Abstract
Malignant peripheral nerve sheath tumors (MPNSTs) are Schwann cell-derived malignancies that arise from plexiform neurofibromas in patients with mutation of the neurofibromin 1 (NF1) gene. We have shown that the growth factor neuregulin-1 (NRG1) also contributes to human neurofibroma and MPNST pathogenesis and that outbred C57BL/6J x SJL/J transgenic mice overexpressing NRG1 in Schwann cells (P0-GGFβ3 mice) recapitulate the process of neurofibroma-MPNST progression. However, it is unclear whether NRG1 acts predominantly within NF1-regulated signaling cascades or instead activates other essential cascades that cooperate with NF1 loss to promote tumorigenesis. We now report that tumorigenesis is suppressed in inbred P0-GGFβ3 mice on a C57BL/6J background. To determine whether NRG1 overexpression interacts with reduced Nf1 or Trp53 gene dosage to “unmask” tumorigenesis in these animals, we followed cohorts of inbred P0-GGFβ3;Nf1+/−, P0-GGFβ3; Trp53+/− and control (P0-GGFβ3, Nf1+/− andTrp53+/−) mice for 1 year. We found no reduction in survival or tumors in control and P0-GGFβ3;Nf1+/− mice. In contrast, P0-GGFβ3; Trp53+/− mice died on average at 226 days, with MPNSTs present in 95% of these mice. MPNSTs in inbred P0-GGFβ3; Trp53+/− mice arose de novo from micro-MPNSTs that uniformly develop intraganglionically. These micro-MPNSTs are of lower grade (WHO grade II-III) than the major MPNSTs (WHO grade III-IV); array comparative genomic hybridization showed that lower grade MPNSTs also had fewer genomic abnormalities. Thus, P0-GGFβ3; Trp53+/− mice represent a novel model of low to high grade MPNST progression. We further conclude that NRG1 promotes peripheral nervous system neoplasia predominantly via its effects on the signaling cascades affected by Nf1 loss.
Keywords: Neurofibromatosis, sarcoma, genetic complementation, mouse models, erbB receptors
Introduction
Neurofibromatosis type 1 (NF1) is the most common genetic disease affecting the human nervous system, occurring in 1 of every 3000–3500 newborn infants [5]. Although individuals with this autosomal dominant tumor susceptibility syndrome can develop a variety of tumor types in their central nervous system and elsewhere, NF1 patients are particularly susceptible to the development of benign tumors in skin (dermal neurofibromas) and large nerves (plexiform neurofibromas). Histologically, dermal and plexiform neurofibromas are identical, being composed of a mixture of Schwann cells, mast cells, macrophages, fibroblasts and other cell types. Nonetheless, the clinical behavior of dermal and plexiform neurofibromas is quite distinct. Of particular note, plexiform neurofibromas, unlike dermal neurofibromas, often progress to become malignant peripheral nerve sheath tumors (MPNSTs), the most common malignancy and the leading cause of death in NF1 patients [11,29]. In both plexiform neurofibromas and MPNSTs, biallelic NF1 loss occurs in intratumoral Schwann cell-like elements [21,32], indicating that the neoplastic component of these tumors is derived from the Schwann cell lineage. In keeping with this, genetically engineered mice with Nf1-null Schwann cells develop plexiform neurofibromas [28,47,53]. Observations in NF1 patients suggest that plexiform neurofibromas progress to become MPNSTs when NF1 loss interacts with the mutation of additional tumor suppressor genes such as TP53 [2,15,25,30]. This suggestion is also supported by studies with genetically engineered mice, as mice with cis-linked Nf1 and Trp53 null mutations (cis-Nf1+/− ;Trp53+/− mice) develop MPNSTs which arise de novo rather than from neurofibromas [6,45].
The growth factor neuregulin-1 (NRG1) has been implicated in the pathogenesis of a variety of cancer types including gliomas [34] and colorectal [1,7,51], ovarian [38], breast [40], and prostate [41] carcinomas. We have hypothesized that this potent Schwann cell mitogen also contributes to the development of plexiform neurofibromas and MPNSTs. In support of this, we have shown that human neurofibromas, MPNSTs and MPNST cell lines express several type II and type III NRG1 isoforms together with the erbB receptors (erbB2, erbB3 and erbB4) mediating NRG1 responses [43]. This coexpression of NRG1 and its receptors in sporadic and NF1-associated peripheral nerve sheath tumors results in constitutive erbB activation, likely via autocrine or paracrine signaling. The importance of this signaling cascade in MPNST biology is underscored by the fact that inhibiting erbB signaling potently impedes MPNST proliferation, invasion and survival [10,22,43].
To directly test the hypothesis that NRG1 overexpression is sufficient for tumorigenesis in the peripheral nervous system (PNS), we generated transgenic mice that overexpress the type II NRG1 isoform glial growth factor-β3 in Schwann cells (P0-GGFβ3 mice). On an outbred C57BL/6J x SJL/J background, P0-GGFβ3 mice develop aggressive spindle cell malignancies with the histologic, immunohistochemical and ultrastructural features of human MPNSTs [16] and recapitulate the process of neurofibroma-MPNST progression seen in NF1 patients [20]. P0-GGFβ3 MPNSTs have abnormalities of Trp53 and in other cell cycle regulatory cascades that parallel those seen in human MPNSTs and, using array comparative genomic hybridization, we have identified multiple additional genomic abnormalities potentially contributing to the pathogenesis of these tumors [20]. Of note, while P0-GGFβ3 MPNSTs have intact Nf1 alleles, they demonstrate the prominent Ras hyperactivation characteristic of NF1 null tumors.
At present, it is unclear how aberrant NRG1/erbB signaling interacts with NF1 and other key tumor suppressors to promote neurofibroma and MPNST pathogenesis. Fortuitously, we have found that tumorigenesis is suppressed when P0-GGFβ3 mice are bred onto a C57BL/6J background. This presented us with an opportunity to clarify the role that NRG1/erbB signaling plays in peripheral nervous system neoplasia. To address this question, we have performed genetic complementation experiments in which we crossed P0-GGFβ3 mice to Nf1+/− or Trp53+/− mice and determined the effect this had on tumorigenesis. These studies clarify the role that NRG1 plays in the pathogenesis of neurofibromas and MPNSTs, provide new evidence as to the source of these tumors and, unexpectedly, produced a new model useful for defining the genomic abnormalities mediating the progression of low grade MPNSTs to higher grade tumors.
Materials and Methods
Antibodies and Other Reagents
Rabbit polyclonal anti-S100β (#Z0311) and mouse monoclonal anti-desmin (clone D33) antibodies were purchased from Dako (Carpinteria, CA). Mouse monoclonal antibodies recognizing nonphosphorylated heavy neurofilaments (clone SMI34) or GAPDH were obtained from Sternberger Monoclonals, Inc. (Lutherville, MD) and Advanced ImmunoChemical (Long Beach, CA), respectively. Mouse monoclonal antibodies against mouse or human nestin (rat-401 and 10C2, respectively) were from Millipore (Billerica, MA). A mouse pan-Ras monoclonal antibody (clone Ras-10) was also purchased from Millipore. A mouse monoclonal anti-Ki67 antibody (#558078) was purchased from BD Biosciences (Franklin Lakes, NJ). Horseradish peroxidase (HRP)-, FITC-, Alexa 488- and Cy3-conjugated secondary antibodies were from Jackson ImmunoResearch Laboratories, Inc. (West Grove, PA). ImmPRESS polymerized reporter staining enzyme systems were purchased from Vector Laboratories (Burlingame, CA). Biotin tyramide signal amplification kits were from Perkin Elmer (Waltham, MA).
Polyclonal IgY antibodies recognizing the amino-terminal kringle domain present in type II NRG1 isoforms and the amino-terminal cysteine-rich domain (CRD) characteristic of sensory and motor-neuron derived factor (SMDF; type III NRG1 isoforms) were raised in chickens (Aves Labs, Inc.; Tigard, Oregon). The sequence of the peptide used to raise the anti-kringle domain antibody was N-CZCGRLKEDSRYIFFMEPD-C. To generate the anti-CRD antibody, a peptide with the sequence N-CZDKIFEYDSPTHLD-C was utilized. For each of these peptide sequences, a cysteine was added to the N-terminus to improve the efficiency of conjugation to keyhole limpet hemocyanin (KLH) and to prevent the formation of structures outside the native protein conformation. An aminocaproic acid (Z) residue was added after the N-terminal cysteine to isolate the cysteines from the sequence of interest and to enhance conjugation of the peptide to KLH. Two hens were immunized with each peptide and immune responses were monitored by performing ELISA analyses with the IgY fraction isolated from eggs. Polyclonal IgY antibodies were affinity purified from egg whites using immobilized immunizing peptide.
The pan-erbB inhibitor canertinib (CI-1033) was purchased from LC Laboratories (Woburn, MA).
Genetically Engineered Mice and Animal Care
Mice were cared for in accordance with the guidelines of the NIH Guide for the Care and Use of Laboratory Animals (Eighth Edition). All animal experiments were approved by the Institutional Animal Care and Use Committee of the University of Alabama at Birmingham. Mice were housed in standard cages with water and food available ad libitum.
Mice carrying a null Trp53 allele (Trp53+/− mice) on a C57BL/6 background were purchased from Taconic (TSG-p53 #P53N4; Germantown, NY, USA). Nf1+/− mice carrying a null allele with a targeted mutation of exon 31 [3] on a C57BL/6 background were obtained from the National Cancer Institute Mouse Repository (stock #01XF3; Frederick, MD). We have previously described the generation and characterization of P0-GGFβ3 mice [16,20]. The P0-GGFβ3 mice used in this study were bred for over 15 generations onto a C57BL/6J background.
Immunoblot Analyses
Tissue and cells were homogenized in HES buffer (40 mM HEPES, 2 mM EDTA, 500 mM sucrose) supplemented with protease (Sigma #P8340) and phosphatase (Sigma #P5726) inhibitor cocktails at a 1:100 ratio. Protein concentrations were determined using a modified Lowry method (DC Protein Assay; Bio-Rad, Hercules, CA). Equivalent quantities of protein lysates were resolved on 8% SDS-polyacrylamide gels, immunoblotted per our previously described protocol [43] and probed. Immunoreactive species were detected by enhanced chemiluminescence (Thermo Scientific Pierce; Rockford, IL). As a loading control, membranes were reprobed with anti-GAPDH antibody (1:20,000 dilution).
Ras Activation Assays
Ras activation assays were performed as per our previously described protocol [20]. Activated Ras proteins pulled down with Raf-1 Ras-binding domain agarose beads were resolved on 12% SDS-PAGE gels, immunoblotted as described above and probed with a pan-Ras antibody that recognizes H-Ras, N-Ras and K-Ras.
Generation of Mouse Cohorts and Necropsies
To establish the cohorts of mice needed to examine a potential interaction between NRG1 overexpression and Trp53 haploinsufficiency, P0-GGFβ3 mice were mated with Trp53+/− mice. The resulting offspring were screened by PCR using genomic DNA isolated from tail snips. Mice carrying the P0-GGFβ3 transgene were identified with our previously described primers [16]. Mice carrying wild-type and mutant Trp53 alleles were identified with the following primers: KO forward (5’-GTGGGAGGGACAAAAGTTCGAGGCC-3’), KO reverse (5’-TTTACGGAGCCCTGGCGCTCGATGT-3’), WT forward (5’-GTGTTTCATTAGTTCCCCACCTTGAC-3’), WT reverse (5’-AGTGGAGGCTGCCAGTCCTAACCC-3’).
P0-GGFβ3 mice were also mated with Nf1+/− mice. Progeny with the P0-GGFβ3 transgene were identified as described above. The presence of wild-type and mutant Nf1 alleles were assessed using the following PCR primers: WT forward (5’-GTATTGAATTGAAGCACCTTTGTTTGG-3’), WT and KO reverse (5’-CTGCCAAGGCTCCCCCAG-3’), KO forward (5’-GCGTGTTCGAATTCGCCAATG-3’).
Cohorts were followed until death or for one year, whichever occurred first; mice which died from issues unrelated to the study (e.g., fighting with other animals) were excluded from the final cohort. All animals were examined daily for evidence of tumors. Endpoints included the discovery of a tumor greater than 1 cm in diameter, hindlimb weakness, lack of normal grooming, abnormal posture or weight loss of 20% compared to non-transgenic animals. Moribund mice or mice with findings suggestive of tumor occurrence were anesthesized with isoflurane and then decapitated. A portion of all tumors identified was used to establish early passage cultures as described below. After removing a small portion that was fixed in glutaraldehyde for possible electron microscopy, the remainder of the tumor and the body was fixed in 4% paraformaldehyde. Complete necropsies were then performed which included a careful examination of the central and peripheral nervous systems for peripheral nerve sheath tumors.
Identification and Diagnosis of Tumors
To facilitate the examination of spinal cords and spinal nerve roots, the vertebral columns from each animal, together with adjacent soft tissue and ribs, were decalcified by immersion in 0.3M EDTA/4% paraformaldehyde (pH 8.0) for 48–72 hours at 4°C. Prior to processing and embedding, fixed tissues were rinsed with PBS. Peripheral nerves were embedded in Histogel (Thermo Scientific HG-4000-12). Tissues were dehydrated by transfers through graded ethanols and xylenes and paraffin-embedded. 5 µm sections were cut, deparaffinized and hematoxylin and eosin (H&E) stained for the initial identification of tumors. All tumor diagnoses were performed by a practicing neuropathologist (S.L.C.) certified by the American Board of Pathology in both Anatomic Pathology and Neuropathology. World Health Organization (WHO) diagnostic criteria [36], as originally proposed by Scheithauer and Woodruff [37], were used for the diagnosis and grading of all identified tumors. In brief, WHO grade II MPNSTs are composed of cells with nuclei >3-fold larger than typical neurofibroma nuclei and demonstrate nuclear hyperchromasia and hypercellularity [37]; these features are assessed independent of mitotic activity, although the presence of mitotic figures provides further support for the diagnosis. WHO grade III MPNSTs show clear cytologic atypia, brisk mitotic activity (4 or more mitoses per 10 high power fields) and prominent hypercellularity. WHO grade IV MPNSTs have all of the features of WHO grade III MPNSTs plus tumor necrosis. We recognize that other investigators have graded mouse peripheral nerve sheath tumors (PNSTs) using an approach described by Stemmer-Rachamimov et al [42]. Under this system, all of the tumors described in our P0-GGFβ3; Trp53+/− model would be classified as genetically engineered mouse (GEM) grade III PNSTs. However, the GEM grading scheme avoids the term “malignant” as at the time it was developed, insufficient evidence was available regarding the clinical behavior of the GEM PNSTs that were being evaluated. Given the highly aggressive behavior of the MPNSTs in our model system, it is clear that these tumors are malignant. This, combined with the need for a grading system that clearly distinguished low, intermediate and high grade MPNSTs, is why we chose to use WHO grading rather than the GEM grading system.
For immunohistochemical assessment of differentiation marker expression, deparaffinized tumor sections were incubated with primary antibodies recognizing S100β (1:600 dilution), desmin (1:50 dilution) or neurofilaments (1:1000), followed by a FITC- or Cy3-conjugated donkey anti-rabbit secondary antibody (Jackson ImmunoResearch). Some tissues were stained with nestin primary antibody (1:1000 dilution for both mouse and human), followed by an ImmPRESS secondary antibody and Cy3 tyramide signal amplification step. Tissue was counterstained with bisbenzamide (0.2mg/mL; Sigma, St. Louis, MO) and images were acquired with a Zeiss (Thornwood, NY) Axioskop fluorescence microscope using AxioVision Rel. 4.8 software (Carl Zeiss Microimaging).
Unna’s methylene blue staining for mast cells was performed per previously published methods [27].
Ki67 and TUNEL Labeling Indices
For Ki67 labeling, antigen retrieval was performed by heating the slides in citrate buffer (9 mL 0.1 M citric acid, 41 mL 0.1 M sodium citrate, 450 mL distilled water) for 20 minutes in a rice cooker and then letting them cool to room temperature. Slides were quenched in 3% fresh hydrogen peroxide in PBS for 10 minutes. Next, they were rinsed in PBS and then incubated in PBS-blocking buffer (PBS-BB; 5% bovine serum albumin in PBS with 1% Tween-20) at room temperature for 1 hour. Sections were incubated with Ki67 antibody diluted 1:25 in PBS-BB overnight at 4°C in a humidity chamber. After three PBS rinses, biotinylated goat anti-mouse IgG (1:250 dilution) was added to slides for 1 hour at room temperature. The sections were then rinsed with PBS and incubated in pre-diluted streptavidin-HRP (cat. # 550946; BD Biosciences) per the manufacturer’s recommendations. TUNEL labeling was performed using the Apoptag Peroxidase In Situ Apoptosis Detection Kit (Millipore), per the manufacturer’s instructions. Ki67 and TUNEL labeled slides were counterstained with hematoxylin, dehydrated through graded alcohols and coverslipped using Cytoseal Mounting Medium (Thomas Scientific; Swedesboro, NJ). Ki67 and TUNEL labeling indices were determined as previously described [4]. At least twelve 40X fields were counted per tumor, with a minimum of two sections per tumor counted.
Establishment of Early Passage Tumor Cultures
To establish early passage tumor cultures, tumor tissue was transferred into Hanks’ balanced salt solution, minced and then incubated in Dulbecco’s minimal essential medium supplemented with 10% fetal calf serum (DMEM10), 2 µM forskolin and 50 nM NRG1β in T25 flasks. Tumor cells were allowed to grow out from explants until confluent. Cultures were then maintained for no more than 5–9 passages in DMEM10 that was not supplemented with forskolin or NRG1β. All cultures were maintained at 37°C in a 5% CO2 atmosphere. RNA isolated from these cultures was reverse transcribed and RT-PCR performed using our previously described panel of PCR primers for key diagnostic and differentiation markers [20]. Schwann cell cultures obtained from C57BL6 wild-type (wt) and transgenic (tg) sciatic nerves were established using the same method, but were continuously treated with NRG1β and forskolin.
qPCR
Real-time quantitative PCR (qPCR) was performed utilizing our previously described protocol [20] with an Applied Biosystems Step One Plus real time PCR system (Life Technologies, Foster City, CA). Assays of Trp53 transcripts were performed using FAM-labeled TaqMan MGB probes (ABI assay Mm00441964_g1). As a control, cDNA encoding18S ribosomal RNA levels were assayed with VIC-labeled Taqman MGB probes (ABI 4319413E). Samples were run in triplicate and test cDNA levels were normalized to 18S rRNA cDNA from the corresponding specimen. Data were analyzed using the Step One Software version 2.2.2.
Copy Number Assays
Taqman Copy Number Assays (Life Technologies) for Trp53 (Mm00370088_cn) were performed as recommended by the manufacturer using genomic DNA isolated from early passage MPNST cultures or C57BL/6J Schwann cell cultures. Mouse Tfrc (part #4458366) was used as the reference assay for these experiments. Data obtained in these experiments were analyzed using CopyCaller Software (Applied Biosystems, Life Technologies).
Nested PCR to identify Trp53 mutations
Total cellular RNA was extracted from early passage tumor cultures with TRIzol per the manufacturer’s recommendations (Invitrogen, Carlsbad, CA). This RNA was used to synthesize cDNA via priming with oligo-dT. Nested PCR to amplify Trp53 sequences was then performed as previously described [20]. Final PCR products were resolved on 1% agarose gels, purified using a Gene Clean Spin Kit (MP Biomedicals, Santa Ana, CA) per manufacturer’s recommendations and sequenced via Sanger sequencing.
Array Comparative Genome Hybridization
A QIAmp DNA mini kit (Qiagen; Valencia, CA) was used as recommended by the manufacturer to isolate genomic DNA from early passage P0-GGFβ3; Trp53+/− MPNST cells and cultured C57BL/6J wild-type Schwann cells. Whole genome array CGH was performed using mouse 4×44K oligonucleotide arrays (Agilent Technologies; Santa Clara, CA); overall median probe spacing in these arrays is approximately 22 kb, with median probe spacing in RefSeq genes being approximately 13 kb. Slide hybridization, washing, scanning and data analysis were performed per our previous described methodology [20]. Genomic breakpoints were mapped using mouse genome build NCBI37/mm9.
Results
Mice Carrying the P0-GGFβ3 Transgene on a C57BL/6J Background Maintain Transgene Expression and Schwann Cell Hyperplasia but Fail to Develop Neurofibromas or MPNSTs
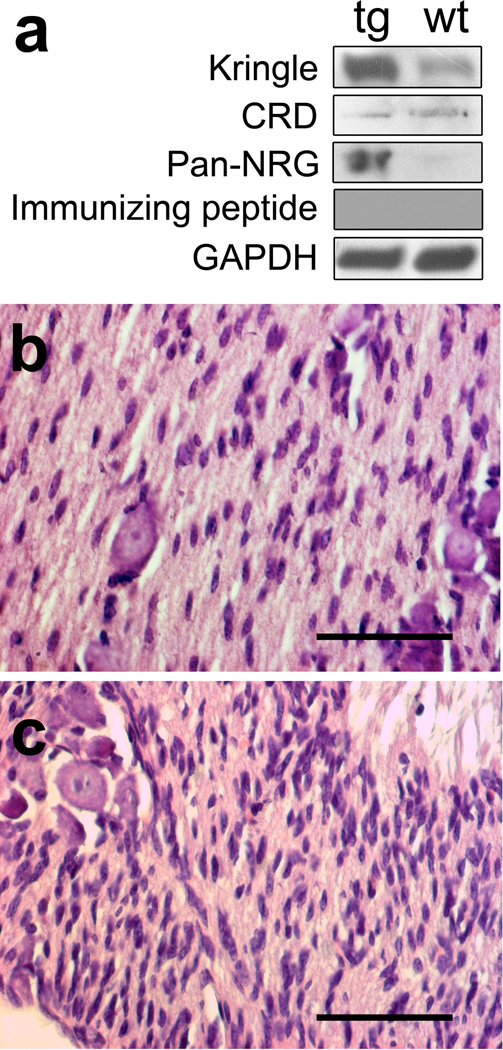
We have previously reported that P0-GGFβ3 mice have a shortened lifespan (average survival, 262 days), develop both neurofibromas and MPNSTs at a high frequency on an outbred C57BL/6J x SJL/J background or when bred onto a C57BL/6J background for 5–8 generations and recapitulate the process of neurofibroma-MPNST progression seen in human NF1 patients [16,20]. However, when we bred P0-GGFβ3 mice on a C57BL/6J background for 15 or more generations (referred to below as an inbred C57BL/6J background), we found that they survived without obvious abnormalities to 1 year of age and no longer developed tumors (see below). To determine why tumorigenesis was lost in mice carrying the P0-GGFβ3 transgene on an inbred C57BL/6J background, we first compared NRG1 expression in the trigeminal nerves of wild-type C57BL/6J mice to that in P0-GGFβ3 mice at 1 month of age, a time which represents the peak of Schwann cell hyperplasia in P0-GGFβ3 mice [16]. Lysates of these nerves were immunoblotted and probed with antibodies recognizing either the kringle domain located at the N-terminus of type II (GGF) NRG1 isoforms, the CRD domain found only in type III NRG1 isoforms or the EGF-like common domain present in all NRG1 proteins (a pan-NRG1 antibody). The anti-CRD domain antibody showed similar levels of type III NRG1 protein in wild-type and P0-GGFβ3 trigeminal nerve (Fig. 1a). In contrast, both the anti-kringle domain and the pan-NRG1 antibodies demonstrated higher levels of expression of GGFβ3 in transgenic animals compared to wild-type controls. Preincubating the anti-kringle domain antibody with immunizing peptide abolished the GGF signal, confirming the specificity of this immunoreactive species. Thus, GGF expression in P0-GGFβ3 trigeminal nerve was elevated compared to wild-type nerve, in keeping with our previous demonstration that GGF mRNA expression is increased in the nerves of P0-GGFβ3 mice carrying the transgene on an outbred C57BL/6J x SJL/J background [16].
Fig. 1.
Transgene expression and Schwann cell hyperplasia is maintained in P0-GGFβ3 mice bred for >15 generations onto a C57BL/6J background. a: Immunoblot analyses of the levels of type II NRG1 protein detected with an anti-kringle domain antibody (Kringle), type III NRG1 protein detected with an anti-CRD domain antibody (CRD) and global NRG1 expression as detected with an antibody recognizing the EGF-like domain present in all biologically active NRG1 isoforms (pan NRG). Incubation of with an immunizing peptide ablates the signal from the anti-kringle domain antibody (Immunizing peptide). To verify equal protein loading, the blots were reprobed with an anti-GAPDH antibody (GAPDH). b: Hematoxylin and eosin stained preparations of trigeminal nerve from 1 month old wild-type and c: P0-GGFβ3 mice. Note the increased cellularity (hyperplasia) in the P0-GGFβ3 nerve relative to the wild-type nerve. Scale bars, 50µm.
Another possible explanation for the differences in tumorigenesis on these different genetic backgrounds is that C57BL/6J and SJL/J mice express different levels of Trp53, as higher levels of this tumor suppressor have been linked to decreased tumor susceptibility in mice [13]. We tested this possibility by performing real-time qPCR on Schwann cell cultures derived from the sciatic nerves of C57BL/6J and SJL/J mice. However, surprisingly we found that Schwann cells derived from the tumor resistant C57BL/6J strain actually had much (approximately 25-fold) lower expression of Trp53 transcripts that was evident in SJL/J Schwann cells (Supplemental Figure 1). Thus, differences in Trp53 expression are unlikely to account for the discrepancy in tumorigenesis between mice carrying the P0-GGFβ3 transgene on a C57BL/6J x SJL/J background and inbred C57BL/6J P0-GGFβ3 mice.
In mice carrying the P0-GGFβ3 transgene on an outbred C57BL/6J x SJL/J background, tumorigenesis is preceded by Schwann cell hyperplasia in nerves and peripheral ganglia throughout the body. To determine whether Schwann cell hyperplasia still occurred in P0-GGFβ3 mice on an inbred C57BL/6J background, we compared the histology of sciatic and trigeminal nerves collected from 1 month old wild-type C57BL/6J mice to that of the same nerves from P0-GGFβ3 mice. We found that Schwann cell hyperplasia was indeed still present in nerves from P0-GGFβ3 mice (Fig. 1b, c). Thus, mice carrying the P0-GGFβ3 transgene on an inbred C57BL/6J background maintain transgene expression at levels similar to what is seen on an outbred C57BL/6J x SJL/J background and still develop Schwann cell hyperplasia. However, on an inbred C57BL/6J genetic background, these preneoplastic peripheral nervous system abnormalities do not advance to tumorigenesis.
Overexpression of Type II NRG1 Interacts with Trp53, but not Nf1, Haploinsufficiency, to Promote MPNST Pathogenesis
NRG1 binding to its erbB receptors results in the simultaneous activation of Ras and multiple other signaling cascades [50]. At present, however, it is not clear whether the activation of Ras (the same event that is facilitated by NF1 loss) is the primary means by which NRG1 overexpression promotes PNS neoplasia or whether NRG1-mediated activation of other essential signaling cascade(s) is instead necessary for tumorigenesis. Our observation that tumorigenesis is suppressed in P0-GGFβ3 mice on an inbred C57BL/6J background gave us the opportunity to test these alternative hypotheses via genetic complementation. We reasoned that if NRG1 promotes tumorigenesis primarily by activating essential signaling cascades that parallel those affected by Nf1 loss, then crossing P0-GGFβ3 mice to Nf1+/− mice could potentially “unmask” tumorigenesis, resulting in the reappearance of neurofibromas and/or MPNSTs in these animals. If, on the other hand, the primary protumorigenic effect of NRG1 overexpression in Schwann cells was modulation of Nf1-regulated signaling cascades, then NRG1 overexpression should substitute for Nf1 loss. In this circumstance, the phenotype of P0-GGFβ3;Trp53+/− mice would be analogous to that of cis-Nf1+/−;Trp53+/− mice which develop MPNSTs de novo rather than from a pre-existing neurofibroma [6,45].
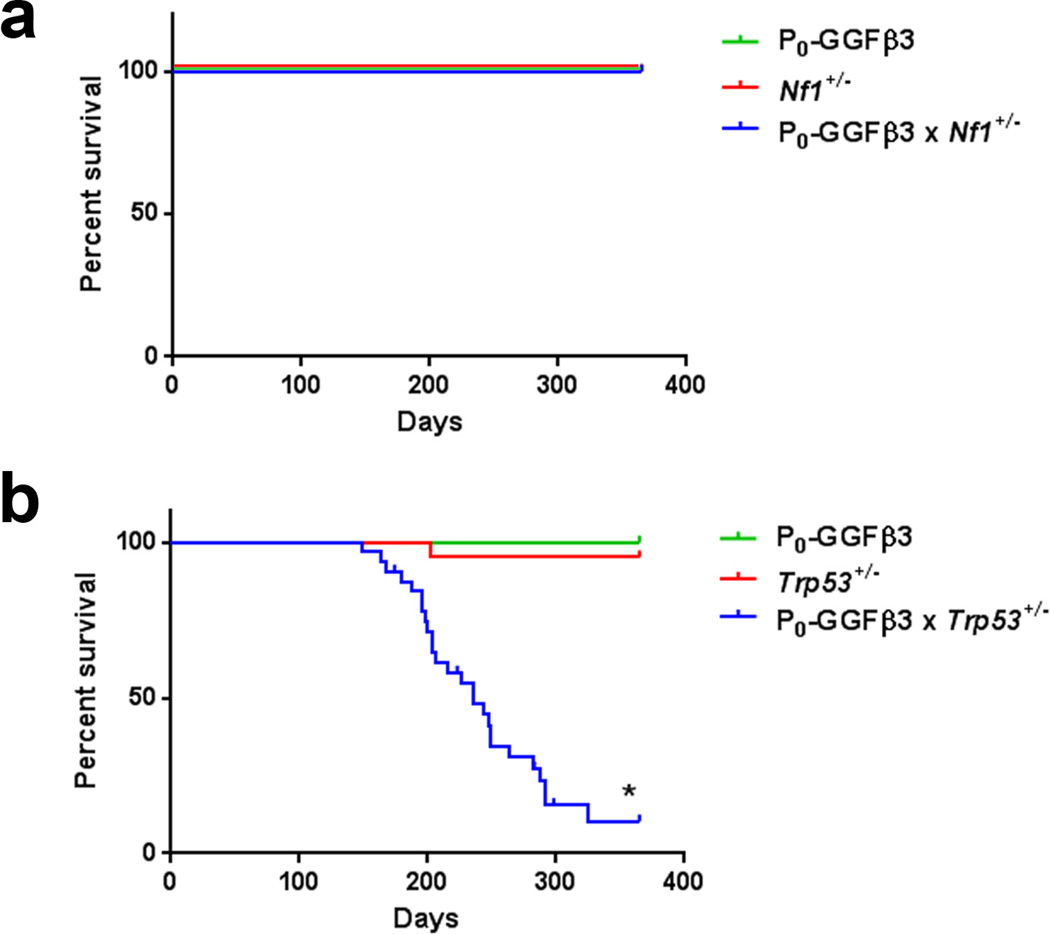
To test these alternative possibilities, we bred mice carrying the P0-GGFβ3 transgene on an inbred C57BL/6J background to animals with the same genetic background that were haploinsufficient for either Trp53 (Trp53+/− mice) or Nf1 (Nf1+/− mice). Cohorts of P0-GGFβ3, Nf1+/−, Trp53+/−, P0-GGFβ3;Nf1+/− and P0-GGFβ3; Trp53+/− mice (20 mice of each genotype, with each cohort containing an equal number of males and females) were then followed until death or one year of age. We found that P0-GGFβ3, Nf1+/−, and P0-GGFβ3; Nf1+/− mice showed no reduction in survival over the first year of life (Fig. 2a). The survival of the Trp53+/− cohort was only slightly and non-significantly lowered due to the death of a single animal (average survival, 357 versus 365 days; Fig. 2b). Strikingly, however, we did observe a significant decrease when comparing the Kaplan-Meier survival curves of P0-GGFβ3; Trp53+/− mice (average survival, 226 days) to those of P0-GGFβ3 and Trp53+/− mice (Fig. 2b; *, p< 0.0001 based on a log-rank test). Indeed, only one P0-GGFβ3; Trp53+/− mouse survived to 1 year of age. When comparing male and female mice within the P0-GGFβ3; Trp53+/− cohort, no significant difference in survival was observed (data not shown).
Fig. 2.
Haploinsufficiency for Trp53, but not Nf1, impairs survival in the presence of NRG 1 overexpression. a: Kaplan-Meier curve indicating the survival rates of P0-GGFβ3, Nf1+/− and P0-GGFβ3;Nf1+/− mice over the first year of life. No decrease in survival is seen for any of these cohorts. b: Kaplan-Meier curve indicating the survival rates of P0-GGFβ3, Trp53+/− and P0-GGFβ3; Trp53+/− mice over the first year of life. While the survival of P0-GGFβ3 and Trp53+/− mice is unimpaired over this interval, the survival of P0-GGFβ3; Trp53+/− mice is significantly shortened (average survival, 226 days). *, p-value<0.0001.
To establish the cause of death in P0-GGFβ3; Trp53+/- mice, we performed complete necropsies, including a detailed microscopic examination of all tissues, on each of the cohorts described above. There was no evidence of neurofibromas, MPNSTs or any other tumor type in any of the P0-GGFβ3, Nf1+/−, or P0-GGFβ3;Nf1+/− mice we examined. Likewise, there was no evidence of neoplasia in any of the Trp53+/− mice except for the single animal that died early; we found a large fibrosarcoma in that mouse. In contrast, 95% (18/19) of the mice in the P0-GGFβ3;Trp53+/− cohort had peripheral nervous system neoplasms (Table 1). The majority of these tumors were associated with trigeminal nerves (11/19; 58%, see Fig. 3a) or dorsal spinal nerve roots (13/19; 68%), with a smaller number of neoplasms identified in the sciatic nerve (2/19; 11%). Within the P0-GGFβ3;Trp53+/− cohort, many of the tumor bearing animals presented with a single tumor. However, 53% (10/19) of the mice had multiple tumors, with a maximum of five neoplasms observed in an individual animal.
Table 1.
Distribution of MPNSTs in P0-GGFβ3;Trp53+/− Mice
| Location | Percentage |
|---|---|
| Mice with tumors at any location | 95% |
| Trigeminal tumors | 58% |
| Spinal dorsal nerve root tumors | 68% |
| Sciatic nerve tumors | 11% |
| Multiple tumors | 53% |
Fig. 3.
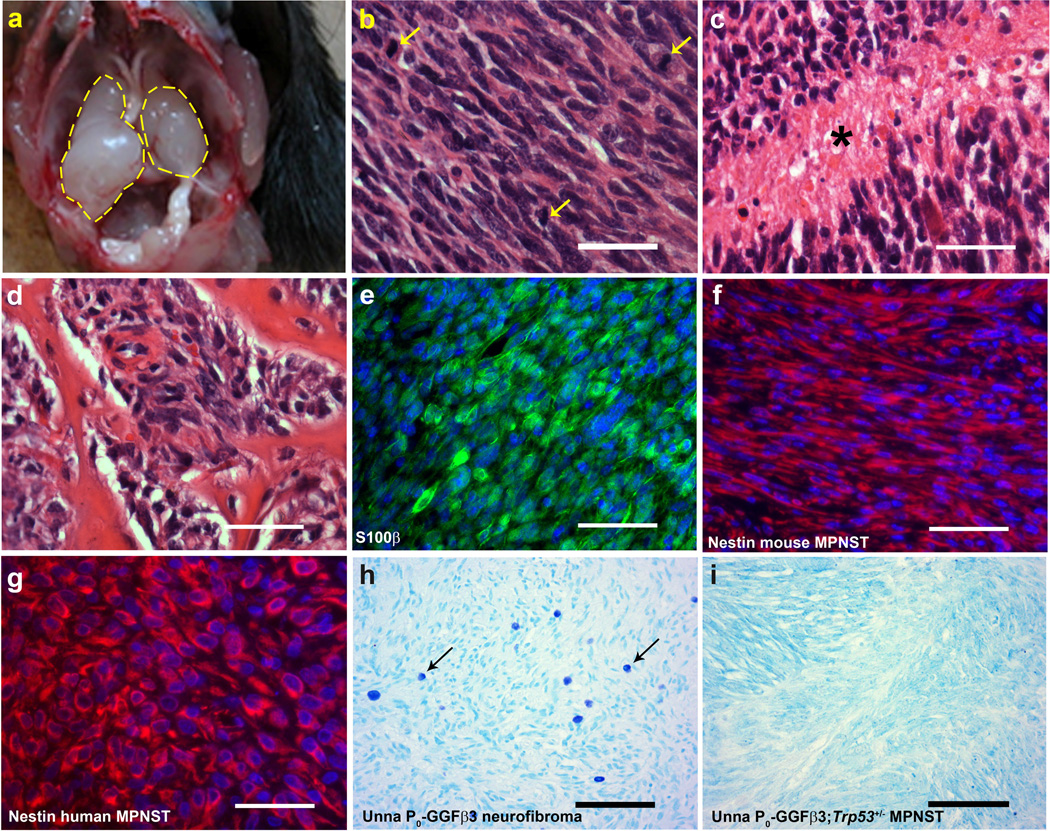
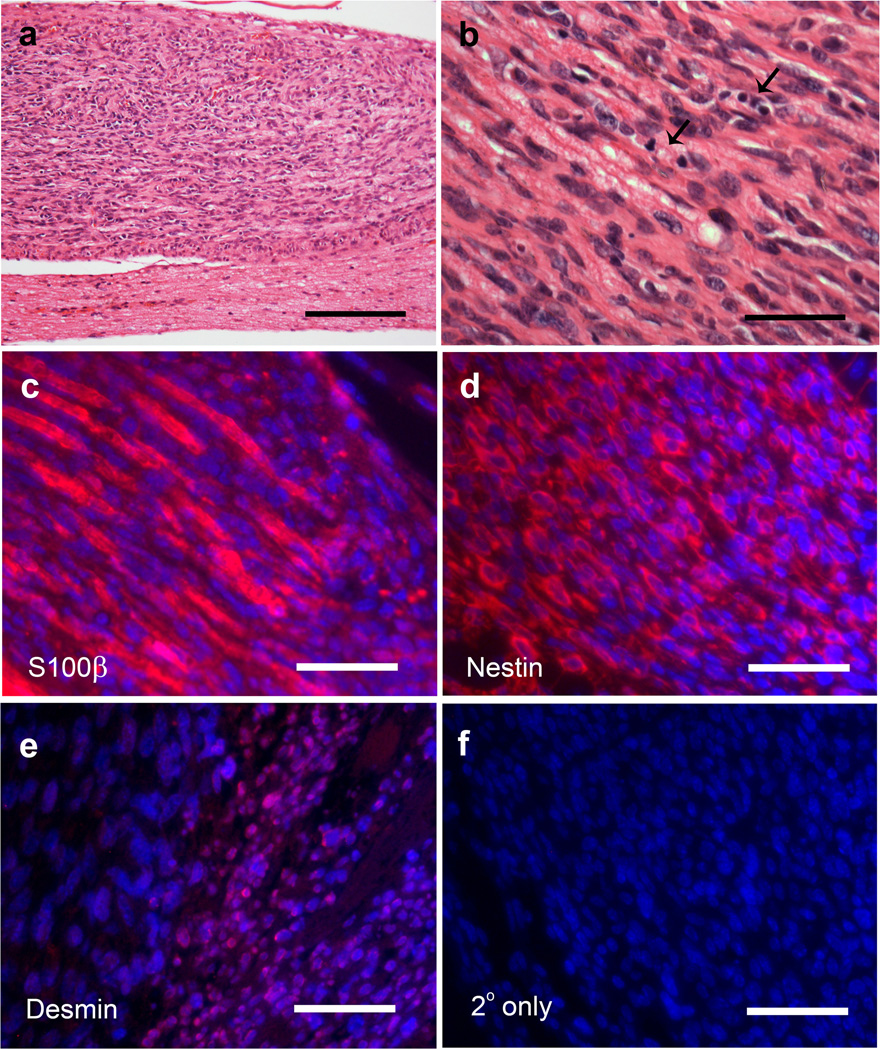
P0-GGFβ3; Trp53+/− mice develop MPNSTs, but not neurofibromas. a: In situ gross image of two independently arising trigeminal tumors. b–d: Hematoxylin and eosin stained sections of MPNSTs arising in P0-GGFβ3; Trp53+/− mice demonstrating the brisk mitotic activity (b, arrows) and tumor necrosis (c, asterisk) typically seen in these tumors. These tumors were highly aggressive, as demonstrated by their tendency to invade bone (d) and other adjacent structures. e: S100β immunoreactivity in an MPNST found in a P0-GGFβ3; Trp53+/− mouse. f–g: Nestin immunoreactivity in a P0-GGFβ3; Trp53+/− MPNST (f) compared to that seen in a human MPNST (g). h–i: Neurofibromas from P0-GGFβ3 parent line contain mast cells (h), while these cells are absent in MPNSTs from the P0-GGFβ3; Trp53+/− mice(i). Scale bars, 50nm (b–g) and 100µm (h and i).
Since Trp53+/− mice can potentially develop lymphomas and other types of sarcomas [8], we performed a pathologic examination of the tumors arising in P0-GGFβ3; Trp53+/− mice to establish whether these lesions were peripheral nerve sheath tumors or some other tumor type. We found that all of the tumors arising in P0-GGFβ3; Trp53+/− mice had a similar histologic appearance. These markedly hypercellular neoplasms lacked the mixture of multiple cell types characteristic of neurofibromas, instead being uniformly composed of closely packed, atypical spindled cells which contained enlarged elongated nuclei with coarse chromatin (Fig. 3b). Brisk mitotic activity was evident in the tumors (Fig. 3b, arrows) and tumor necrosis was commonly seen (Fig. 3c, asterisk). These neoplasms were also highly aggressive, often invading adjacent bone (Fig. 3d) and soft tissues. The tumors found in P0-GGFβ3; Trp53+/− mice were immunoreactive for S100β (Fig. 3e), consistent with an origin from the Schwann cell lineage. As immunoreactivity for both S100β and nestin distinguishes MPNSTs from other histologically similar sarcomas [31,39], we also examined nestin expression in P0-GGFβ3; Trp53+/− tumors. We found that the mouse tumors (Fig. 3f), like human MPNSTs (Figure 3g), were strongly nestin positive. Further, unlike the neurofibromas isolated from outbred P0-GGFβ3 mice (Fig. 3h), the tumors in P0-GGFβ3; Trp53+/− mice lacked mast cell infiltration (Fig. 3i) which is consistent with our identification of these tumors as MPNSTs.
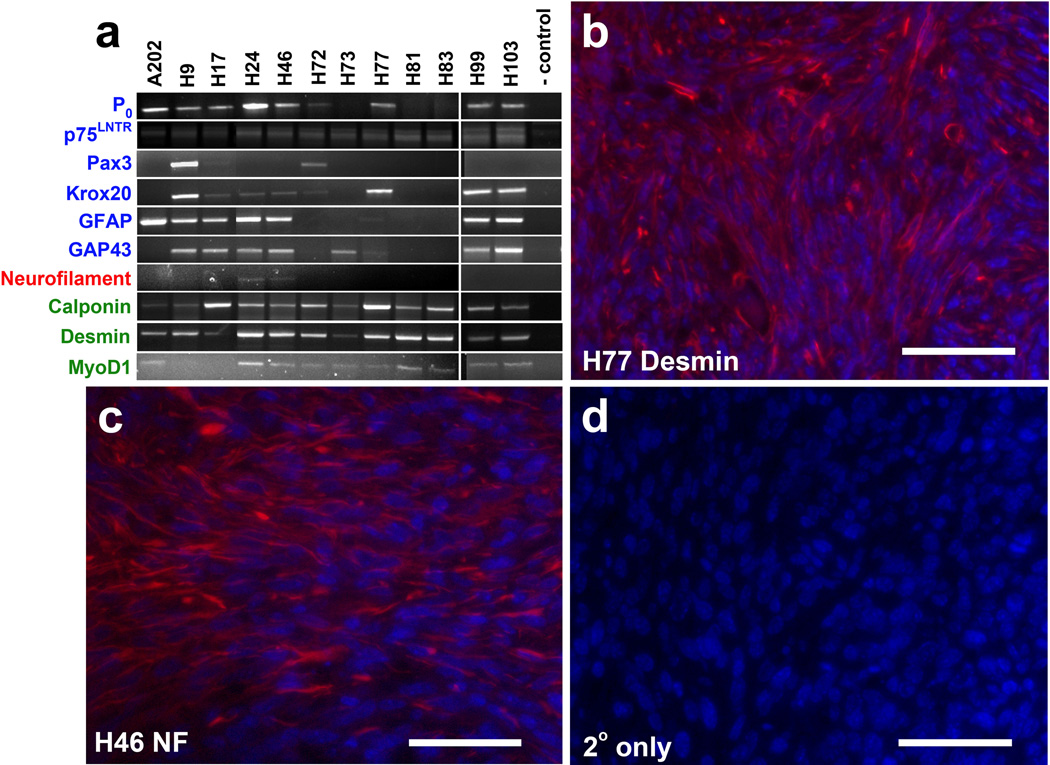
A subset of human MPNSTs (e.g., the group of neoplasms referred to as Triton tumors) are capable of divergent differentiation, which is demonstrable by the expression of muscle or neural markers. A similar potential for divergent differentiation has also been observed in the MPNSTs that develop in cis-Nf1+/−;Trp53+/− mice [45] and in the MPNSTs that arise in mice carrying the P0-GGFβ3 transgene on an outbred background [20]. To determine whether this feature was also evident in P0-GGFβ3; Trp53+/− MPNSTs, we used reverse transcription-PCR to examine the expression of a panel of Schwann cell, neuronal and muscle markers in early passage cultures established from twelve independently arising tumors (Figure 4a). As expected for MPNSTs, multiple transcripts encoding Schwann cell markers (P0, p75LNTR, Pax3, Krox20, glial fibrillary acidic protein and GAP43) were expressed to varying degrees in the P0-GGFβ3;Trp53+/− MPNST cultures (Fig. 4a). Expression of light (68 kDa) neurofilament mRNA, a marker we have previously detected in a small subset of P0-GGFβ3 MPNSTs, was similarly present in a restricted subset (two of twelve cultures) of P0-GGFβ3; Trp53+/− MPNSTs. In contrast, transcripts encoding muscle markers were commonly detected in P0-GGFβ3; Trp53+/− MPNSTs, with desmin and calponin mRNAs present in the overwhelming majority of the tumors. Interestingly, MyoD transcripts were also widely expressed in P0-GGFβ3; Trp53+/− MPNSTs—this marker was present in a relatively small subset of P0-GGFβ3 MPNSTs (3/18 tumors) in our earlier study [20]. To verify that the expression of neuronal and muscle markers in P0-GGFβ3; Trp53+/− MPNST cultures was not a culture artifact, we immunostained sections of the tumors from which the cultures were derived for representative markers of each class. Immunoreactivity for both desmin (Fig. 4b) and neurofilaments (Fig. 4c) was evident in these tumors, confirming that our observations in the early passage cultures accurately reflected the immunophenotype of the parent tumors. Considered collectively, the histologic features and immunophenotype of the tumors arising in P0-GGFβ3; Trp53+/− mice are thus highly similar to that of human MPNSTs, the MPNSTs found in cis-Nf1+/−;Trp53+/− mice and the MPNSTs we have previously described in outbred P0-GGFβ3 mice [16,20]. We conclude that NRG1 overexpression interacts with Trp53 haploinsufficiency, thereby promoting MPNST pathogenesis and shortening the lifespan of P0-GGFβ3; Trp53+/− mice. In contrast, we found no evidence that NRG1 overexpression interacted with decreased Nf1 gene dosage to promote tumorigenesis.
Fig. 4.
Early passage P0-GGFβ3; Trp53+/− MPNST cultures consistently express Schwann cell markers and variably express neuronal and muscle markers. a: RT-PCR of Schwann cell (blue), neuronal (red), and muscle (green) markers expressed in passage 8–9 cultures from P0-GGFβ3; Trp53+/− MPNSTs. b–c: Immunostains validating in vivo expression of desmin and neurofilaments in mouse tumors H77 and H46, respectively. d: Cy3-secondary only control. Scale bars, 50µm.
MPNSTs in P0-GGFβ3;Trp53+/− Mice Arise De Novo Rather than from a Neurofibroma Precursor
Although MPNSTs were common in P0-GGFβ3;Trp53+/− mice, neurofibromas were extraordinarily rare in these animals; indeed, we found only a single neurofibroma in one mouse within this cohort. This is quite different from outbred P0-GGFβ3 mice, where we identified multiple neurofibromas in the dorsal spinal nerve roots, trigeminal nerves and sympathetic nervous system of almost all of the mice we examined [20]. The fact that we did not find large numbers of neurofibromas in P0-GGFβ3; Trp53+/− mice suggested that the MPNSTs occurring in these animals arose de novo rather than from pre-existing plexiform neurofibromas. As the trigeminal nerves and dorsal spinal nerve roots were the most common sites where we found MPNSTs in P0-GGFβ3; Trp53+/− mice, we carefully examined these nerves and their associated ganglia to determine whether small nascent tumors were present within these structures. We found that 47% of our (9/19) P0-GGFβ3; Trp53+/− mice had one or more microtumors in their trigeminal nerves or dorsal spinal nerve roots. Strikingly, these microtumors were uniformly located within the trigeminal or dorsal root ganglia (Fig. 5a). The microtumors had a histologic appearance similar to that of the larger MPNSTs described above, being evident as moderately hypercellular proliferations of atypical spindled cells with enlarged hyperchromatic nuclei (Fig. 5b). As in the larger MPNSTs, mitotic activity (Fig. 5b, arrows) was readily identified in the microtumors. The identity of these lesions was confirmed by their immunoreactivity for S100β (Fig. 5c) and nestin (Fig. 5d). Like the major tumors, the microtumors also demonstrated focal desmin immunoreactivity (Fig. 5e) which was abolished when primary antibody was not included (Fig. 5f). In addition, Unna stains showed no evidence of mast cells (data not shown). Given the pathologic features of the microtumors and the virtually complete absence of neurofibromas in P0-GGFβ3; Trp53+/− mice, we conclude that MPNSTs in P0 GGFβ3; Trp53+/− mice, like those in cis-Nf1+/−;Trp53+/− mice [45], arise de novo rather than from a neurofibroma precursor.
Fig. 5.
MPNSTs occurring in P0-GGFβ3; Trp53+/− mice develop within peripheral nervous system ganglia. a: A markedly hypercellular microtumor developing within the trigeminal ganglion of a P0-GGFβ3; Trp53+/− mouse. Scale bar, 100µm. b: Higher power view of the tumor shown in A. Note the increased nuclear size and atypia within this lesion and the frequent occurrence of mitotic figures (arrows). c: Immunostain demonstrating S100β immunoreactivity in the microtumor cells and Schwann cells associated with entrapped axons in the lesion. d: Like the major MPNSTs, the microtumors also show nestin immunoreactivity. e: Desmin immunoreactivity is also seen focally in the microtumors. f: Cy3-secondary only control. Scale bars in B–D, 50µm.
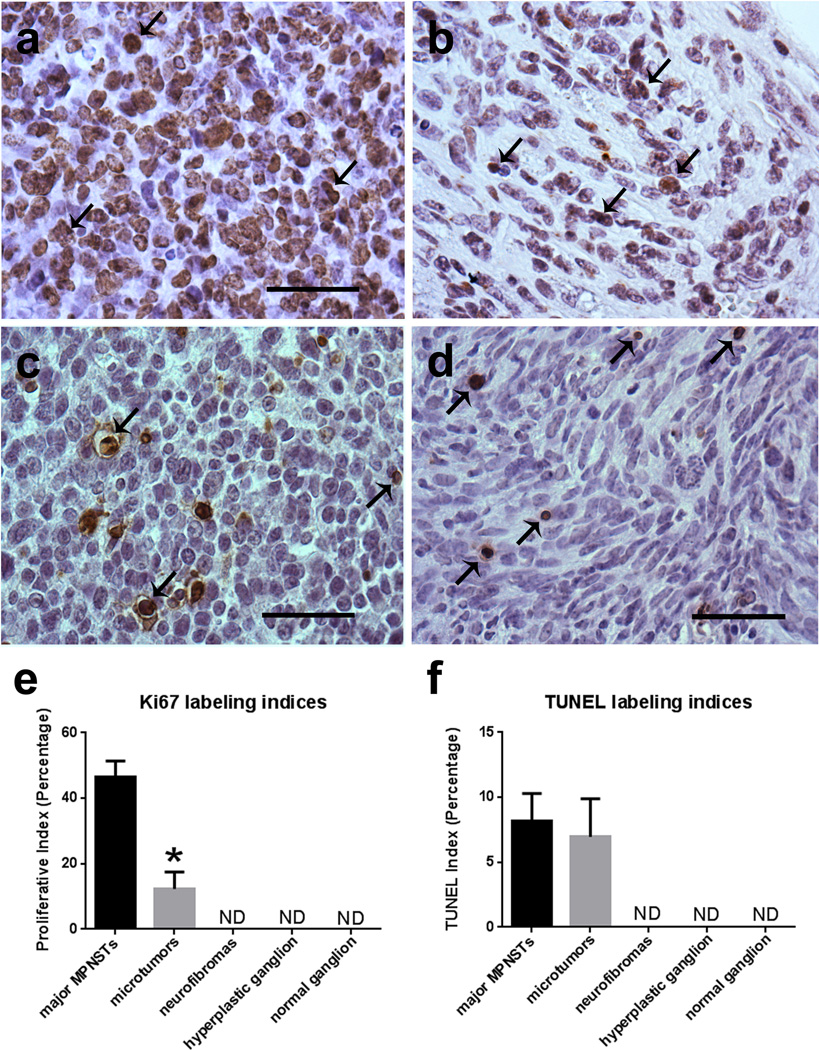
Although the microtumors we identified in P0-GGFβ3; Trp53+/− mice clearly had the pathologic features of MPNSTs, we also noted that the cellularity and frequency of mitotic figures appeared to be lower in the microtumors than in the larger MPNSTs found in these animals. To determine whether the microtumors were lower grade MPNSTs and to clearly distinguish them from neurofibromas, we quantified expression of the Ki67 proliferation marker and performed TUNEL labeling in three major MPNSTs and three microtumors derived from P0-GGFβ3;Trp53+/− mice, three neurofibromas identified in outbred P0-GGFβ3 mice, three hyperplastic trigeminal ganglia from 1 month old P0-GGFβ3 mice and three normal trigeminal ganglia from 1 month old C57BL/6J wild-type mice. We found extensive nuclear Ki67 immunoreactivity in the major MPNSTs (Fig. 6a), with Ki67 labeling indices that averaged 46.4% (Fig. 6e). Ki67 labeling was also readily detected in the microtumors (Fig. 6b). However, the Ki67 labeling in the microtumors (average labeling index, 12.3%) was significantly less than we observed in the major MPNSTs (Fig. 6e). On the other hand, Ki67 labeling in the microtumors was significantly higher than that in neurofibromas, hyperplastic transgenic ganglia or wild-type ganglia; indeed, Ki67 labeling was difficult to detect in these three latter types of specimens. These latter indices are consistent with those observed in human neurofibromas, which have labeling indices reported as either undetectable or around 1–2% [23,52]. TUNEL labeling was also readily detected in the major MPNSTs (Fig. 6c) and microtumors (Fig. 6d), demonstrating the occurrence of a baseline level of apoptosis. However, TUNEL labeling was not significantly increased in the major MPNSTs relative to the microtumors (Fig. 6F; Fig. 8.2% versus 7%). TUNEL labeling was not detectable in the neurofibromas, hyperplastic transgenic ganglia or wild-type ganglia.
Fig. 6.
Ki67 labeling indices in the microtumors occurring in P0-GGFβ3; Trp53+/− mice are higher than seen in neurofibromas and non-neoplastic ganglia but lower than is seen in the larger tumors present in these animals. a, b: Major MPNST (a) and micro-MPNST (b) stained for the proliferative marker Ki67 shown side-by-side for comparison. c, d: DNA fragmentation in major MPNSTs (c) and microtumors (d), as labeled via TUNEL. e: Quantification of Ki67 labeling in major MPNSTs, micro-MPNSTs (microtumors), neurofibromas, neoplastic ganglia, and nonneoplastic ganglia demonstrates a significant difference between major MPNSTs and micro-MPNSTs (p<0.0001; n=3 animals per specimen type). f: Quantification of TUNEL labeling in major MPNSTs and micro-MPNSTs, neurofibromas, neoplastic ganglia, and non-neoplastic ganglia. Scale bars in A–D, 50µm.
Fig. 8.
Array CGH ideograms demonstrate increasingly complex copy number variations in the tumors with increasing grade. Grade III tumors surveyed either clustered with WHO grade II tumors or WHO grade IV tumors in terms of complexity.
If the microtumors are the progenitor lesions that develop into the major MPNSTs, we would anticipate that the microtumors would tend to be lower grade malignancies while the major MPNSTs would predominantly be higher grade lesions. To evaluate this possibility, we graded all of the microtumors and the major MPNSTs identified in our P0-GGFβ3; Trp53+/− cohort using WHO criteria for the grading of human MPNSTs [36]. As shown in Supplemental Table 1, 59% (10/17) of the microtumors met diagnostic criteria for WHO grade II MPNSTs, while 41% (7/17) of the microtumors were classified as WHO grade III MPNSTs. None of the microtumors had the features of grade IV MPNSTs. In contrast, the majority of the major MPNSTs were high grade tumors, with 38% (8/21) of the major MPNSTs having the diagnostic features seen in human WHO grade IV MPNSTs, 52% (11/21) of these tumors being WHO grade III MPNSTs and only 10% (2/21) of the major MPNSTs (2/21) meeting WHO grade II diagnostic criteria. Considered collectively, the observations noted above suggest that MPNSTs initially develop de novo as low grade intraganglionic malignancies in P0-GGFβ3; Trp53+/− mice. As these low grade MPNSTs expand, they presumably accumulate additional genomic abnormalities, resulting in their progression to become high grade (WHO grade III-IV) MPNSTs.
P0-GGFβ3; Trp53+/− MPNSTs Demonstrate ErbB-Dependent Ras Hyperactivation While Remaining Trp53 Haploinsufficient
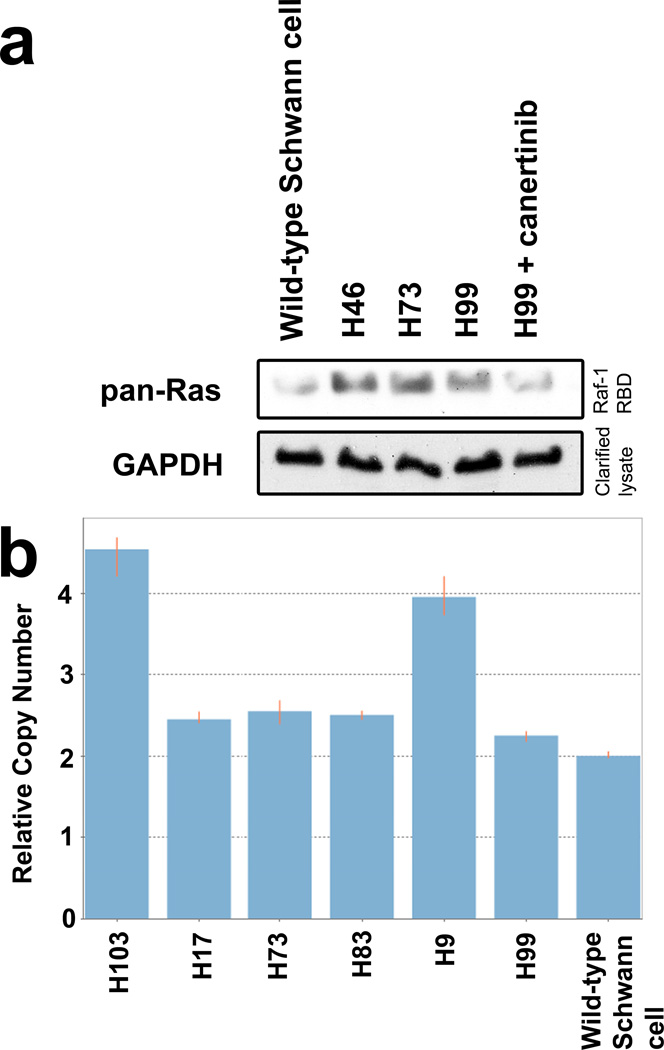
Our genetic complementation experiments demonstrated that NRG1 overexpression could substitute for Nf1 loss to promote de novo MPNST pathogenesis in a Trp53+/− genetic background, which suggests that NRG1 promotes tumorigenesis via its effects on the same signaling pathway that is affected by Nf1 loss. If this interpretation is correct, we would predict that NRG1 overexpression would induce biochemical abnormalities analogous to those resulting from Nf1 loss such as Ras hyperactivation. To test this, we compared the relative levels of activated Ras in non-neoplastic Schwann cells to that in three P0-GGFβ3; Trp53+/− MPNST early passage cultures. In these experiments, activated Ras protein was pulled down from cell lysates using a GST fusion protein containing the Raf-1 Ras binding domain (Raf-1 RBD; Fig. 7a), which only binds activated Ras. Proteins captured with the Raf-1 RBD were then immunoblotted and probed with a pan-Ras antibody. We found that the levels of activated Ras were increased in all three MPNST cultures relative to non-neoplastic Schwann cells. Further, treatment with the pan-erbB inhibitor, canertinib (CI-1033), reduced the levels of activated Ras to the baseline levels evident in non-neoplastic Schwann cells. Considered collectively, these findings demonstrate that NRG-1 signaling mediated by its erbB3 and/or erbB4 receptors, like Nf1 loss, induces Ras hyperactivation in P0-GGFβ3; Trp53+/− MPNSTs.
Fig. 7.
ErbB-dependent Ras hyperactivation and persistent Trp53 haploinsufficiency is evident in P0-GGFβ3;Trp53+/− MPNSTs. a: Ras activity is increased in the MPNSTs compared to Schwann cells and this activity is attenuated by treatment with the pan-erbB inhibitor canertinib. b: Copy number assays demonstrate that P0-GGFβ3; Trp53+/− MPNSTs retain a wild-type copy of Trp53.
Mutations of TP53 occur commonly in human MPNSTs [2,15,25,30,44]. However, it has also been noted that biallelic TP53 inactivation is rare in MPNSTs [26], which has led to the suggestion that TP53 haploinsufficiency may be sufficient for neurofibroma-MPNST progression [44]. To determine if P0-GGFβ3; Trp53+/− MPNSTs lose their remaining wild-type copy of Trp53 during malignant transformation, we performed copy number assays (CNAs) on genomic DNA isolated from six MPNSTs (H9, H73, H81, H83, H99, and H103). In these assays, a duplex real-time PCR reaction was performed which queried the genomic copy number for Trp53 and then normalized that value to that of a gene of known copy number (transferrin receptor c). Using this technique, we found that four of the six tumors retained their wild-type copy of Trp53 (Fig. 7b; two copies of Trp53 are evident in these tumors because the Trp53 knockout targets exon 5 whereas the CNA primers overlap the exon 1-intron 1 junction). The other two MPNSTs carried four copies of the Trp53 gene, which was confirmed via array comparative genome hybridization (aCGH, see below). Since it was possible that the non-targeted Trp53 gene in P0-GGFβ3; Trp53+/− MPNSTs might have focal (missense, nonsense or frameshift) mutations rather than gene deletion, we next performed nested PCR to amplify the Trp53 coding sequences from four representative tumors and sequenced these products in their entirety. None of these tumors had point or frameshift mutations of the Trp53 gene. We conclude that Trp53 haploinsufficiency is sufficient for MPNST pathogenesis in P0-GGFβ3; Trp53+/− mice.
Whole Genome Array Comparative Genomic Hybridization Indicates that Copy Number Variations in Low Grade P0-GGFβ3; Trp53+/− MPNSTs Are Less Extensive Than Those in P0-GGFβ3 MPNSTs
Using aCGH, we have previously shown that high grade (predominantly WHO grade IV) P0-GGFβ3 MPNSTs carry multiple whole chromosome gains and losses and focal copy number variations (CNVs) [20]. If the P0-GGFβ3; Trp53+/− MPNSTs we have graded as WHO grade II and III lesions truly are lower grade tumors, we would predict that the genomes of these tumors would have fewer CNVs than those of P0-GGFβ3 MPNSTs. Further, we would anticipate that if low grade P0-GGFβ3; Trp53+/− MPNSTs progress to become higher grade MPNSTs analogous to those seen in P0-GGFβ3 mice, at least some of the focal CNVs observed in low grade P0-GGFβ3; Trp53+/− MPNSTs would correspond to those we have previously identified in high grade P0-GGFβ3 MPNSTs. To test these postulates, we isolated genomic DNA from seven P0-GGFβ3;Trp53+/− early passage MPNST cultures (one grade II and six grade III tumors) and wild-type Schwann cells and labeled these DNAs with Cy5-dUTP or Cy3-dUTP, respectively. Equivalent amounts of MPNST and Schwann cell DNA were then hybridized to high density aCGH microarrays and regions of unbalanced gains or losses were identified by comparing the relative ratios of the signals from the Schwann cell and MPNST DNA samples.
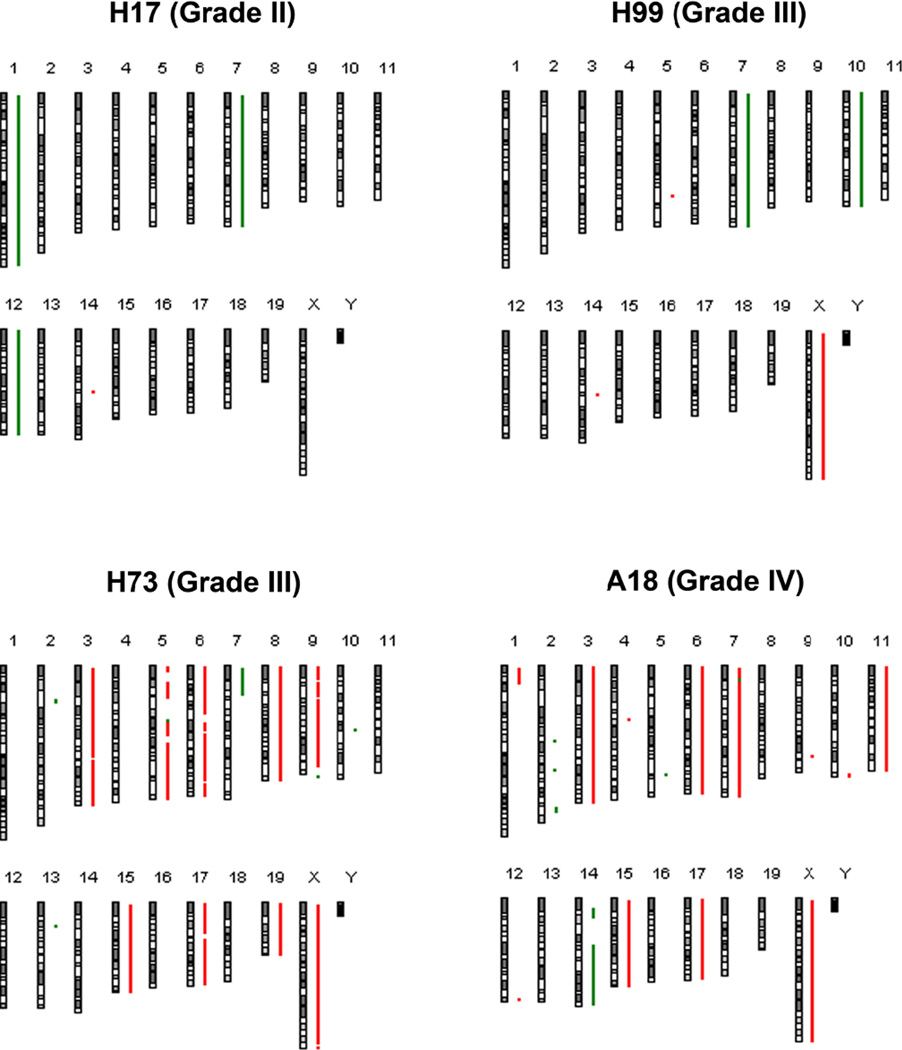
In our initial assessment of the CNVs in these P0-GGFβ3; Trp53+/− MPNSTs, we identified two predominant patterns of genomic abnormalities. The WHO grade II tumor (H17) in this series had relatively few unbalanced CNVs (Figure 8), which was distinctly different from the much more complex pattern we have previously observed in WHO grade IV P0-GGFβ3 MPNSTs (e.g., see A18 in Figure 8). Curiously, however, the grade III P0-GGFβ3; Trp53+/− MPNSTs tumors did not show genetic abnormalities intermediate between those seen in grade II P0-GGFβ3; Trp53+/− MPNSTs and grade IV P0-GGFβ3 MPNSTs. Instead, these WHO grade III tumors clustered into two distinct groups, with approximately half of the tumors having relatively few gains and losses, much as was seen in our WHO grade II MPNSTs, while the other half had complex genomic abnormalities quite similar to those observed in grade IV P0-GGFβ3 MPNSTs.
Copy number variations (CNVs) affecting whole chromosomes or chromosome arms occurred frequently in P0-GGFβ3; Trp53+/− MPNSTs, with an overall average of 5.4 alterations per tumor genome (median 3, range 0–11; in Supplemental Table 2, italic labels denote tumors with relatively few CNVs and bold labels indicate MPNSTs with complex genomic abnormalities). The WHO grade II/simple grade III tumors averaged 1.75 abnormalities per tumor (range 0–3) with 6 chromosomal losses and one gain in total in this group. In contrast, the complex WHO grade III tumors averaged 10.3 abnormalities per tumors (range 10–11); this included an average of 7.3 gains (range 2–11) and 3 losses (range 0–8) per tumor. The most commonly observed alterations were losses of chromosome 7 (4/7 tumors) and 12 (3/7 tumors) and gains of the X chromosome (3/7 tumors). All three of these abnormalities were evident in both WHO II/simple III tumors and complex WHO III tumors. Additionally, a loss of chromosome 10, an abnormality that was universally present in the high grade P0-GGFβ3 MPNSTs we previously studied [20], was present in one simple and one complex grade III tumor. Overall, the whole chromosome and chromosome arm CNVs present in complex grade III tumors were highly similar to those we have previously observed in P0-GGFβ3 MPNSTs.
We have previously shown that focal CNVs in P0-GGFβ3 MPNSTs commonly include genes that have been shown to promote the pathogenesis of a variety of human cancer types [20]. To determine whether focal CNVs in P0-GGFβ3; Trp53+/− MPNSTs similarly affected driver genes implicated in the development of other tumor types, we identified the chromosomal regions encompassed by the focal CNVs in these tumors and compared the genes within these intervals to those listed in the comprehensive Bushman Laboratory Cancer Gene List (http://www.bushmanlab.org/links/genelists). We identified 34 focal regions of unbalanced chromosomal gain or loss in the seven P0-GGFβ3; Trp53+/− MPNSTs we examined (Supplemental Table 3), which included 13 unbalanced gains and 21 unbalanced losses scattered across 16 chromosomes. Several of these focal CNVs were present in multiple tumors. For example, a gain on chromosome 14 (chr14: 69,877,095-70,083,114) which contains the Loxl2 gene was found in 5/7 tumors, including both simple and complex WHO grade II and III tumors. Interestingly, a nearly identical copy number gain was evident in 5/11 P0-GGFβ3 MPNSTs in our earlier study (although this CNV did not include Loxl2 in our P0-GGFβ3 MPNSTs [20]). Similarly, a loss on chromosome 13 (chr13: 27,351,198-27,897,114) was seen in 4/7 tumors (both simple and complex WHO grade III). We identified multiple genes previously implicated in human tumorigenesis in 16 of the 34 focal CNVs (Supplemental Table 3, bolded and underlined). In total, 27 cancer-associated genes were identified in these regions, with 1–4 genes occurring in each focal CNV that contained such a gene. The candidate driver genes identified in the focal CNVs had numerous functions potentially contributing to MPNST pathogenesis (Table 2). The affected regions encode proteins involved in transcriptional regulation (Zeb2, Foxp3, Gata1), proliferation (Myc, Reg3a) and cell adhesion (Cd53, Cntn4, Cntn6, Pxn). Numerous genes encoding key cytoplasmic signaling molecules were also affected, including Prkcdbp, Mapk1, Rap1gds1, Arap2 and Crkl. Notably, several of the candidate driver genes within the focal CNVs (e.g., Pvt1, Myc) that we identified in P0-GGFβ3; Trp53+/− MPNSTs were also evident in P0-GGFβ3 MPNSTs in our earlier study [20]. However, other candidate driver genes that were commonly affected in P0-GGFβ3 MPNSTs (e.g., Cdkn2a, Cdkn2b) were not affected in any of the P0-GGFβ3; Trp53+/− MPNSTs we examined, suggesting that these changes occur at later stages of MPNST progression.
Table 2.
Functions of candidate driver genes in focal regions of unbalanced gain and loss
| Gene symbol | Gene name | Function |
|---|---|---|
| Cd53 | Leukocyte surface antigen | Cell surface glycoprotein, signal transduction |
| Akap9 | A-kinase anchor protein 9 | Scaffolding protein for protein kinases and phosphatases, assembly on centrosome and Golgi apparatus |
| Steap1 | Six-transmembrane epithelial antigen of the prostate 1 | Cell surface antigen expressed in cell-cell junctions |
| Steap2 | Six-transmembrane epithelial antigen of the prostate 2 | Metalloreductase |
| Cdk6 | Cyclin-dependent kinase 6 | Cell cycle progression |
| Pxn | Paxillin | Cytoskeletal protein, cell adhesion |
| Triap1 | TP53 regulated inhibitor of apoptosis1 | Cell survival |
| Loxl2 | Lysyl oxidase-like 2 | Crosslinking of collagen and elastin, regulator of sprouting angiogenesis, epithelial-mesenchymal transition |
| Zeb2 | Zinc-finger E-box binding homeobox 2 | Transcritional repressor |
| Rap1gds1 | RAP1, GTP-GDP dissociation stimulator 1 | Stimulates GTP-GDP exchange of many small G-proteins |
| Arap2 | Arf-GAP with Rho-GAP domain | Binds to Rho-A GTP |
| Reg3a | Regenerating islet-derived 3-alpha | Cell proliferation and differentiation |
| Cntn6 | Contactin 6 | Cell adhesion and ligand of Notch |
| Cntn4 | Contactin 4 | Cell adhesion |
| Crbn | Cereblon | Part of E3 ubiquitin ligase complex |
| Cd69 | Cluster of differentiation 69 | Calcium-dependent transmembrane receptor |
| Prkcdbp | Protein kinase C, delta binding protein | Binds protein kinase C, possible tumor suppressor |
| Gm3258 | Predicted gene 3258 | Function unknown |
| Nkain2 | Na+/K+ transporting ATPase interacting 2 | Transmembrane protein interacting with β-subunit of Na+/K+ ATPase |
| Myc | Myelocytomatosis oncogene | Nuclear phosphoprotein regulating cell cycle progression, apoptosis and transformation |
| Pvt1 | Plasmacytoma variant translocation-1 | Encodes several miRNAs |
| MAPK1 | Mitogen-activated protein kinase 1 | Serine/threonine kinase |
| Crkl | V-crk sarcoma virus CT10 oncogene homolog (avian)-like | Protein kinase activating Ras and Jun kinase pathways |
| Foxp3 | Forkhead box P3 | Transcriptional regulation |
| Pim2 | Proviral integrations of moloney virus 2 | Serine/threonine kinase, protooncogene, cell cycle progression |
| Gata1 | GATA binding protein 1 | Transcriptional activator |
DISCUSSION
We have presented evidence that NRG1 receptors are constitutively activated in human neurofibromas and MPNSTs [43] and that the activation of these receptors is necessary for the migration [10] and proliferation [43] of MPNST cells. We have also previously shown that outbred transgenic mice overexpressing NRG1 in Schwann cells (P0-GGFβ3 mice) develop numerous neurofibromas which progress to become MPNSTs, developing malignancies with genomic abnormalities that parallel those seen in human MPNSTs [16,20]. While these findings indicate that NRG1 signaling promotes the pathogenesis of neurofibromas and MPNSTs, it has been unclear which NRG1 regulated signaling pathways are necessary for PNS tumorigenesis. Our fortuitous observation that tumorigenesis is suppressed in P0-GGFβ3 mice bred onto a inbred C57BL/6J background has now presented us with the opportunity to determine whether NRG1 promotes neoplasia predominantly through its effects on Nf1-regulated signaling cascades or instead activates other essential signaling cascades that cooperate with those affected by Nf1 loss. Strikingly, while we found no evidence that NRG1 overexpression in the presence of Nf1 haploinsufficiency led to tumorigenesis, NRG1 and Trp53 haploinsufficiency cooperatively promoted de novo MPNST pathogenesis. Inbred P0-GGFβ3; Trp53+/− mice thus recapitulate the phenotype of cis-Nf1+/−;Trp53+/− mice [ 6,45], demonstrating that NRG1 overexpression and Nf1 loss are interchangeable in these animals. Our findings therefore suggest that NRG1 promotes Schwann cell neoplasia primarily via its effects on Nf1-regulated signaling cascades, an observation that has important implications for other NRG1-dependent tumor types. Further, our demonstration that the MPNSTs in P0-GGFβ3; Trp53+/− mice uniformly arise within PNS ganglia reinforces a hypothesis we have previously proposed regarding the source of these tumors and the mechanisms underlying their development. Finally, we have presented evidence indicating that the MPNSTs in P0-GGFβ3; Trp53+/− mice begin as low grade malignancies and then progress to become high grade tumors. Consequently, P0-GGFβ3; Trp53+/− mice represent a novel model system that will be highly useful for identifying the genomic changes mediating the progression of MPNSTs from WHO grade II to WHO grade IV lesions.
Tumorigenesis is suppressed in P0-GGFβ3 mice bred for >15 generations onto a C57BL/6J genetic background. This is not due to an artifact such as transgene silencing as transgene expression is maintained in these mice and they still develop Schwann cell hyperplasia. Additionally, we have observed hindlimb paralysis in some older inbred P0-GGFβ3 mice (not shown) in the absence of tumors, which indicates that these animals also continue to develop the peripheral neuropathies we described in our initial studies of this model [16]. Consequently, the most parsimonious explanation for our observations is that there are modifier genes in the C57BL/6J genetic background that impede NRG1-induced neurofibroma pathogenesis. In keeping with this suggestion, studies in NF1 patients indicate that modifier genes are present in human populations that influence the number of dermal neurofibromas a NF1 patient will develop [9]. Genetic background also influences nervous system tumorigenesis in genetically engineered mice. For example, cis-Nf1+/−; Trp53+/− mice on a C57BL/6J background develop astrocytomas and MPNSTs with high penetrance, while mice carrying these mutations on a 129S4/SvJae background are susceptible to MPNST pathogenesis and resistant to gliomagenesis [14,33,38,40]. It remains to be determined whether the modifier gene(s) that suppress neurofibroma pathogenesis in C57BL/6J mice are the same modifier genes that influence neurofibroma development in humans.
We have previously demonstrated that human neurofibromas, MPNSTs and MPNST cell lines express class II (GGF) and III (cysteine-rich domain) isoforms of NRG1 as well as its receptors, erbB3 and erbB4 [43]. The erbB receptors are constitutively phosphorylated in MPNSTs and treatment with pan-erbB inhibitors reduces MPNST proliferation. We have also shown that NRG1β, but not NRG1α augments MPNST migration by acting as a chemotactic factor [10]. ErbB3 and erbB4 are also present in the invadopodia of MPNSTs and inhibition of these receptors results in decreased migration. Considered collectively, these data indicate that NRG1 signaling is important in MPNST pathogenesis. NRG1, like many growth factors that act through membrane tyrosine kinases, potently activates Ras signaling. In keeping with this, we have previously shown that Ras is hyperactivated in P0-GGFβ3 MPNSTs, an observation which led us to propose that NRG1 promotes PNS neoplasia at least in part by activating Nf1-regulated signaling cascades [20]. However, NRG1 is also capable of activating a variety of other signaling molecules such as phosphatidylinositol-3 kinase [51], Grb2 [12], Grb7 [12], Abl2 [19], Cbl [19], NF-κB [41], Src [34] and Myc [7]. Consequently, it has not been clear whether NRG1 promotes neurofibroma and MPNST pathogenesis by modulating the activity of Nf1-regulated signaling cascades, by modulating the activity of other essential non-Nf1 regulated signaling pathways or a combination of both mechanisms. We have shown that NRG1 overexpression does not interact with Nf1 haploinsufficiency to promote tumorigenesis. Thus, reduced Nf1 gene dosage, which renders many cell types hyperresponsive to growth factors [17,18,48,49], occurring in concert with the activation of NRG1 regulated signaling cascades is not sufficient for PNS tumorigenesis. In contrast, we found that P0-GGFβ3; Trp53+/− mice developed MPNSTs with high penetrance. Further, the MPNSTs in P0-GGFβ3; Trp53+/− mice arise de novo rather than from pre-existing neurofibromas, much as is seen in cis-Nf1+/−;Trp53+/− mice. These observations demonstrate that NRG1 overexpression can substitute for Nf1 loss, a finding which argues that NRG1 promotes PNS neoplasia primarily by its effects on Nf1-regulated pathways rather than by activating essential parallel signaling cascades that act cooperatively with Nf1-regulated pathways. In support of this, Ras activity is increased in these tumors compared to wild-type Schwann cells, and this increase in activity is diminished by inhibiting the NRG1 receptors. As NRG1 has been implicated in the pathogenesis of a variety of other tumor types including gliomas [34] and colorectal [1,7,51], ovarian [38], breast [40], and prostate [41] carcinomas, it will be of interest to determine whether NRG1 similarly promotes the pathogenesis of these other tumor types via its effects on NF1-regulated signaling cascades.
Given the promitogenic effects of NRG1 on Schwann cells and the proproliferative effects of Ras activation, it is tempting to postulate that MPNST pathogenesis in P0-GGFβ3; Trp53+/− mice results from enhanced DNA synthesis driven by NRG1/Ras signaling that is occurring in cells whose genomes are unstable as a result of Trp53 loss. However, this is not the only way in which NRG1 overexpression and Nf1 loss could interact to promote tumorigenesis as neurofibromin contains functional domains in addition to its Ras GAP-related domain. For instance, neurofibromin contains a region that interacts with focal adhesion kinase (FAK) [24] and glycerophospholipids (a Sec14p homology domain) [46] and thus potentially affects signaling events involving these molecules. Further, it is conceivable that other regions within the very large and still poorly understood neurofibromin protein have functions that have not yet been discovered. Consequently, NRG1 overexpression may promote tumorigenesis via activation of Ras, interaction with other Nf1-regulated but Ras-independent signaling cascades or both mechanisms.
We have previously pointed out that the neurofibromas and MPNSTs arising in outbred P0-GGFβ3 mice tend to develop within sensory (spinal dorsal nerve root), sympathetic and motor (trigeminal) ganglia [16], an observation which is consistent with the fact that other tumors of schwannian origin such as schwannomas also arise preferentially in ganglia. Of note, neurofibromas developing on spinal nerve roots in humans also typically occur in dorsal nerve roots (i.e., centered on the DRG) rather than on ventral nerve roots. These observations are consistent with the hypothesis that neurofibromas and MPNSTs are derived from a progenitor which is enriched in ganglia, that the intraganglionic microenvironment is particularly conducive to tumorigenesis or a combination of both factors. These suggestions are given additional support by the phenotype of P0-GGFβ3; Trp53+/− mice. In our previous studies of P0-GGFβ3 mice, the neurofibromas we found were typically centered on a ganglion but also extended well into adjacent nerve root, making it difficult to pinpoint the site where the tumors originated. In contrast, in P0-GGFβ3; Trp53+/− mice, we identified multiple microtumors which were completely contained within ganglia and surrounded by normal, uninvolved peripheral nerve. Given these observations, it is now clearly evident that NRG1-induced peripheral nerve sheath tumors originate within ganglia.
In previous studies, we demonstrated that P0-GGFβ3 mice recapitulate the process of neurofibroma-MPNST progression seen in human NF1 patients and that the MPNSTs developing in these animals have genomic abnormalities that parallel those seen in human MPNSTs [20]. This indicated that P0-GGFβ3 mice were a valuable model in which could be used to identify the genomic abnormalities driving neurofibroma pathogenesis and neurofibroma-MPNST progression. However, the MPNSTs we found in P0-GGFβ3 mice were almost all high-grade (WHO grade IV) lesions and so this model cannot be easily used to study the intermediate steps by which MPNSTs progress from low to high grade. Unexpectedly, we have found that the MPNSTs developing in P0-GGFβ3; Trp53+/− mice encompass the full spectrum of low to high grade MPNSTs seen in humans. In keeping with these pathologic observations, aCGH performed on P0-GGFβ3; Trp53+/− MPNSTs indicates the unbalanced gains and losses in these tumors became progressively more complex with increasing tumor grade. However, in contrast to the three-step WHO grading system, P0-GGFβ3; Trp53+/− MPNSTs tended to fall into only two groups. The first of these groups included grade II MPNSTs and a subset of WHO grade III tumors that had a relatively limited number of CNVs, while the second group included a set of grade III MPNSTs that had much more complex genomic abnormalities that were more similar to those seen in the grade IV P0-GGFβ3 MPNSTs we have previously studied. These observations suggest that a genomic crisis occurs at some point in grade III evolution which dramatically complicates the complexity of higher grade MPNSTs. It is also notable that some pathologists prefer to simply stratify MPNSTs as low and high grade malignancies. The bimodal distribution of genomic abnormalities evident in P0-GGFβ3; Trp53+/− MPNSTs gives some support to this idea. However, we would caution that the genomes of a much larger collection of P0-GGFβ3 and P0-GGFβ3; Trp53+/− MPNSTs should be examined before firmly arriving at this conclusion.
Upon analyzing the aCGH dataset described in this manuscript, we identified CNVs on chromosomes 13 and 14 that occurred repeatedly in both low grade and higher-grade MPNSTs, which suggests that this interval contains an important, but as yet unknown, driver gene which is affected very early in MPNST evolution. In addition, we found 34 focal CNVs in these tumors, including 16 CNVs which contained a total of 27 genes previously implicated in the pathogenesis of other cancer types. In the MPNSTs with limited genomic abnormalities, we identified six intervals which contained a total of eight genes (Steap1, Steap2, Akap9, Cdk6, Pxn, Triap1, Cdc53 and Loxl2) that regulate cell cycle progression, apoptosis, cell adhesion and intracellular signaling. This suggests that changes affecting the signaling pathways that include these genes are essential for the initial stages of MPNST pathogenesis. As noted above, our data also indicate that approximately half of the major MPNSTs isolated from P0-GGFβ3; Trp53+/− mice have only limited genomic abnormalities, with the remainder have complex genomic abnormalities similar to those seen in P0-GGFβ3 MPNSTs. Based on these observations, we would predict the microtumors in P0-GGFβ3; Trp53+/− mice will also have relatively limited genomic abnormalities which will include key genomic changes essential for the earliest stages of MPNST development. Defining these abnormalities will provide two key pieces of information. First, as was pointed out in a recent review [35], the current WHO grading system offers little in the way of prognostic information. This limitation likely reflects the fact that the WHO grading system is based on subjective morphologic criteria rather than an assessment of key genomic abnormalities. The identification of molecular markers that can be used to redefine MPNST tumor grades thus has the potential to markedly improve our ability to predict patient outcomes. Second, defining the genomic abnormalities associated with specific stages in MPNST pathogenesis will identify the signaling cascades that should be targeted therapeutically at different stages in the evolution of these sarcomas.
In summary, we have found that tumorigenesis is suppressed in transgenic mice overexpressing NRG1 in Schwann cells, which suggests that a modifier gene or genes capable of suppressing neurofibroma pathogenesis is present in the C57BL/6J genetic background. Genetic complementation experiments in which inbred P0-GGFβ3 mice were crossed to Nf1+/− or Trp53+/− mice showed that, while tumorigenesis was absent in the P0-GGFβ3;Nf1+/− animals, P0-GGFβ3;Trp53+/− mice developed MPNSTs. Further, the MPNSTs seen in P0-GGFβ3; Trp53+/− mice developed de novo rather than from neurofibromas as in seen in the parent P0-GGFβ3 line. These observations clarify the role that NRG1 plays in PNS neoplasia as they indicate that NRG1 promotes tumorigenesis primarily via its effects on the signaling cascades that are affected by neurofibromin loss. In keeping with our earlier suggestion that neurofibromas and MPNSTs originate intraganglionically, we have also now shown that the MPNSTs in P0-GGFβ3; Trp53+/− mice originate from microtumors that develop within PNS ganglia. Unexpectedly, P0-GGFβ3; Trp53+/− mice have proven to be a new model of PNS neoplasia that is potentially useful for identifying key mutations mediating the progression of low grade MPNSTs to higher grade tumors. Consequently, partnering genomic analyses of the various MPNST grades occurring in P0-GGFβ3; Trp53+/− mice with similar analyses of the neurofibromas and MPNSTs that develop in P0-GGFβ3 mice will give us a global understanding of the evolution of neurofibromas and MPNSTs and will identify important new therapeutic targets in these tumors.
Supplementary Material
Supplemental Figure 1: C57BL/6J mice have lower expression of Trp53 transcripts compared to SJL/J mice.
Acknowledgments
This work was supported by grants from the National Institute of Neurological Diseases and Stroke (R01 NS048353 to S.L.C., F30 NS063626 to N.M.B. and F31 NS081824 to S.N.B.), the National Cancer Institute (R01 CA122804 to S.L.C. and R01 CA134773 to K.A.R. and S.L.C.) and the Department of Defense (X81XWH-09-1-0086 and W81XWH-12-1-0164 to S.L.C.). We thank the Alabama Neuroscience Blueprint Core Center (P30 NS57098) and the UAB Neuroscience Core Center (P30 NS47466) for technical assistance. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the Department of Defense.
Footnotes
Conflict of Interest
The authors declare that they have no conflicts of interest.
References
- 1.Beji A, Horst D, Engel J, Kirchner T, Ullrich A. Toward the prognostic significance and therapeutic potential of HER3 receptor tyrosine kinase in human colon cancer. Clin Cancer Res. 2012;18:956–968. doi: 10.1158/1078-0432.CCR-11-1186. [DOI] [PubMed] [Google Scholar]
- 2.Birindelli S, Perrone F, Oggionni M, et al. Rb and TP53 pathway alterations in sporadic and NF1-related malignant peripheral nerve sheath tumors. Lab Invest. 2001;81:833–844. doi: 10.1038/labinvest.3780293. [DOI] [PubMed] [Google Scholar]
- 3.Brannan CI, Perkins AS, Vogel KS, et al. Targeted disruption of the neurofibromatosis type-1 gene leads to developmental abnormalities in heart and various neural crest-derived tissues. Genes Dev. 1994;8:1019–1029. doi: 10.1101/gad.8.9.1019. [DOI] [PubMed] [Google Scholar]
- 4.Byer SJ, Eckert JM, Brossier NM, et al. Tamoxifen inhibits malignant peripheral nerve sheath tumor growth in an estrogen receptor-independent manner. Neurooncology. 2011;13:28–41. doi: 10.1093/neuonc/noq146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carroll SL. Molecular mechanisms promoting the pathogenesis of Schwann cell neoplasms. Acta Neuropathol. 2012;123:321–348. doi: 10.1007/s00401-011-0928-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cichowski K, Shih TS, Schmitt E, et al. Mouse models of tumor development in neurofibromatosis type 1. Science. 1999;286:2172–2176. doi: 10.1126/science.286.5447.2172. [DOI] [PubMed] [Google Scholar]
- 7.De Boeck A, Pauwels P, Hensen K, et al. Bone marrow-derived mesenchymal stem cells promote colorectal cancer progression through paracrine neuregulin 1/HER3 signalling. Gut. 2013;62:550–560. doi: 10.1136/gutjnl-2011-301393. [DOI] [PubMed] [Google Scholar]
- 8.Donehower LA, Harvey M, Slagle BL, et al. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 9.Easton DF, Ponder MA, Huson SM, Ponder BA. An analysis of variation in expression of neurofibromatosis (NF) type 1 (NF1): evidence for modifying genes. Am J Hum Genet. 1993;53:305–313. [PMC free article] [PubMed] [Google Scholar]
- 10.Eckert JM, Byer SJ, Clodfelder-Miller BJ, Carroll SL. Neuregulin-1 beta and neuregulin-1 alpha differentially affect the migration and invasion of malignant peripheral nerve sheath tumor cells. Glia. 2009;57:1501–1520. doi: 10.1002/glia.20866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Evans DG, Baser ME, McGaughran J, Sharif S, Howard E, Moran A. Malignant peripheral nerve sheath tumours in neurofibromatosis 1. J Med Genet. 2002;39:311–314. doi: 10.1136/jmg.39.5.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fiddes RJ, Campbell DH, Janes PW, et al. Analysis of Grb7 recruitment by heregulin-activated erbB receptors reveals a novel target selectivity for erbB3. J Biol Chem. 1998;273:7717–7724. doi: 10.1074/jbc.273.13.7717. [DOI] [PubMed] [Google Scholar]
- 13.Garcia-Cao I, Garcia-Cao M, Martin-Caballero J, et al. "Super p53" mice exhibit enhanced DNA damage response, are tumor resistant and age normally. Embo J. 2002;21:6225–6235. doi: 10.1093/emboj/cdf595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hawes JJ, Tuskan RG, Reilly KM. Nf1 expression is dependent on strain background: implications for tumor suppressor haploinsufficiency studies. Neurogenetics. 2007;8:121–130. doi: 10.1007/s10048-006-0078-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holtkamp N, Atallah I, Okuducu AF, et al. MMP-13 and p53 in the progression of malignant peripheral nerve sheath tumors. Neoplasia. 2007;9:671–677. doi: 10.1593/neo.07304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huijbregts RP, Roth KA, Schmidt RE, Carroll SL. Hypertrophic neuropathies and malignant peripheral nerve sheath tumors in transgenic mice overexpressing glial growth factor beta3 in myelinating Schwann cells. J Neurosci. 2003;23:7269–7280. doi: 10.1523/JNEUROSCI.23-19-07269.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ingram DA, Hiatt K, King AJ, et al. Hyperactivation of p21(ras) and the hematopoietic-specific Rho GTPase, Rac2, cooperate to alter the proliferation of neurofibromin-deficient mast cells in vivo and in vitro. J Exp Med. 2001;194:57–69. doi: 10.1084/jem.194.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ingram DA, Yang FC, Travers JB, et al. Genetic and biochemical evidence that haploinsufficiency of the Nf1 tumor suppressor gene modulates melanocyte and mast cell fates in vivo. J Exp Med. 2000;191:181–188. doi: 10.1084/jem.191.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaushansky A, Gordus A, Budnik BA, Lane WS, Rush J, MacBeath G. System-wide investigation of ErbB4 reveals 19 sites of Tyr phosphorylation that are unusually selective in their recruitment properties. Chem Biol. 2008;15:808–817. doi: 10.1016/j.chembiol.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kazmi SJ, Byer SJ, Eckert JM, et al. Transgenic mice overexpressing neuregulin-1 model neurofibroma-malignant peripheral nerve sheath tumor progression and implicate specific chromosomal copy number variations in tumorigenesis. Am J Pathol. 2013;182:646–667. doi: 10.1016/j.ajpath.2012.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kluwe L, Friedrich R, Mautner VF. Loss of NF1 allele in Schwann cells but not in fibroblasts derived from an NF1-associated neurofibroma. Genes Chromosomes Cancer. 1999;24:283–285. doi: 10.1002/(sici)1098-2264(199903)24:3<283::aid-gcc15>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 22.Kohli L, Kaza N, Lavalley NJ, et al. The pan erbB inhibitor PD168393 enhances lysosomal dysfunction-induced apoptotic death in malignant peripheral nerve sheath tumor cells. Neuro-oncology. 2012;14:266–277. doi: 10.1093/neuonc/nor226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kourea HP, Cordon-Cardo C, Dudas M, Leung D, Woodruff JM. Expression of p27(kip) and other cell cycle regulators in malignant peripheral nerve sheath tumors and neurofibromas: the emerging role of p27(kip) in malignant transformation of neurofibromas. Am J Pathol. 1999;155:1885–1891. doi: 10.1016/S0002-9440(10)65508-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kweh F, Zheng M, Kurenova E, Wallace M, Golubovskaya V, Cance WG. Neurofibromin physically interacts with the N-terminal domain of focal adhesion kinase. Mol Carcinog. 2009;48:1005–1017. doi: 10.1002/mc.20552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Legius E, Dierick H, Wu R, et al. TP53 mutations are frequent in malignant NF1 tumors. Genes Chromosomes Cancer. 1994;10:250–255. doi: 10.1002/gcc.2870100405. [DOI] [PubMed] [Google Scholar]
- 26.Lothe RA, Smith-Sorensen B, Hektoen M, et al. Biallelic inactivation of TP53 rarely contributes to the development of malignant peripheral nerve sheath tumors. Genes Chromosomes Cancer. 2001;30:202–206. [PubMed] [Google Scholar]
- 27.Luna L. Unna's Method for Mast Cells. In: Luna L, editor. Manual of Histologic Staining Methods of the Armed Forces Institute of Pathology. City: McGraw-Hill Book Company; 1968. pp. 115–116. [Google Scholar]
- 28.Mayes DA, Rizvi TA, Cancelas JA, et al. Perinatal or Adult Nf1 Inactivation Using Tamoxifen-Inducible PlpCre Each Cause Neurofibroma Formation. Cancer Res. 2011;71:4675–4685. doi: 10.1158/0008-5472.CAN-10-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCaughan JA, Holloway SM, Davidson R, Lam WW. Further evidence of the increased risk for malignant peripheral nerve sheath tumour from a Scottish cohort of patients with neurofibromatosis type 1. J Med Genet. 2007;44:463–466. doi: 10.1136/jmg.2006.048140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Menon AG, Anderson KM, Riccardi VM, et al. Chromosome 17p deletions and p53 gene mutations associated with the formation of malignant neurofibrosarcomas in von Recklinghausen neurofibromatosis. Proc Natl Acad Sci U S A. 1990;87:5435–5439. doi: 10.1073/pnas.87.14.5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Olsen SH, Thomas DG, Lucas DR. Cluster analysis of immunohistochemical profiles in synovial sarcoma, malignant peripheral nerve sheath tumor, and Ewing sarcoma. Mod Pathol. 2006;19:659–668. doi: 10.1038/modpathol.3800569. [DOI] [PubMed] [Google Scholar]
- 32.Perry A, Roth KA, Banerjee R, Fuller CE, Gutmann DH. NF1 deletions in S-100 protein-positive and negative cells of sporadic and neurofibromatosis 1 (NF1)-associated plexiform neurofibromas and malignant peripheral nerve sheath tumors. Am J Pathol. 2001;159:57–61. doi: 10.1016/S0002-9440(10)61673-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reilly KM, Broman KW, Bronson RT, et al. An imprinted locus epistatically influences Nstr1 and Nstr2 to control resistance to nerve sheath tumors in a neurofibromatosis type 1 mouse model. Cancer Res. 2006;66:62–68. doi: 10.1158/0008-5472.CAN-05-1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ritch PA, Carroll SL, Sontheimer H. Neuregulin-1 enhances motility and migration of human astrocytic glioma cells. J Biol Chem. 2003;278:20971–20978. doi: 10.1074/jbc.M213074200. [DOI] [PubMed] [Google Scholar]
- 35.Rodriguez FJ, Folpe AL, Giannini C, Perry A. Pathology of peripheral nerve sheath tumors: diagnostic overview and update on selected diagnostic problems. Acta Neuropathol. 2012;123:295–319. doi: 10.1007/s00401-012-0954-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scheithauer BWLDN, Hunter S, Woodruff JM, Antonescu CR. Malignant peripheral nerve sheath tumour (MPNST) In: Louis DNOH, Wiestler OD, Cavenee WK, editors. WHO Classification of Tumours of the Central Nervous System. City: IARC; 2007. pp. 160–162. [Google Scholar]
- 37.Scheithauer BWWJM, Erlandson RA. Primary malignant tumors of peripheral nerve. In: Scheithauer BWWJM, Erlandson RA, editors. Tumors of the Peripheral Nervous System. City: Armed Forces Institute of Pathology; 1999. pp. 303–365. [Google Scholar]
- 38.Sheng Q, Liu X, Fleming E, et al. An activated ErbB3/NRG1 autocrine loop supports in vivo proliferation in ovarian cancer cells. Cancer Cell. 2010;17:298–310. doi: 10.1016/j.ccr.2009.12.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shimada S, Tsuzuki T, Kuroda M, et al. Nestin expression as a new marker in malignant peripheral nerve sheath tumors. Pathol Int. 2007;57:60–67. doi: 10.1111/j.1440-1827.2006.02059.x. [DOI] [PubMed] [Google Scholar]
- 40.Smirnova T, Zhou ZN, Flinn RJ, et al. Phosphoinositide 3-kinase signaling is critical for ErbB3-driven breast cancer cell motility and metastasis. Oncogene. 2012;31:706–715. doi: 10.1038/onc.2011.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Soler M, Mancini F, Meca-Cortes O, et al. HER3 is required for the maintenance of neuregulin-dependent and -independent attributes of malignant progression in prostate cancer cells. Int J Cancer. 2009;125:2565–2575. doi: 10.1002/ijc.24651. [DOI] [PubMed] [Google Scholar]
- 42.Stemmer-Rachamimov AO, Louis DN, Nielsen GP, et al. Comparative pathology of nerve sheath tumors in mouse models and humans. Cancer Res. 2004;64:3718–3724. doi: 10.1158/0008-5472.CAN-03-4079. [DOI] [PubMed] [Google Scholar]
- 43.Stonecypher MS, Byer SJ, Grizzle WE, Carroll SL. Activation of the neuregulin-1/ErbB signaling pathway promotes the proliferation of neoplastic Schwann cells in human malignant peripheral nerve sheath tumors. Oncogene. 2005;24:5589–5605. doi: 10.1038/sj.onc.1208730. [DOI] [PubMed] [Google Scholar]
- 44.Upadhyaya M, Kluwe L, Spurlock G, et al. Germline and somatic NF1 gene mutation spectrum in NF1-associated malignant peripheral nerve sheath tumors (MPNSTs) Hum Mutat. 2008;29:74–82. doi: 10.1002/humu.20601. [DOI] [PubMed] [Google Scholar]
- 45.Vogel KS, Klesse LJ, Velasco-Miguel S, Meyers K, Rushing EJ, Parada LF. Mouse tumor model for neurofibromatosis type 1. Science. 1999;286:2176–2179. doi: 10.1126/science.286.5447.2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Welti S, Fraterman S, D'Angelo I, Wilm M, Scheffzek K. The sec14 homology module of neurofibromin binds cellular glycerophospholipids: mass spectrometry and structure of a lipid complex. J Mol Biol. 2007;366:551–562. doi: 10.1016/j.jmb.2006.11.055. [DOI] [PubMed] [Google Scholar]
- 47.Wu J, Williams JP, Rizvi TA, et al. Plexiform and dermal neurofibromas and pigmentation are caused by Nf1 loss in desert hedgehog-expressing cells. Cancer Cell. 2008;13:105–116. doi: 10.1016/j.ccr.2007.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang FC, Chen S, Clegg T, et al. Nf1+/− mast cells induce neurofibroma like phenotypes through secreted TGF-beta signaling. HumMolGenet. 2006;15:2421–2437. doi: 10.1093/hmg/ddl165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang FC, Ingram DA, Chen S, et al. Neurofibromin-deficient Schwann cells secrete a potent migratory stimulus for Nf1+/− mast cells. J Clin Invest. 2003;112:1851–1861. doi: 10.1172/JCI19195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 51.Yonezawa M, Wada K, Tatsuguchi A, et al. Heregulin-induced VEGF expression via the ErbB3 signaling pathway in colon cancer. Digestion. 2009;80:215–225. doi: 10.1159/000229775. [DOI] [PubMed] [Google Scholar]
- 52.Zhou H, Coffin CM, Perkins SL, Tripp SR, Liew M, Viskochil DH. Malignant peripheral nerve sheath tumor: a comparison of grade, immunophenotype, and cell cycle/growth activation marker expression in sporadic and neurofibromatosis 1-related lesions. Am J Surg Pathol. 2003;27:1337–1345. doi: 10.1097/00000478-200310000-00006. [DOI] [PubMed] [Google Scholar]
- 53.Zhu Y, Ghosh P, Charnay P, Burns DK, Parada LF. Neurofibromas in NF1:Schwann cell origin and role of tumor environment. Science. 2002;296:920–922. doi: 10.1126/science.1068452. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1: C57BL/6J mice have lower expression of Trp53 transcripts compared to SJL/J mice.