin M, Zhao Y, Zhang L, Zhou Z. Structural mechanism of CCM3 heterodimerization with GCKIII kinases. Structure. 2013;21(4):680–688. doi: 10.1016/j.str.2013.02.015. [DOI] [PubMed] [Google Scholar]
- 62.Xu X, Wang X, Zhang Y, Wang DC, Ding J. Structural basis for the unique heterodimeric assembly between cerebral cavernous malformation 3 and germinal center kinase III. Structure. 2013;21(6):1059–1066. doi: 10.1016/j.str.2013.04.007. [DOI] [PubMed] [Google Scholar]
- 63.Hilder TL, Malone MH, Bencharit S, Colicelli J, Haystead TA, Johnson GL, Wu CC. Proteomic identification of the cerebral cavernous malformation signaling complex. J Proteome Res. 2007;6(11):4343–4355. doi: 10.1021/pr0704276. [DOI] [PubMed] [Google Scholar]
- 64.Costa B, Kean MJ, Ast V, Knight JD, Mett A, Levy Z, Ceccarelli DF, Badillo BG, Eils R, Konig R, Gingras AC, Fainzilber M. STK25 protein mediates TrkA and CCM2 protein-dependent death in pediatric tumor cells of neural origin. J Biol Chem. 2012;287(35):29285–29289. doi: 10.1074/jbc.C112.345397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stahl S, Gaetzner S, Voss K, Brackertz B, Schleider E, Surucu O, Kunze E, Netzer C, Korenke C, Finckh U, Habek M, Poljakovic Z, Elbracht M, Rudnik-Schoneborn S, Bertalanffy H, Sure U, Felbor U. Novel CCM1, CCM2, and CCM3 mutations in patients with cerebral cavernous malformations: in-frame deletion in CCM2 prevents formation of a CCM1/CCM2/CCM3 protein complex. Hum Mutat. 2008;29(5):709–717. doi: 10.1002/humu.20712. [DOI] [PubMed] [Google Scholar]
- 66.Kean MJ, Ceccarelli DF, Goudreault M, Sanches M, Tate S, Larsen B, Gibson LC, Derry WB, Scott IC, Pelletier L, Baillie GS, Sicheri F, Gingras AC. Structure-function analysis of core STRIPAK proteins: a signaling complex implicated in Golgi polarization. J Biol Chem. 2011;286(28):25065–25075. doi: 10.1074/jbc.M110.214486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Berman JR, Kenyon C. Germ-cell loss extends C. elegans life span through regulation of DAF-16 by kri-1 and lipophilic-hormone signaling. Cell. 2006;124(5):1055–1068. doi: 10.1016/j.cell.2006.01.039. [DOI] [PubMed] [Google Scholar]
- 68.Frische EW, Zwartkruis FJ. Rap1, a mercenary among the Ras-like GTPases. Dev Biol. 2010;340(1):1–9. doi: 10.1016/j.ydbio.2009.12.043. [DOI] [PubMed] [Google Scholar]
- 69.Boettner B, Van Aelst L. Control of cell adhesion dynamics by Rap1 signaling. Curr Opin Cell Biol. 2009;21(5):684–693. doi: 10.1016/j.ceb.2009.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Glading A, Han J, Stockton RA, Ginsberg MH. KRIT-1/CCM1 is a Rap1 effector that regulates endothelial cell cell junctions. J Cell Biol. 2007;179(2):247–254. doi: 10.1083/jcb.200705175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu JJ, Stockton RA, Gingras AR, Ablooglu AJ, Han J, Bobkov AA, Ginsberg MH. A mechanism of Rap1-induced stabilization of endothelial cell–cell junctions. Mol Biol Cell. 2011;22(14):2509–2519. doi: 10.1091/mbc.E11-02-0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Glading AJ, Ginsberg MH. Rap1 and its effector KRIT1/CCM1 regulate beta-catenin signaling. Dis Model Mech. 2010;3(1–2):73–83. doi: 10.1242/dmm.003293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Faurobert E, Rome C, Lisowska J, Manet-Dupe S, Boulday G, Malbouyres M, Balland M, Bouin AP, Keramidas M, Bouvard D, Coll JL, Ruggiero F, Tournier-Lasserve E, Albiges-Rizo C. CCM1-ICAP-1 complex controls beta1 integrin-dependent endothelial contractility and fibronectin remodeling. J Cell Biol. 2013;202(3):545–561. doi: 10.1083/jcb.201303044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Millon-Fremillon A, Brunner M, Abed N, Collomb E, Ribba AS, Block MR, Albiges-Rizo C, Bouvard D. CaMKII-mediated intramolecular opening of integrin cytoplasmic domain associated protein-1 (ICAP-1alpha) negatively regulates beta1 integrins. J Biol Chem. 2013;288(28):20248–20260. doi: 10.1074/jbc.M113.455956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chang DD, Wong C, Smith H, Liu J. ICAP-1, a novel beta1 integrin cytoplasmic domain-associated protein, binds to a conserved and functionally important NPXY sequence motif of beta1 integrin. J Cell Biol. 1997;138(5):1149–1157. doi: 10.1083/jcb.138.5.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang XA, Hemler ME. Interaction of the integrin beta1 cytoplasmic domain with ICAP-1 protein. J Biol Chem. 1999;274(1):11–19. doi: 10.1074/jbc.274.1.11. [DOI] [PubMed] [Google Scholar]
- 77.Calderwood DA, Tai V, Di Paolo G, De Camilli P, Ginsberg MH. Competition for talin results in trans-dominant inhibition of integrin activation. J Biol Chem. 2004;279(28):28889–28895. doi: 10.1074/jbc.M402161200. [DOI] [PubMed] [Google Scholar]
- 78.Chang DD, Hoang BQ, Liu J, Springer TA. Molecular basis for interaction between Icap1 alpha PTB domain and beta 1 integrin. J Biol Chem. 2002;277(10):8140–8145. doi: 10.1074/jbc.M109031200. [DOI] [PubMed] [Google Scholar]
- 79.Bouvard D, Vignoud L, Dupe-Manet S, Abed N, Fournier HN, Vincent-Monegat C, Retta SF, Fassler R, Block MR. Disruption of focal adhesions by integrin cytoplasmic domain-associated protein-1 alpha. J Biol Chem. 2003;278(8):6567–6574. doi: 10.1074/jbc.M211258200. [DOI] [PubMed] [Google Scholar]
- 80.Brunner M, Millon-Fremillon A, Chevalier G, Nakchbandi IA, Mosher D, Block MR, Albiges-Rizo C, Bouvard D. Osteoblast mineralization requires {beta}1 integrin/ICAP-1-dependent fibronectin deposition. J Cell Biol. 2011;194(2):307–322. doi: 10.1083/jcb.201007108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Millon-Fremillon A, Bouvard D, Grichine A, Manet-Dupe S, Block MR, Albiges-Rizo C. Cell adaptive response to extracellular matrix density is controlled by ICAP-1-dependent beta1-integrin affinity. J Cell Biol. 2008;180(2):427–441. doi: 10.1083/jcb.200707142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zawistowski JS, Serebriiskii IG, Lee MF, Golemis EA, Marchuk DA. KRIT1 association with the integrin-binding protein ICAP-1: a new direction in the elucidation of cerebral cavernous malformations (CCM1) pathogenesis. Hum Mol Genet. 2002;11(4):389–396. doi: 10.1093/hmg/11.4.389. [DOI] [PubMed] [Google Scholar]
- 83.Bouvard D, Aszodi A, Kostka G, Block MR, Albiges-Rizo C, Fassler R. Defective osteoblast function in ICAP-1-deficient mice. Development. 2007;134(14):2615–2625. doi: 10.1242/dev.000877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang J, Basu S, Rigamonti D, Dietz HC, Clatterbuck RE. Krit1 modulates beta 1-integrin-mediated endothelial cell proliferation. Neurosurgery. 2008;63(3):571–578. doi: 10.1227/01.NEU.0000325255.30268.B0. [DOI] [PubMed] [Google Scholar]
- 85.Wustehube J, Bartol A, Liebler SS, Brutsch R, Zhu Y, Felbor U, Sure U, Augustin HG, Fischer A. Cerebral cavernous malformation protein CCM1 inhibits sprouting angiogenesis by activating DELTA-NOTCH signaling. Proc Natl Acad Sci USA. 2010;107(28):12640–12645. doi: 10.1073/pnas.1000132107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Brutsch R, Liebler SS, Wustehube J, Bartol A, Herberich SE, Adam MG, Telzerow A, Augustin HG, Fischer A. Integrin cytoplasmic domain-associated protein-1 attenuates sprouting angiogenesis. Circ Res. 2010;107(5):592–601. doi: 10.1161/CIRCRESAHA.110.217257. [DOI] [PubMed] [Google Scholar]
- 87.You C, Sandalcioglu IE, Dammann P, Felbor U, Sure U, Zhu Y. Loss of CCM3 impairs DLL4-Notch signalling: implication in endothelial angiogenesis and in inherited cerebral cavernous malformations. J Cell Mol Med. 2013;17(3):407–418. doi: 10.1111/jcmm.12022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hamada K, Shimizu T, Matsui T, Tsukita S, Hakoshima T. Structural basis of the membrane-targeting and unmasking mechanisms of the radixin FERM domain. EMBO J. 2000;19(17):4449–4462. doi: 10.1093/emboj/19.17.4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lu TJ, Lai WY, Huang CY, Hsieh WJ, Yu JS, Hsieh YJ, Chang WT, Leu TH, Chang WC, Chuang WJ, Tang MJ, Chen TY, Lu TL, Lai MD. Inhibition of cell migration by autophosphorylated mammalian sterile 20-like kinase 3 (MST3) involves paxillin and protein-tyrosine phosphatase-PEST. J Biol Chem. 2006;281(50):38405–38417. doi: 10.1074/jbc.M605035200. [DOI] [PubMed] [Google Scholar]
- 90.Ceccarelli DF, Laister RC, Mulligan VK, Kean MJ, Goudreault M, Scott IC, Derry WB, Chakrabartty A, Gingras AC, Sicheri F. CCM3/PDCD10 heterodimerizes with germinal center kinase III (GCKIII) proteins using a mechanism analogous to CCM3 homodimerization. J Biol Chem. 2011;286(28):25056–25064. doi: 10.1074/jbc.M110.213777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fidalgo M, Fraile M, Pires A, Force T, Pombo C, Zalvide J. CCM3/PDCD10 stabilizes GCKIII proteins to promote Golgi assembly and cell orientation. J Cell Sci. 2010;123(Pt 8):1274–1284. doi: 10.1242/jcs.061341. [DOI] [PubMed] [Google Scholar]
- 92.Preisinger C, Short B, De Corte V, Bruyneel E, Haas A, Kopajtich R, Gettemans J, Barr FA. YSK1 is activated by the Golgi matrix protein GM130 and plays a role in cell migration through its substrate 14-3-3zeta. J Cell Biol. 2004;164(7):1009–1020. doi: 10.1083/jcb.200310061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ling P, Lu TJ, Yuan CJ, Lai MD. Biosignaling of mammalian Ste20-related kinases. Cell Signal. 2008;20(7):1237–1247. doi: 10.1016/j.cellsig.2007.12.019. [DOI] [PubMed] [Google Scholar]
- 94.Zheng X, Xu C, Di Lorenzo A, Kleaveland B, Zou Z, Seiler C, Chen M, Cheng L, Xiao J, He J, Pack MA, Sessa WC, Kahn ML. CCM3 signaling through sterile 20-like kinases plays an essential role during zebrafish cardiovascular development and cerebral cavernous malformations. J Clin Invest. 2010;120(8):2795–2804. doi: 10.1172/JCI39679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Richardson BT, Dibble CF, Borikova AL, Johnson GL. Cerebral cavernous malformation is a vascular disease associated with activated RhoA signaling. Biol Chem. 2013;394(1):35–42. doi: 10.1515/hsz-2012-0243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Goudreault M, D’Ambrosio LM, Kean MJ, Mullin MJ, Larsen BG, Sanchez A, Chaudhry S, Chen GI, Sicheri F, Nesvizhskii AI, Aebersold R, Raught B, Gingras AC. A PP2A phosphatase high density interaction network identifies a novel striatin-interacting phosphatase and kinase complex linked to the cerebral cavernous malformation 3 (CCM3) protein. Mol Cell Proteomics. 2009;8(1):157–171. doi: 10.1074/mcp.M800266-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Stockton RA, Shenkar R, Awad IA, Ginsberg MH. Cerebral cavernous malformations proteins inhibit Rho kinase to stabilize vascular integrity. J Exp Med. 2010;207(4):881–896. doi: 10.1084/jem.20091258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Borikova AL, Dibble CF, Sciaky N, Welch CM, Abell AN, Bencharit S, Johnson GL. Rho kinase inhibition rescues the endothelial cell cerebral cavernous malformation phenotype. J Biol Chem. 2010;285(16):11760–11764. doi: 10.1074/jbc.C109.097220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Uhlik MT, Abell AN, Johnson NL, Sun W, Cuevas BD, Lobel-Rice KE, Horne EA, Dell’Acqua ML, Johnson GL. Rac-MEKK3-MKK3 scaffolding for p38 MAPK activation during hyperosmotic shock. Nat Cell Biol. 2003;5(12):1104–1110. doi: 10.1038/ncb1071. [DOI] [PubMed] [Google Scholar]
- 100.Zhou X, Izumi Y, Burg MB, Ferraris JD. Rac1/osmosensing scaffold for MEKK3 contributes via phospholipase C-gamma1 to activation of the osmoprotective transcription factor NFAT5. Proc Natl Acad Sci USA. 2011;108(29):12155–12160. doi: 10.1073/pnas.1108107108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Goitre L, Balzac F, Degani S, Degan P, Marchi S, Pinton P, Retta SF. KRIT1 regulates the homeostasis of intracellular reactive oxygen species. PLoS ONE. 2010;5(7):e11786. doi: 10.1371/journal.pone.0011786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Guazzi P, Goitre L, Ferro E, Cutano V, Martino C, Trabalzini L, Retta SF. Identification of the Kelch family protein Nd1-L as a novel molecular interactor of KRIT1. PLoS ONE. 2012;7(9):e44705. doi: 10.1371/journal.pone.0044705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bacigaluppi S, Retta SF, Pileggi S, Fontanella M, Goitre L, Tassi L, La Camera A, Citterio A, Patrosso MC, Tredici G, Penco S. Genetic and cellular basis of cerebral cavernous malformations: implications for clinical management. Clin Genet. 2013;83(1):7–14. doi: 10.1111/j.1399-0004.2012.01892.x. [DOI] [PubMed] [Google Scholar]
- 104.Maddaluno L, Rudini N, Cuttano R, Bravi L, Giampietro C, Corada M, Ferrarini L, Orsenigo F, Papa E, Boulday G, Tournier-Lasserve E, Chapon F, Richichi C, Retta SF, Lampugnani MG, Dejana E. EndMT contributes to the onset and progression of cerebral cavernous malformations. Nature. 2013;498(7455):492–496. doi: 10.1038/nature12207. [DOI] [PubMed] [Google Scholar]
- 105.Zhang Y, Tang W, Zhang H, Niu X, Xu Y, Zhang J, Gao K, Pan W, Boggon TJ, Toomre D, Min W, Wu D. A network of interactions enables CCM3 and STK24 to coordinate UNC13D-driven vesicle exocytosis in neutrophils. Dev Cell. 2013;27:215–226. doi: 10.1016/j.devcel.2013.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Song Y, Eng M, Ghabrial AS. Focal defects in single-celled tubes mutant for cerebral cavernous malformation 3, GCKIII, or NSF2. Dev Cell. 2013;25(5):507–519. doi: 10.1016/j.devcel.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gunel M, Laurans MS, Shin D, DiLuna ML, Voorhees J, Choate K, Nelson-Williams C, Lifton RP. KRIT1, a gene mutated in cerebral cavernous malformation, encodes a microtubule-associated protein. Proc Natl Acad Sci USA. 2002;99(16):10677–10682. doi: 10.1073/pnas.122354499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lampugnani MG, Orsenigo F, Rudini N, Maddaluno L, Boulday G, Chapon F, Dejana E. CCM1 regulates vascular-lumen organization by inducing endothelial polarity. J Cell Sci. 2010;123(Pt 7):1073–1080. doi: 10.1242/jcs.059329. [DOI] [PubMed] [Google Scholar]
- 109.Fournier HN, Dupe-Manet S, Bouvard D, Luton F, Degani S, Block MR, Retta SF, Albiges-Rizo C. Nuclear translocation of integrin cytoplasmic domain-associated protein 1 stimulates cellular proliferation. Mol Biol Cell. 2005;16(4):1859–1871. doi: 10.1091/mbc.E04-08-0744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Czubayko M, Knauth P, Schluter T, Florian V, Bohnensack R. Sorting nexin 17, a non-self-assembling and a PtdIns(3)P high class affinity protein, interacts with the cerebral cavernous malformation related protein KRIT1. Biochem Biophys Res Commun. 2006;345(3):1264–1272. doi: 10.1016/j.bbrc.2006.04.129. [DOI] [PubMed] [Google Scholar]
- 111.Brahme NN, Calderwood DA. Cell adhesion: a FERM grasp of the tail sorts out integrins. Curr Biol. 2012;22(17):R692–R694. doi: 10.1016/j.cub.2012.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bottcher RT, Stremmel C, Meves A, Meyer H, Widmaier M, Tseng HY, Fassler R. Sorting nexin 17 prevents lysosomal degradation of beta1 integrins by binding to the beta1-integrin tail. Nat Cell Biol. 2012;14(6):584–592. doi: 10.1038/ncb2501. [DOI] [PubMed] [Google Scholar]
- 113.Dibble CF, Horst JA, Malone MH, Park K, Temple B, Cheeseman H, Barbaro JR, Johnson GL, Bencharit S. Defining the functional domain of programmed cell death 10 through its interactions with phosphatidylinositol-3,4,5-trisphosphate. PLoS ONE. 2010;5(7):e11740. doi: 10.1371/journal.pone.0011740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.McDonald DA, Shi C, Shenkar R, Stockton RA, Liu F, Ginsberg MH, Marchuk DA, Awad IA. Fasudil decreases lesion burden in a murine model of cerebral cavernous malformation disease. Stroke. 2012;43(2):571–574. doi: 10.1161/STROKEAHA.111.625467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Li DY, Whitehead KJ. Evaluating strategies for the treatment of cerebral cavernous malformations. Stroke. 2010;41(10 Suppl):S92–S94. doi: 10.1161/STROKEAHA.110.594929. [DOI] [PMC free article] [PubMed] [Google Scholar]