Abstract
Aberrant Neuregulin 1-ErbB4 signalling has been implicated in schizophrenia. We previously identified a novel schizophrenia-associated missense mutation (valine to leucine) in the NRG1 transmembrane domain. This variant inhibits formation of the NRG1 intracellular domain (ICD) and causes decreases in dendrite formation. To assess the global effects of this mutation we used lymphoblastoid cell lines from unaffected heterozygous carriers (Val/Leu) and non carriers (Val/Val). Transcriptome data showed 367 genes differentially expressed between the two groups (Val/Val N=6, Val/Leu N=5, T test, FDR (1%), alpha = 0.05, −log10 p value > 1.5). Ingenuity pathway (IPA) analyses showed inflammation and NRG1 signalling as the top pathways altered. Within NRG1 signalling, Protein Kinase C (PKC)–eta (PRKCH) and non-receptor tyrosine kinase (SRC) were down-regulated in heterozygous carriers. Novel kinome profiling (Serine/Threonine) was performed after stimulating cells (V/V N=6, V/L N=6) with ErbB4, to induce release of the NRG1 ICD, and revealed significant effects of treatment on the phosphorylation of 35 peptides. IPA showed neurite outgrowth (6 peptides) as the top annotated function. Phosphorylation of these peptides was significantly decreased in ErbB4-treated Val/Val but not in Val/Leu cells. These results show that perturbing NRG1 ICD formation has major effects on cell signalling, including inflammatory and neurite formation pathways, and may contribute significantly to schizophrenia pathophysiology.
Keywords: Neuregulin-1, transcriptome, kinome, schizophrenia, lymphoblastoid cells
Introduction
NRG1 is a well -established schizophrenia candidate gene (Harrison, Law 2006; Tosato et al. 2005; Greenwood et al. 2012). The NRG1 protein regulates many important functions in the nervous system by interacting with cognate receptors belonging to the ErbB family, of which the NRG1-ErbB4 interaction has been shown to be particularly relevant to nervous system function (Shamir et al. 2012; Mei, Xiong 2008). The downstream targets of this pathway include ERK, AKT, and PKC. Altered phosphorylation of these targets, particularly AKT (Keri et al. 2009) and ERK (Funk et al. 2012; Kyosseva et al. 1999) has been reported in schizophrenia. Of the numerous single nucleotide polymorphisms (SNPs) identified within the NRG1 gene to be associated with schizophrenia worldwide, only one is known to have a direct role in regulating NRG1 function (Talmage 2008; Weickert et al. 2012). This variant, which causes a change from valine (GTG) to leucine (TTG) (V>L) in the transmembrane domain of the NRG1 protein, was first identified by our laboratory, and is associated with schizophrenia in the population of the Central Valley of Costa Rica (CVCR) (Walss-Bass et al. 2006). We have further found that this variant is associated with immune dysregulation, indicated by increased levels of pro-inflammatory cytokines and autoantibodies in carriers of the variant (Marballi et al. 2010). This is of immense importance, given the large body of studies showing dysregulation of the immune system (Potvin et al. 2008; Strous, Shoenfeld 2006), including elevated levels of pro-inflammatory cytokines and autoantibodies in schizophrenia. Other groups subsequently showed that the V>L change impedes formation of the NRG1 intracellular domain (ICD) by blocking gamma secretase-mediated intracellular cleavage of membrane bound isoforms of NRG1, such as NRG1 type III (Dejaegere et al. 2008), leading to decreased dendrite formation in cortical neurons (Chen et al. 2010). Interestingly, high levels of pro-inflammatory cytokines decrease dendrite formation in vitro (Gilmore et al. 2004).
The NRG1 ICD, generated by gamma secretase intracellular cleavage, migrates to the nucleus and regulates expression of Bcl-XL, Bak, Rip and Oct-3 genes (Bao et al. 2003). In order to further explore the impact of the V>L change on gene expression and cell signalling, specifically NRG1-ErbB4 signalling, we utilized lymphoblastoid cell lines (LCLs) from unaffected individuals from the CVCR that were either heterozygous carriers (Val/Leu) or homozygous non-carriers (Val/Val) to perform whole genome expression (V/L N=5, V/V N=6) and whole kinome profiling (V/L N=6, V/V N=6) studies. LCLs are ideal for the study of the effects of genetic variants on cell function, as they avoid confounding environmental effects such as psychotropic drugs used by patients, and allow for focus solely on mechanistic aspects of genetic perturbation. We hypothesized that the V>L change that perturbs formation of the ICD would impact gene expression and signalling in pathways important for schizophrenia development.
Materials and Methods
Ethics statement
Peripheral leucocytes were isolated from blood of subjects from the CVCR, at the time of recruitment, as previously described (Walss-Bass et al. 2006) in accordance with the principles of the Declaration of Helsinki with approval from the Institutional Review Boards of the University of Costa Rica and the University of Texas Health Science Center at San Antonio. All participants provided written informed consent.
Lymphoblastoid cell lines-generation and maintenance
Lymphoblastoid cell lines (LCLs) were generated from leucocytes using LeucoPREP brand cell separation tubes (Becton Dickinson Labware, Franklin Lakes, NJ, USA) and transformed using Epstein–Barr virus (EBV). Cells were grown in RPMI 1640 medium with 2 mM L-glutamine and 15% bovine growth serum, 1% penicillin streptomycin at 37°C in a humidified 5% CO2 incubator. As previously described (Marballi et al. 2010), given that the goal of this study was to determine the association of the NRG1 V>L mutation with alterations in gene expression and cell signalling, independent of psychiatric diagnosis, cell lines used in this study were from unaffected, unrelated individuals who had one first degree relative with psychosis: homozygous wild type (GG, Val/Val, N=6; three males, three females, average age 51.4±20.54 yr) and heterozygous T allele carriers (GT, Val/Leu, N=5; two males, three females, average age 58.5±19.15 yr). While conducting the experiments the investigators were blinded to the experimental groups. Cell viability was determined by neutral red assay (Sigma, St.Louis, MO, USA). Only viable cells are capable of incorporating neutral red dye by active transport. Briefly, after 24 hours of incubation of equal number of cells, cells were rinsed with PBS and incubated in media containing 0.033% neutral red for two hours. Cells were then washed several times with PBS and the incorporated neutral red dye was solubilized by gentle rocking for 10 minutes with a solution of 1% acetic acid and 50% ethanol. After 10 min, the solution was collected and the amount of incorporated neutral red dye was determined spectrophotometrically by measuring the absorbance of the solution at 540 nm.
Whole Genome Expression Studies
RNA was extracted from cells using the Trizol method. RNA samples were cleaned up using RNEasy plus micro kit and run on an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) to ensure RNA integrity for whole genome studies. cRNA was prepared, hybridized to Cy3, loaded as singlicates on to Human WG-6 v3 beadchips (Illumina, Inc, San Diego, CA, USA), and scanned using an Illumina iScan reader. All data were quantile normalized, exported into text files using Bead studio software and data analyses were performed using box plots in JMP genomics (Version 3.0, SAS Institute Inc., Cary, NC, USA). Student’s T test was used in JMP to analyze values, with a Benjamini Hochberg False Discovery Rate (FDR) correction of 1%, alpha=0.05, and a cutoff of −log10 (p value)>1.5. The list of significant genes obtained was imported into Ingenuity pathway analyses (IPA, Ingenuity® Systems, Redwood City, CA, USA) for identification of altered pathways/disease.
Validation of microarray data using real time PCR (qPCR)
Two μg of RNA was converted to cDNA using the High Capacity cDNA Reverse Transcription kit (Life Technologies, Carlsbad, CA, USA). One hundred ng of cDNA was used along with Taqman assays (Life Technologies, Carlsbad, CA, USA) for PRKCH (Protein Kinase C-eta isoform -Hs00178933_m1) and SRC (00178494_m1). The human large ribosomal subunit gene (RPLPO) was used as the housekeeping gene. Real time PCR was run using standard settings on a 7900HT sequence detection system (Life Technologies, Carlsbad, CA, USA). Each cell line was run in technical duplicates. Delta Ct (ΔCt) values were calculated by subtracting housekeeping gene Ct from the gene of interest Ct values. 2−ΔCt values were used for plotting data.
Cell line ErbB4 treatment and preparation for kinome assay
For these studies we added an additional sample to the Val/Leu group to bring the total to N=6 (three males, three females, average age 62.8±19.17 yr). For each cell line, approximately 1x107 cells from actively growing cultures were pelleted, washed once with PBS and seeded on 6 well plates with 2 ml of serum- free media. Cells were serum-starved overnight, and then treated with 10μg/ml ErbB4 (R&D Systems, Minneapolis, MN, USA) or PBS (vehicle) in complete media for 4 hr. ErbB4 treatment has been previously shown to stimulate the NRG1 back signalling pathway, i.e., ICD formation (Hancock et al. 2011). Lysates were prepared using the M-PER Mammalian extraction buffer (Thermo Fisher Scientific Inc., Rockford, IL, USA) with added protease and phosphatase inhibitors. Briefly, cells were pelleted at 4°C, washed twice with ice cold PBS, followed by lysis in the extraction buffer for 15 minutes on ice and centrifugation at 10,000 x g for 15 minutes at 4°C. Supernatants were collected, aliquots were flash frozen in liquid nitrogen and stored at −80°C. Protein quantitation was performed using nanodrop (Thermo Fisher Scientific Inc., Rockford, IL, USA) and BCA assay (Thermo Fisher Scientific Inc., Rockford, IL, USA).
PamStation kinomic analysis
Kinomic profiling was performed using a PamStation® 96 microarray (PamGene International Cambridge, MA, USA) with STK PamChip® containing 140 consensus and 4 control phosphopeptide sequences in each well representing the Serine/Threonine kinome. Each well of the PamChip® was first blocked in 2% bovine serum albumin (BSA). Following protein concentration determination (by BCA assay), 1.5μg of protein were loaded per well of the PamChip® along with standard kinase buffer (PamGene), 400 μM ATP, and FITC-labeled anti-phospho Ser and Thr antibodies (PamGene). Lysate from each cell line was run as a singlicate (Total N=24). The assay mix, containing the active kinases in the sample lysates, was pumped through the PamChip® wells to facilitate interaction with the specific peptide substrates immobilized in the chip. The degree of phosphorylation was measured in real time via the kinetic phosphoserine/threonine antibodies binding to each phosphorylated peptide substrate every six seconds for the length of the program (60 min). Following 60 min incubation, the post-wash signals (multiple exposure times) were integrated for each spot and a log transformation of the data was carried out. The signal intensities for each peptide were analyzed using BioNavigator Software (PamGene) as previously described (Jarboe et al. 2012).
Statistical analyses
Data analyses were carried out using IBM SPSS version 2.0 (IBM Corp, Armonk, NY, USA) using log transformed intensity values. Student’s T test was used for all experiments comparing Val/Val to Val/Leu individuals (whole genome array, qPCR). For IPA analysis, the software identifies gene networks by mapping the connectivity between genes, and computes significant biological/cellular functions that are overrepresented based on input data using a Right tailed Fisher’s exact test to calculate p values of identified pathways, based on the number of significant molecules in the pathway. IPA also provides canonical pathways significant to the data, based on known metabolic and signalling pathways from the literature. For the kinome data, multivariate analyses (2 way ANOVA) were carried out to assess changes in phosphorylation of 140 different peptides and assess effects of genotype and treatment on phosphorylation. Since we only had 2 groups per condition [treatment (vehicle or ErbB4)]; genotype (Val/Val or Val/Leu)], post-hoc testing was not possible in this two way model. Based on our previous studies, we had an a priori hypothesis that we would observe changes between Val/Val and Val/Leu groups, therefore paired T tests were used for comparisons of vehicle and ErbB4-treated Val/Val and Val/Leu cells. In all cases statistical significance was applied at p<0.05. Putative upstream kinases for significant peptides were ascertained using the kinexus database (http://www.phosphonet.ca/).
Results
Differential gene expression between Val/Val and Val/Leu groups
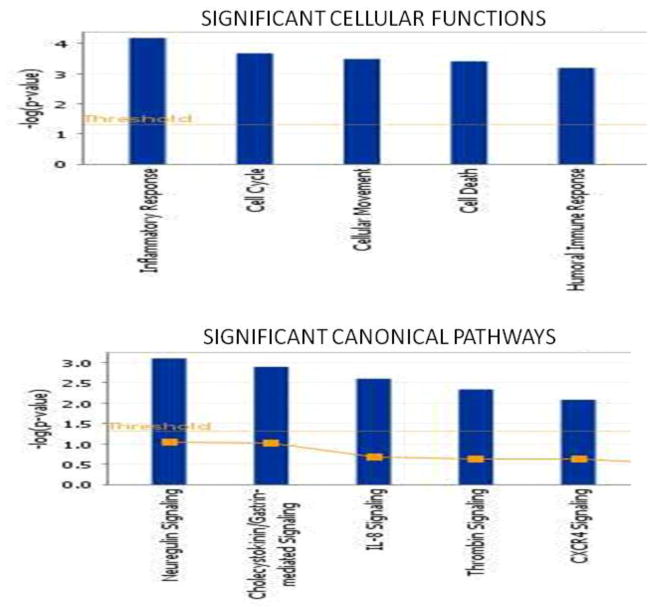
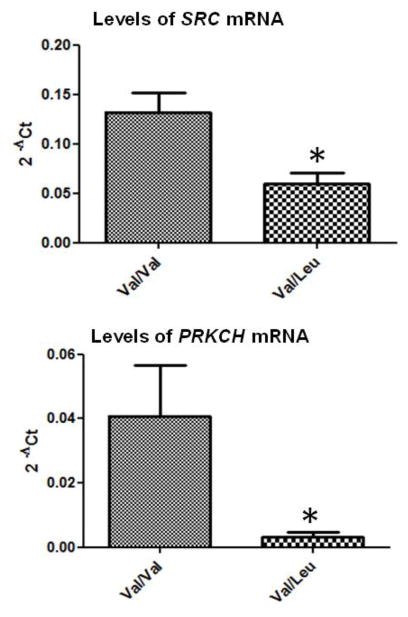
Whole genome expression analyses showed 367 genes to be significantly different between groups using Student’s T test and 1% FDR with alpha = 0.05 and a cutoff of −log10 > 1.5. 204 genes were downregulated while 163 genes were upregulated in V>L carriers. Ingenuity Pathway analysis (IPA) showed these genes to be involved in 18 different networks based on connections with other genes, the top 6 of which are shown in Table 1. Of the biological functions within the 18 networks, inflammatory response and cell cycle were the most overrepresented (Figure 1). No significant differences in cell proliferation or viability were observed when comparing cells from Val/Val individuals compared to cells from Val/Leu heterozygous subjects (data not shown). IPA identified the canonical pathway of NRG1 signalling as having the greatest significance, with 5 genes being involved in this pathway, including a disintegrin and metalloproteinase 17 (ADAM 17), neuregulin 2 (NRG2), megakaryocyte associated tyrosine kinase (MATK), protein Kinase C eta (PRKCH) and the non-receptor tyrosine kinase SRC. PKC and Src are important secondary messengers downstream of NRG1-ErbB4 signalling, and have both been implicated in schizophrenia. PRKCH and SRC mRNA levels were 6.6 and 1.6 fold lower, respectively, in Leu carriers compared to wild-type (T test, p<0.05), and this down-regulation was validated by qPCR (Figure 2). Of genes within the NRG family, only NRG2 was differentially expressed between Val/Val and Val/Leu groups, with increased expression in Leu carriers. No differences in expression levels of ErbB2, ErbB3 or ErbB4 were observed between groups.
Table 1.
Ingenuity pathway analyses of gene expression data arranged by relevance score
| ID | Molecules in Network | Score | Top Functions |
|---|---|---|---|
| 1 | AACS↑, ADAM17↑, AP2A2↓, Calcineurin protein(s), Calpain, CCR4, CD97↓, E2f, ERK1/2, HIST1H2BJ ↑(includes EG:8970), Hsp90, IFN Beta, IL12 (complex), Immunoglobulin, Interferon alpha, LDL, LPP, MARK2↓, MATR3, NFKB1↓, NRG2↑, P38 MAPK, Pkc(s), PPAP2B, PPP3R2, PRKCH↓, RLN3, RPL23A, S100A9, SNAP23↓, SREBF2↑, STX11↓, Tgf beta, TNNC2, Vegf | 40 | Reproductive System Development and Function, Cell Cycle, Hematological Disease |
| 2 | Ap1, CD27↑, CD180, DOK2, ERK, Fibrinogen, HAX1↑, IKK (complex), IL1F9, Integrin, LCP2↓, LY96, MATK, Mlc, MUC2, NCK, Nfat (family), NFkB (complex), NFKB1↓, NPHS2, PDGF BB, PLC gamma, Rac, Ras, Ras homolog, RHOB, RIN3, ROCK1↓, SH3BP2, SLC3A1, SRC↓, SRCIN1, SSH1↓, TCR, VAV, WISP2 | 29 | Inflammatory Response, Cell Death, Cellular Assembly and Organization |
| 3 | ALDH1A1, ALDH8A1, BAI1, BAIAP3 (includes EG:8938), C2ORF44↑, C9ORF64↓, CBR3, DHRS4↓, DLG4, DNAJA2, DNAJA3, DNAJB4, DNAJB9, DNAJC14, DNAJC17↑, DTWD1, FEZF2, FYCO1↑, HNF4A, HPS3, HPS6, HPS5 (includes EG:11234), IL15, MRPL33↑, ONECUT1, RUVBL2, SARS2↑, SLC25A32, SREBF1, SUCLG1, TGFB1, TP53, ZNF317↑ | 23 | Genetic Disorder, Drug Metabolism, Lipid Metabolism |
| 4 | ABI3BP, ACMSD↑, AGFG1 (includes EG:3267), AR, BCL7C↓, c-Myc/N-Myc, CDH1, CLEC2A, CSNK1G2,↓ Erk1/2 dimer, ESR1, FBXW8↑, GTF3C4, KCTD6, MAGEA11, MIR101, MIR214 (includes EG:406996), MPP5, MYB (includes EG:293405), MYC, MYCN, MYO9A, NADSYN1↓, PHF5A, PLSCR4↑, retinoic acid, REXO4↑, RN5S, RPL41, RPL13A↑, RUVBL2, SCPEP1, SLC25A19, SMARCA4, SNX20↓ | 21 | Cell Cycle, Cancer, Dermatological Diseases and Conditions |
| 5 | 26s Proteasome, ADA↓, ADRBK2, Akt, BCAS3, Caspase, CDKN2A↑, DYRK3, ELP4, ELP6, ELP3 (includes EG:55140), ERMAP, FABP3↓, FOXG1, FOXO6, FSH, Histone h3, Histone h4, Insulin, JMJD6, Jnk, MAD2L2↓, MAF, Mapk, OPN1LW (includes EG:5956), OPN1SW, PARP10, PAX7↑, PHC2, PI3K, PPP1R1B, PQBP1, RNA polymerase II, SEC14L2, TFF1 | 20 | Neurological Disease, Genetic Disorder, Ophthalmic Disease |
| 6 | ANXA8, ART2A, BAP1, beta-estradiol, BRCA1, C11ORF82, C13ORF27, CCL23↑, CIC↓, CREB1, DEFB1, GABPB2, GBP6, GBP8, GBP4 (includes EG:17472), GIP2, GVIN1, HERC2, IFNB1, IFNG, IRGM, KIR2DS1, LY6, MNS1↑, NLRC5↓ (includes EG:84166), PCBP4↑, PRB2, PTPRV, RPL27, RPL36AL↑, RTP4, SNCA, TMEM164↓, TP53, TRIM22 | 19 | Cell Cycle, Cell Death, Connective Tissue Development and Function |
Table shows top 6 of 18 networks altered between Val/Val and Val/Leu individuals. Genes with increased (↑) or decreased (↓) expression in Val//Leu compared to Val/Val groups and present in networks are bolded, related molecules in network are not bolded. Underlined in the top functions column, are the functions of Cell cycle, Inflammatory response and Cell death, which are overrepresented amongst the top 6 networks. These functions are shown in Figure 1.
Fig. 1.
Functional analyses of whole genome data by Ingenuity pathway analysis (IPA). Cells from Val/Val (n=6) and Val Leu (n=5) groups were subjected to whole genome analyses using Illumina Human WG-6 v3 beadchips. Data was analyzed by JMP Genomics software using Student’s T test with a Benjamini Hochberg False Discovery Rate (FDR) correction of 1%, alpha=0.05, and a cutoff of −log10 (p value)>1.5.; output generated was imported into IPA. Panels show cellular functions (top) and canonical pathways (bottom) associated with differentially expressed genes and networks (Table 1). Right-tailed Fisher’s exact test was used to calculate p-values determining the significance of each biological function and/or pathway. Threshold= −log (pvalue)>1.5.
Fig. 2.
Quantitative PCR validation of whole genome data using single gene real time PCR assays. RNA was extracted from the Val/Val (n=6) and Val/Leu (n=5) cells, converted to cDNA, and subjected to qPCR. Relative expression levels are shown with 2−ΔCt values (* represents statistical significance T test p<0.05), SRC (upper panel) and PRKCH (lower panel), with the human large ribosomal protein (RPLPO) as an endogenous control.
Val>Leu variant affects phosphorylation of targets regulating neurite outgrowth
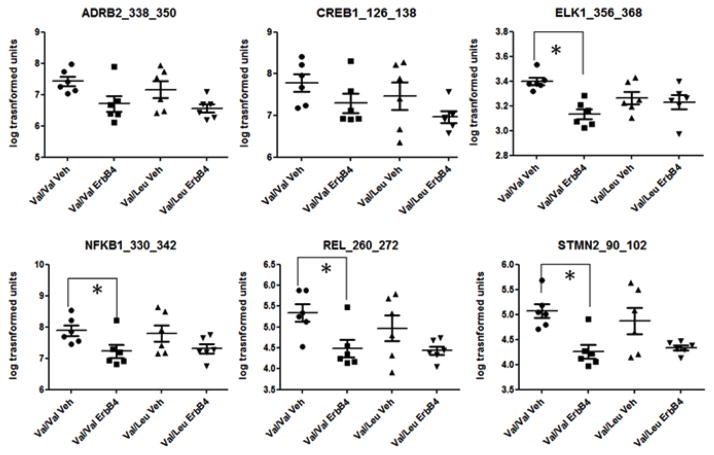
In order to test the effects of the Val>Leu variant on phosphorylation of targets within NRG1-ErbB4 pathways, we treated the LCLs with recombinant ErbB4. This treatment stimulates the intracellular cleavage of NRG1 and formation of the intracellular domain (Bao et al. 2003; Hancock et al. 2011) which is perturbed in presence of the Val>Leu variant (Dejaegere et al. 2008). Comparisons across the four groups (Val/Val vehicle, Val/Leu vehicle, Val/Val ErbB4, Val/Leu ErbB4) using multivariate analyses (two way ANOVA) revealed effects of ErbB4 treatment on 35 peptides, effect of genotype on 2 peptides, and effect of genotype and treatment on 1 peptide (Supplementary Table S1). IPA analyses revealed 10 of the 35 peptides affected by ErbB4 treatment to be involved in cell morphology and nervous system development and function. In both these functions, “outgrowth of neurites” was the top common pathway altered, with 6 molecules being involved: beta 2 adrenergic receptor (ADRB2), cyclic AMP response element binding protein (CREB1), Nuclear factor NF-kappa-B p105 subunit (NFkB1), C-Rel proto-oncogene protein (C-REL), ETS domain containing protein (ELK1) and Stathmin 2 (STMN2) (Figure 3). In most cases (4 out of 6), treatment with ErbB4 caused a significant decrease in phosphorylation in the Val/Val group (T test, p<0.05), while no significant differences were seen in the Val/Leu group after treatment (Figure 3). ADRB2, CREB1, and NFkB1 share PKA-alpha as an upstream kinase. ADRB2 and CREB1 also share AKT1 as an upstream kinase (information from Kinexus database, http://www.phosphonet.ca/). ELK1 is phosphorylated at numerous serine/threonine residues by ERK1/2 (Cruzalegui et al. 1999), two of which are represented in the array (T363, T368) (Table 2). CREB1 is phosphorylated by ERK1/2 at Ser 133 (Davis et al. 2000), also represented in the array (Table 2). Both ERK1/2 and AKT1 are secondary messengers downstream of NRG1/ErbB4 signalling (Hahn et al. 2006) and are regulated by PKC (Puente et al. 2006; Uht et al. 2007) and Src (Hu et al. 2009; Lodeiro et al. 2009) (Figure 4). ELK1 is also a substrate for Srcasm (Src-activating and signaling molecule), which is a downstream substrate of Src (Li et al. 2005). Integrated pathway analysis of molecules identified by the transcriptome and kinome data show these molecules share a common pathway (Figure 4).
Fig. 3.
Differential phosphorylation of molecules involved in neurite formation. Cells from Val/Val (n=6) and Val/Leu (n=6) groups were serum-starved and subsequently treated with vehicle or 10μg/ml recombinant ErbB4 for 4 hours; lysates extracted were used for the kinome array. Average log transformed intensity values for each respective peptide for each cell line were used to calculate average intensity values for peptide phosphorylation in each group. * Significant T test (p<0.05) between Val/Val Veh (Vehicle-treated) and Val/Val ErbB4-treated cell lines.
Table 2.
Kinome array peptides regulating neurite formation & altered by ErbB4 treatment
| ID | Description | Sequence | * Ser | * Thr |
|---|---|---|---|---|
| ADRB2_338_350 | Beta-2 adrenergic receptor Beta-2 adrenoceptor) (Beta-2adrenoreceptor). |
ELLCLRRSSLKAY | [345, 346] | |
| CREB1_126_138 | cAMP response element-binding protein (CREB). | EILSRRPSYRKIL | [129, 133] | |
| ELK1_356_368 | ETS domain-containing protein Elk-1. | LLPTHTLTPVLLT | [359, 361, 363, 368] | |
| NFKB1_330_342 | Nuclear factor NF-kappa-B p105 subunit (DNA-binding factor KBF1) | FVQLRRKSDLETS | [337, 342] | [341] |
| REL_260_272 | C-Rel proto-oncogene protein (C-Rel protein). | KMQLRRPSDQEVS | [267, 272] | |
| STMN2_90_102 | Stathmin-2 (Protein SCG10) (Superior cervical ganglion-10 protein). | AAGERRKSQEAQV | [97] |
Positions of Ser (Serine) and Threonine (Thr) residues; blank boxes indicate no Ser/Thr residues.
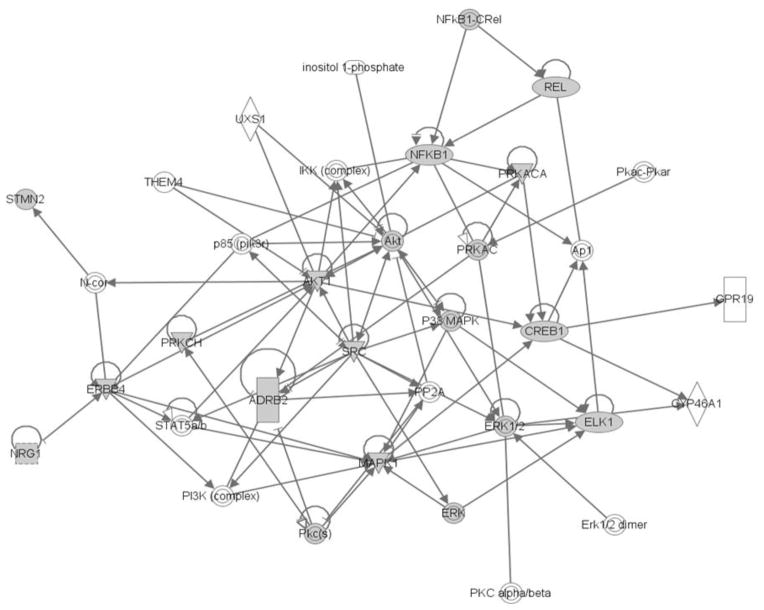
Fig. 4.
NRG1 signalling and neurite formation mediators. Highlighted in grey are molecules within the NRG1-ERBB4 signalling pathway found to be altered in expression (PRKCH and SRC) or phosphorylation (NFkB1, REL, CREB1, ELK1, STMN2 and ADRB2), or kinases directly phosphorylating these molecules (AKT, ERK, MAPK). Other molecules not highlighted in grey are also part of the pathway/network but not found to be altered in our study. Triangles represent kinases; rectangles represent ligand-gated nuclear receptor; concentric circles represent enzyme families; ellipse represents transcription regulators. Figure was generated using IPA pathway analysis software. This software uses a Right tailed Fisher’s exact test to calculate p values of identified pathways, based on the number of significant molecules in the pathway and the fold change of each molecule.
Discussion
NRG1 is one of the most biologically plausible schizophrenia candidate susceptibility genes (Kanakry et al. 2007, Talmage, 2008). Extensive studies in NRG1 function have shown that it impacts different aspects of the nervous system such as myelination (Taveggia et al. 2008, Velanac et al. 2012), radial glia migration (Poluch, Juliano 2007) and neurotransmitter receptor expression (Stefansson et al. 2002; Corfas et al. 2004). Many of these functions are mediated through external interaction of NRG1 with the receptor ErbB4. However, research in the last few years has shown that internal cleavage of NRG1, on the cytoplasmic side of the cell membrane, is equally important (Hancock et al. 2011; Hancock et al. 2008). This cleavage, regulated by gamma secretase, leads to formation of the intracellular domain (ICD), which regulates expression of several different genes (Bao et al. 2003). Our laboratory found a functional transmembrane mutation (V>L) (Walss-Bass et al. 2006) that has been shown by other groups to block the NRG1 intracellular cleavage event (Dejaegere et al. 2008) and caused dendrite formation deficits (Chen et al. 2010), a process implicated in schizophrenia development (Glausier, Lewis 2012). The V>L variant was associated with schizophrenia in the CVCR population (Walss-Bass et al. 2006) and, more recently, also associated with elevated levels of auto-antibodies and pro-inflammatory cytokines in plasma and LCL’s from heterozygous carriers from the CVCR (Marballi et al. 2010). Elevated levels of inflammatory cytokines and autoantibodies have been consistently reported in schizophrenia patients (Potvin et al. 2008). These studies led us to hypothesize that the Val>Leu amino-acid change may cause dysregulation of gene expression and cell signaling events in carriers of this variant, and this may contribute to development of schizophrenia. Whole genome expression analyses showed 367 genes to be differentially expressed in heterozygous carriers compared to wild-type. Inflammatory response was a top function altered, which validates our previous results where we found high levels of pro-inflammatory cytokines in the presence of the Val>Leu variant (Marballi et al. 2010). Cell cycle and cell death were also top functions found to be altered. Examination of cell proliferation and viability did not show significant differences between Val/Val and Val/Leu groups at baseline. However, it is possible that differences would be found under conditions of stress, and this remains to be investigated. To this respect, it is known that many of the symptoms shown in schizophrenia are manifested and exacerbated when patients are under conditions of stress (Corcoran et al. 2003).
We also found the NRG1 signaling pathway to be the top canonical pathway altered in Val/Leu carriers. The fact that expression levels of NRG1 and ErbB2,3,4 were not found to be significantly different between genotype groups supports the hypothesis that the observed differences in global gene expression are due to alterations in NRG1 protein cleavage.
Overall, slightly more genes were down-regulated than up-regulated in Val/Leu carriers. However, focusing on the top 6 pathways outlined by IPA analysis (Table 1), there is no generalized trend for up or down regulation. While changes in expression levels in some cases may be directly due to inhibited NRG1 ICD formation, other changes may be due to compensatory mechanisms in attempts to counteract the effects of altered NRG1 cleavage. Therefore, a detailed analysis of each of the genes in each pathway is required to specifically assess the effects of alteration in expression on specific cell functions. For this study, however, we focused on genes directly relevant to the NRG1-ErbB4 signaling pathway, PRKCH and SRC. PRKCH, was the top overall gene in terms of fold change (6.6 fold decreased expression in Leu carriers). This isoform of PKC regulates keratinocyte differentiation, T cell antigen presentation, AKT signaling (Shahaf et al. 2012), and interacts with the ERK pathway to regulate the nose-touch response in C. elegans (Hyde et al. 2011). The non-receptor tyrosine kinase (SRC) had the next highest fold change in Val/Leu compared to Val/Val groups. Importantly, PKC and Src are not only important effectors of the NRG1-ErbB4 pathway, but also play crucial roles in regulating NMDA receptor (NMDAR) function. PKC regulates NMDAR trafficking through a SNARE dependent-exocytosis mechanism on the surface of dendrites (Lan et al. 2001). Src has been reported to play a role in activation of the NMDAR by regulating its phosphorylation (Kalia et al. 2006; Xu et al. 2012). NRG1-ErbB4 signaling inhibits Src tyrosine kinase activity (Pitcher et al. 2011), which may lead to NMDAR hypofunction. Dysfunction of the NMDAR has been hypothesized to be relevant in schizophrenia (Weickert et al. 2012; Hamm et al. 2012), and contributes to cognitive dysfunction in schizophrenia patients (Pitcher et al. 2011). Enhanced NRG1-mediated ErbB4 signaling and subsequent decreased activation of NMDARs was found in postmortem human brains (Hahn et al. 2006). We have previously found that the NRG1 ICD is decreased in postmortem brains from schizophrenia patients (Marballi et al. 2012). Our current results strengthen the “Src link in schizophrenia” (Hahn 2011) and show that altering NRG1 intracellular cleavage may regulate SRC expression, contributing to NMDAR dysfunction. Given that NRG1 signaling was the top canonical pathway altered between Val/Val and Val/Leu groups, we were further interested in finding whether other kinases downstream of SRC and PKC were affected in V>L carriers. For this, we performed whole kinome profiling in cells treated with ErbB4, in order to stimulate release of the NRG1 ICD (Hancock et al. 2011). Kinome profiling revealed that phosphorylation of thirty five substrates was altered by ErbB4 treatment, with six of these involved in regulation of neurite formation: ADRB2, CREB1, ELK1, STMN2, REL, and NFkB1. Interestingly, 5 of the 6 peptides share a common Basic-Basic-X-Ser motif (Table 2), consistent with possible PKA or PKC involvement. Overall, within the neurite formation molecules, treatment with ErbB4 caused a significant decrease in phosphorylation in the Val/Val cell lines, but no differences in the Val/Leu group (Figure 3). This was significant in 4 out of the 6 peptides. This suggests that the ICD may be involved in regulating these phosphorylation events, which are blocked in the presence of the V>L variant. Decreases in dendritic spine density have been observed in schizophrenia patients (Jaaro-Peled et al. 2010) and in NRG1 type III-knockout mice (Chen et al. 2008). Importantly, the NRG1 V>L variant is reported to cause decreased dendrite formation in a mouse model (Chen et al. 2010) and our present results in humans validate these findings. Also, high levels of proinflammatory cytokines have been shown to decrease dendrite formation in vitro (Gilmore et al. 2004). Our current results suggest that these deficits in dendrite formation may be mediated by deficits in NRG1 intracellular cleavage, through inflammatory pathways and differential phosphorylation of neurite formation mediators. It is important to note that NFkB1, a key regulator of expression of pro-inflammatory cytokines (Tak, Firestein 2001), is one of the substrates altered in both expression and phosphorylation (Table 1 and Figure 3), supporting the genome-wide expression results of altered inflammatory pathways, and our previous findings of immune system dysregulation in carriers of the V>L variant (Marballi et al. 2010). Also of interest is that the phosphorylation of Elk1 is controlled by a downstream substrate of Srcasm, which in turn is a target of Src (Li et al. 2005). Given that SRC mRNA levels are decreased in Val/Leu carriers, this suggests a direct link between the V>L variant, SRC, and its downstream kinase activity.
Using the Kinexus database, we identified PKA, ERK and AKT as the putative upstream kinases that were altered in activity. PKA has been implicated in neurite growth and has been shown to interact with the MAPK pathway in regulating neuronal differentiation (Vogt Weisenhorn et al. 2001; Yao et al. 1998). ERK is a crucial regulator of neurite growth (Sarina et al. 2013; Auer et al. 2012) and is involved in maintaining balance between gliogenesis and neurogenesis during development (Chang et al. 2011). Protein Kinase C mediates activation of ERK-regulated dendritic spine density (Goldin, Segal 2003). The AKT pathway regulates dendritic size and complexity (Kumar et al. 2005). Importantly, PKC and SRC are regulators of both ERK and AKT (Puente et al. 2006; Uht et al. 2007; Hu et al. 2009; Lodeiro et al. 2009) and they also regulate neurite formation (Garcia et al. 2013; Liao et al. 2012; Zhao et al. 2009; Kotani et al. 2007). Taken together, our preliminary results suggest that the gene expression changes (PRKCH, SRC) we found in the presence of the V>L variant may lead to alterations in activity of PKA, ERK and AKT kinases, leading to deficits in dendrite formation via altered phosphorylation of neurite formation mediators. Integration of the transcriptome and kinome findings by IPA show that the main molecules altered by gene expression and phosphorylation are indeed part of a common pathway (Figure 4). The NRG1 ICD is released via ErbB4 treatment and migrates to the nucleus where it both represses and up regulates expression in a gene-specific context in neurons (Bao et al. 2003; Hancock et al 2011). Therefore, blocking ICD formation could potentially cause widespread effects on multiple pathways. Based on our results, we hypothesize that the NRG1 ICD may be responsible for regulating levels of kinases involved in phosphorylation of neurite formation mediators, and this may contribute to constitutive/fine control of dendrite formation. The NRG1 V>L variant has been shown inhibit ICD formation (Dejaegere et al. 2008) and to cause dendrite formation deficits (Chen et al. 2010). Given that the Val/Leu individuals in our study possess one mutant allele, the levels of ICD are likely decreased in these individuals and they do not respond properly to ErbB4 treatment. This could cause changes in kinase expression, such as PKC and SRC, leading to ultimately blocking changes in phosphorylation of neurite formation mediators.
Our study was limited in sample size based on the use of cells from individuals without a psychiatric diagnosis, and matched for age and gender. Therefore, we emphasize that the present results, while important, are preliminary and must be interpreted with caution. These results must be replicated using additional and larger cohorts. Furthermore, it is of immense importance to validate the putative targets by functional biochemical and/or molecular assays to understand specifically how the activity of the identified kinases is modified via NRG1 intracellular signaling. Whether the trend towards lower kinase activity in V/L vs. V/V cells is indeed due to disrupted NRG1 intracellular signalling, or due to inhibition of extracellular autocrine signalling is an important question that remains to be answered. However, this work is beyond the scope of the present study, which was meant to be exploratory and aimed to identify putative pathways regulated by NRG1 intracellular signalling. We encourage further studies focusing on the pathways identified here. Another limitation of the study is the use of lymphoblastoid cell lines. As is the case with immortalized cells, these cell lines have certain limitations such as differences in baseline growth rates, EBV copy numbers and ATP levels (Choy et al. 2008). However, the procedure for immortalization was carried out in an identical manner for both wild-type and mutation carriers and we did not observe significant differences in baseline growth rates or cell viability between groups. Due to the difficulty of obtaining neuronal cells from patients, LCLs have been used as a functional model system to study various mental disorders such as schizophrenia (Cheng et al. 2012) and autism (Granese et al. 2013). This reiterates their utility as an appropriate surrogate for brain tissue. A future line of study could involve use of iPS-derived neuronal cells from Val/Val and Val/Leu subjects to validate the findings obtained with LCLs.
In conclusion, we have performed an exploratory study to identify novel cell signaling targets of the NRG1 ICD using lymphoblastoid cell lines. While the small sample sizes we used is a limitation, this study is novel in that we have used two different global approaches, which led us to the same pathways altered, suggesting convergent validity for our findings. Genome wide expression studies showed alterations in expression of PRKCH and SRC (involved in NRG1 signaling) in the presence of the V>L variant. A novel kinomics approach identified altered phosphorylation of neurite formation mediators, downstream of PKC and Src kinases (Figure 4). We have for the first time demonstrated the use of kinomics in peripheral blood cells and integrated two high throughput techniques to examine the biological effects of the V>L mutation. These findings shed light on the role of the NRG1 ICD in cell signaling and ultimately its contribution to the development of schizophrenia.
Supplementary Material
Additional file Table S1 – Peptides differentially phosphorylated in Val/Val and Val/Leu cell lines
Acknowledgments
This work was supported in part by K01MH077777 and NARSAD: Brain and Behavior Research Foundation grants awarded to CWB; UT system grant: translational science training across disciplines awarded to KKM. The authors thank Dr. Teresa Johnson-Pais, Dr. Carolina Livi, Yasmin Ench and Mandy Rolando (Genomics core-University of Texas Health Science Center at San Antonio, UTHSCSA) and Dr. Christopher Willey and Dr. Joshua Anderson (Kinome core-University of Alabama, Birmingham) for their services. We thank the families from the CVCR; this research would not be possible without them.
Footnotes
Conflict of Interest
The authors declare they have no conflict of interest.
Contributor Information
Ketan K Marballi, Email: marballi@livemail.uthscsa.edu.
Robert E McCullumsmith, Email: smithrob@uab.edu.
Stefani Yates, Email: syates@uab.edu.
Michael A Escamilla, Email: m.escamilla@ttuhsc.edu.
Robin J Leach, Email: leach@uthscsa.edu.
Henriette Raventos, Email: hravento@racsa.co.cr.
Consuelo Walss-Bass, Email: walss@uthscsa.edu.
References
- Auer M, Schweigreiter R, Hausott B, Thongrong S, Holtje M, Just I, Bandtlow C, Klimaschewski L. Rho-independent stimulation of axon outgrowth and activation of the ERK and Akt signaling pathways by C3 transferase in sensory neurons. Frontiers in cellular neuroscience. 2012;6:43. doi: 10.3389/fncel.2012.00043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao J, Wolpowitz D, Role LW, Talmage DA. Back signaling by the Nrg-1 intracellular domain. The Journal of cell biology. 2003;161 (6):1133–1141. doi: 10.1083/jcb.200212085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CY, Ma KH, Wang JK, Tung YL, Chueh SH. Inhibition of protein kinase C promotes differentiation of neuroblastoma x glioma NG108–15 hybrid cells. The European journal of neuroscience. 2011;34 (7):1074–1084. doi: 10.1111/j.1460-9568.2011.07835.x. [DOI] [PubMed] [Google Scholar]
- Chen Y, Hancock ML, Role LW, Talmage DA. Intramembranous valine linked to schizophrenia is required for neuregulin 1 regulation of the morphological development of cortical neurons. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2010;30 (27):9199–9208. doi: 10.1523/JNEUROSCI.0605-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YJ, Johnson MA, Lieberman MD, Goodchild RE, Schobel S, Lewandowski N, Rosoklija G, Liu RC, Gingrich JA, Small S, Moore H, Dwork AJ, Talmage DA, Role LW. Type III neuregulin-1 is required for normal sensorimotor gating, memory-related behaviors, and corticostriatal circuit components. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2008;28 (27):6872–6883. doi: 10.1523/JNEUROSCI.1815-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng MC, Chuang YA, Lu CL, Chen YJ, Luu SU, Li JM, Hsu SH, Chen CH. Genetic and functional analyses of early growth response (EGR) family genes in schizophrenia. Progress in Neuropsychopharmacology and Biological Psychiatry. 2012;39 (1):149–155. doi: 10.1016/j.pnpbp.2012.06.004. [DOI] [PubMed] [Google Scholar]
- Choy E, Yelensky R, Bonakdar S, Plenge RM, Saxena R, De Jager PL, Shaw SY, Wolfish CS, Slavik JM, Cotsapas C, Rivas M, Dermitzakis ET, Cahir-McFarland E, Kieff E, Hafler D, Daly MJ, Altshuler D. Genetic analysis of human traits in vitro: drug response and gene expression in lymphoblastoid cell lines. PLoS Genetics. 2008;4 (11):e1000287. doi: 10.1371/journal.pgen.1000287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran C, Walker E, Huot R, Mittal V, Tessner K, Kestler L, et al. The stress cascade and schizophrenia: etiology and onset. Schizophrenia Bulletin. 2003;29(4):671–92. doi: 10.1093/oxfordjournals.schbul.a007038. [DOI] [PubMed] [Google Scholar]
- Corfas G, Roy K, Buxbaum JD. Neuregulin 1-erbB signaling and the molecular/cellular basis of schizophrenia. Nature neuroscience. 2004;7 (6):575–580. doi: 10.1038/nn1258. [DOI] [PubMed] [Google Scholar]
- Cruzalegui FH, Cano E, Treisman R. ERK activation induces phosphorylation of Elk-1 at multiple S/T-P motifs to high stoichiometry. Oncogene. 1999;18 (56):7948–7957. doi: 10.1038/sj.onc.1203362. [DOI] [PubMed] [Google Scholar]
- Davis S, Vanhoutte P, Pages C, Caboche J, Laroche S. The MAPK/ERK cascade targets both Elk-1 and cAMP response element-binding protein to control long-term potentiation-dependent gene expression in the dentate gyrus in vivo. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2000;20 (12):4563–4572. doi: 10.1523/JNEUROSCI.20-12-04563.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejaegere T, Serneels L, Schafer MK, Van Biervliet J, Horre K, Depboylu C, Alvarez-Fischer D, Herreman A, Willem M, Haass C, Hoglinger GU, D’Hooge R, De Strooper B. Deficiency of Aph1B/C-gamma-secretase disturbs Nrg1 cleavage and sensorimotor gating that can be reversed with antipsychotic treatment. Proceedings of the National Academy of Sciences of the United States of America. 2008;105 (28):9775–9780. doi: 10.1073/pnas.0800507105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk AJ, McCullumsmith RE, Haroutunian V, Meador-Woodruff JH. Abnormal activity of the MAPK- and cAMP-associated signaling pathways in frontal cortical areas in postmortem brain in schizophrenia. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 2012;37 (4):896–905. doi: 10.1038/npp.2011.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia L, Castillo C, Carballo J, Rodriguez Y, Forsyth P, Medina R, Martinez JC, Longart M. ErbB receptors and PKC regulate PC12 neuronal-like differentiation and sodium current elicitation. Neuroscience. 2013;236:88–98. doi: 10.1016/j.neuroscience.2013.01.026. [DOI] [PubMed] [Google Scholar]
- Gilmore JH, Fredrik Jarskog L, Vadlamudi S, Lauder JM. Prenatal infection and risk for schizophrenia: IL-1beta, IL-6, and TNFalpha inhibit cortical neuron dendrite development. Neuropsychopharmacology: official publication of the American College of Neuropsychopharmacology. 2004;29 (7):1221–1229. doi: 10.1038/sj.npp.1300446. [DOI] [PubMed] [Google Scholar]
- Glausier JR, Lewis DA. Dendritic spine pathology in schizophrenia. Neuroscience. 2012 doi: 10.1016/j.neuroscience.2012.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldin M, Segal M. Protein kinase C and ERK involvement in dendritic spine plasticity in cultured rodent hippocampal neurons. The European journal of neuroscience. 2003;17 (12):2529–2539. doi: 10.1046/j.1460-9568.2003.02694.x. [DOI] [PubMed] [Google Scholar]
- Granese B, Scala I, Spatuzza C, Valentino A, Coletta M, Vacca RA, De Luca P, Andria G. Validation of microarray data in human lymphoblasts shows a role of the ubiquitin-proteasome system and NF-kB in the pathogenesis of Down syndrome. BMC Medical Genomics. 2013;6:24. doi: 10.1186/1755-8794-6-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood TA, Light GA, Swerdlow NR, Radant AD, Braff DL. Association analysis of 94 candidate genes and schizophrenia-related endophenotypes. PloS one. 2012;7 (1):e29630. doi: 10.1371/journal.pone.0029630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn CG. A Src link in schizophrenia. Nature medicine. 2011;17 (4):425–427. doi: 10.1038/nm0411-425. [DOI] [PubMed] [Google Scholar]
- Hahn CG, Wang HY, Cho DS, Talbot K, Gur RE, Berrettini WH, Bakshi K, Kamins J, Borgmann-Winter KE, Siegel SJ, Gallop RJ, Arnold SE. Altered neuregulin 1-erbB4 signaling contributes to NMDA receptor hypofunction in schizophrenia. Nature medicine. 2006;12 (7):824–828. doi: 10.1038/nm1418. [DOI] [PubMed] [Google Scholar]
- Hamm JP, Gilmore CS, Clementz BA. Augmented gamma band auditory steady-state responses: support for NMDA hypofunction in schizophrenia. Schizophrenia research. 2012;138 (1):1–7. doi: 10.1016/j.schres.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock ML, Canetta SE, Role LW, Talmage DA. Presynaptic type III neuregulin1-ErbB signaling targets {alpha}7 nicotinic acetylcholine receptors to axons. The Journal of cell biology. 2008;181 (3):511–521. doi: 10.1083/jcb.200710037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock ML, Nowakowski DW, Role LW, Talmage DA, Flanagan JG. Type III neuregulin 1 regulates pathfinding of sensory axons in the developing spinal cord and periphery. Development. 2011;138 (22):4887–4898. doi: 10.1242/dev.072306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison PJ, Law AJ. Neuregulin 1 and schizophrenia: genetics, gene expression, and neurobiology. Biological psychiatry. 2006;60 (2):132–140. doi: 10.1016/j.biopsych.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Hu X, Wu X, Xu J, Zhou J, Han X, Guo J. Src kinase up-regulates the ERK cascade through inactivation of protein phosphatase 2A following cerebral ischemia. BMC neuroscience. 2009;10:74. doi: 10.1186/1471-2202-10-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde R, Corkins ME, Somers GA, Hart AC. PKC-1 acts with the ERK MAPK signaling pathway to regulate Caenorhabditis elegans mechanosensory response. Genes, brain, and behavior. 2011;10 (3):286–298. doi: 10.1111/j.1601-183X.2010.00667.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaaro-Peled H, Ayhan Y, Pletnikov MV, Sawa A. Review of pathological hallmarks of schizophrenia: comparison of genetic models with patients and nongenetic models. Schizophrenia bulletin. 2010;36 (2):301–313. doi: 10.1093/schbul/sbp133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarboe JS, Jaboin JJ, Anderson JC, Nowsheen S, Stanley JA, Naji F, Ruijtenbeek R, Tu T, Hallahan DE, Yang ES, Bonner JA, Willey CD. Kinomic profiling approach identifies Trk as a novel radiation modulator. Radiotherapy and oncology : journal of the European Society for Therapeutic Radiology and Oncology. 2012;103 (3):380–387. doi: 10.1016/j.radonc.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalia LV, Pitcher GM, Pelkey KA, Salter MW. PSD-95 is a negative regulator of the tyrosine kinase Src in the NMDA receptor complex. The EMBO journal. 2006;25 (20):4971–4982. doi: 10.1038/sj.emboj.7601342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanakry CG, Li Z, Nakai Y, Sei Y, Weinberger DR. Neuregulin-1 regulates cell adhesion via an ErbB2/phosphoinositide-3 kinase/Akt-dependent pathway: potential implications for schizophrenia and cancer. PloS one. 2007;2 (12):e1369. doi: 10.1371/journal.pone.0001369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keri S, Seres I, Kelemen O, Benedek G. Neuregulin 1-stimulated phosphorylation of AKT in psychotic disorders and its relationship with neurocognitive functions. Neurochemistry international. 2009;55 (7):606–609. doi: 10.1016/j.neuint.2009.06.002. [DOI] [PubMed] [Google Scholar]
- Kotani T, Morone N, Yuasa S, Nada S, Okada M. Constitutive activation of neuronal Src causes aberrant dendritic morphogenesis in mouse cerebellar Purkinje cells. Neuroscience research. 2007;57 (2):210–219. doi: 10.1016/j.neures.2006.10.007. [DOI] [PubMed] [Google Scholar]
- Kumar V, Zhang MX, Swank MW, Kunz J, Wu GY. Regulation of dendritic morphogenesis by Ras-PI3K-Akt-mTOR and Ras-MAPK signaling pathways. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2005;25 (49):11288–11299. doi: 10.1523/JNEUROSCI.2284-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyosseva SV, Elbein AD, Griffin WS, Mrak RE, Lyon M, Karson CN. Mitogen-activated protein kinases in schizophrenia. Biological psychiatry. 1999;46 (5):689–696. doi: 10.1016/s0006-3223(99)00104-3. [DOI] [PubMed] [Google Scholar]
- Lan JY, Skeberdis VA, Jover T, Grooms SY, Lin Y, Araneda RC, Zheng X, Bennett MV, Zukin RS. Protein kinase C modulates NMDA receptor trafficking and gating. Nature neuroscience. 2001;4 (4):382–390. doi: 10.1038/86028. [DOI] [PubMed] [Google Scholar]
- Li W, Marshall C, Mei L, Dzubow L, Schmults C, Dans M, Seykora J. Srcasm modulates EGF and Src-kinase signaling in keratinocytes. Journal of Biological Chemistry. 2005;280 (7):6036–6046. doi: 10.1074/jbc.M406546200. [DOI] [PubMed] [Google Scholar]
- Liao KK, Wu MJ, Chen PY, Huang SW, Chiu SJ, Ho CT, Yen JH. Curcuminoids promote neurite outgrowth in PC12 cells through MAPK/ERK- and PKC-dependent pathways. Journal of agricultural and food chemistry. 2012;60 (1):433–443. doi: 10.1021/jf203290r. [DOI] [PubMed] [Google Scholar]
- Lodeiro M, Theodoropoulou M, Pardo M, Casanueva FF, Camina JP. c-Src regulates Akt signaling in response to ghrelin via beta-arrestin signaling-independent and -dependent mechanisms. PloS one. 2009;4 (3):e4686. doi: 10.1371/journal.pone.0004686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marballi K, Cruz D, Thompson P, Walss-Bass C. Differential neuregulin 1 cleavage in the prefrontal cortex and hippocampus in schizophrenia and bipolar disorder: preliminary findings. PloS one. 2012;7 (5):e36431. doi: 10.1371/journal.pone.0036431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marballi K, Quinones MP, Jimenez F, Escamilla MA, Raventos H, Soto-Bernardini MC, Ahuja SS, Walss-Bass C. In vivo and in vitro genetic evidence of involvement of neuregulin 1 in immune system dysregulation. Journal of molecular medicine. 2010;88 (11):1133–1141. doi: 10.1007/s00109-010-0653-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei L, Xiong WC. Neuregulin 1 in neural development, synaptic plasticity and schizophrenia. Nature reviews Neuroscience. 2008;9 (6):437–452. doi: 10.1038/nrn2392. d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrie RJ, Zhao B, Bedford F, Lamarche-Vane N. Compartmentalized DCC signalling is distinct from DCC localized to lipid rafts. Biology of the cell / under the auspices of the European Cell Biology Organization. 2009;101 (2):77–90. doi: 10.1042/BC20070108. [DOI] [PubMed] [Google Scholar]
- Pitcher GM, Kalia LV, Ng D, Goodfellow NM, Yee KT, Lambe EK, Salter MW. Schizophrenia susceptibility pathway neuregulin 1-ErbB4 suppresses Src upregulation of NMDA receptors. Nature medicine. 2011;17 (4):470–478. doi: 10.1038/nm.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poluch S, Juliano SL. A normal radial glial scaffold is necessary for migration of interneurons during neocortical development. Glia. 2007;55 (8):822–830. doi: 10.1002/glia.20488. [DOI] [PubMed] [Google Scholar]
- Potvin S, Stip E, Sepehry AA, Gendron A, Bah R, Kouassi E. Inflammatory cytokine alterations in schizophrenia: a systematic quantitative review. Biological psychiatry. 2008;63 (8):801–808. doi: 10.1016/j.biopsych.2007.09.024. [DOI] [PubMed] [Google Scholar]
- Puente LG, He JS, Ostergaard HL. A novel PKC regulates ERK activation and degranulation of cytotoxic T lymphocytes: Plasticity in PKC regulation of ERK. European journal of immunology. 2006;36 (4):1009–1018. doi: 10.1002/eji.200535277. [DOI] [PubMed] [Google Scholar]
- Sarina, Yagi Y, Nakano O, Hashimoto T, Kimura K, Asakawa Y, Zhong M, Narimatsu S, Gohda E. Induction of neurite outgrowth in PC12 cells by artemisinin through activation of ERK and p38 MAPK signaling pathways. Brain research. 2013;1490:61–71. doi: 10.1016/j.brainres.2012.10.059. [DOI] [PubMed] [Google Scholar]
- Shahaf G, Rotem-Dai N, Koifman G, Raveh-Amit H, Frost SA, Livneh E. PKCeta is a negative regulator of AKT inhibiting the IGF-I induced proliferation. Experimental cell research. 2012;318 (7):789–799. doi: 10.1016/j.yexcr.2012.01.018. [DOI] [PubMed] [Google Scholar]
- Shamir A, Kwon OB, Karavanova I, Vullhorst D, Leiva-Salcedo E, Janssen MJ, Buonanno A. The importance of the NRG-1/ErbB4 pathway for synaptic plasticity and behaviors associated with psychiatric disorders. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2012;32 (9):2988–2997. doi: 10.1523/JNEUROSCI.1899-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefansson H, Sigurdsson E, Steinthorsdottir V, Bjornsdottir S, Sigmundsson T, Ghosh S, Brynjolfsson J, Gunnarsdottir S, Ivarsson O, Chou TT, Hjaltason O, Birgisdottir B, Jonsson H, Gudnadottir VG, Gudmundsdottir E, Bjornsson A, Ingvarsson B, Ingason A, Sigfusson S, Hardardottir H, Harvey RP, Lai D, Zhou M, Brunner D, Mutel V, Gonzalo A, Lemke G, Sainz J, Johannesson G, Andresson T, Gudbjartsson D, Manolescu A, Frigge ML, Gurney ME, Kong A, Gulcher JR, Petursson H, Stefansson K. Neuregulin 1 and susceptibility to schizophrenia. American journal of human genetics. 2002;71 (4):877–892. doi: 10.1086/342734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strous RD, Shoenfeld Y. Schizophrenia, autoimmunity and immune system dysregulation: a comprehensive model updated and revisited. Journal of autoimmunity. 2006;27 (2):71–80. doi: 10.1016/j.jaut.2006.07.006. [DOI] [PubMed] [Google Scholar]
- Tak PP, Firestein GS. NF-kappaB: a key role in inflammatory diseases. The Journal of clinical investigation. 2001;107 (1):7–11. doi: 10.1172/JCI11830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talmage DA. Mechanisms of neuregulin action. Novartis Foundation symposium. 2008;289:74–84. doi: 10.1002/9780470751251.ch6. discussion 84–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taveggia C, Thaker P, Petrylak A, Caporaso GL, Toews A, Falls DL, Einheber S, Salzer JL. Type III neuregulin-1 promotes oligodendrocyte myelination. Glia. 2008;56 (3):284–293. doi: 10.1002/glia.20612. [DOI] [PubMed] [Google Scholar]
- Tosato S, Dazzan P, Collier D. Association between the neuregulin 1 gene and schizophrenia: a systematic review. Schizophrenia bulletin. 2005;31 (3):613–617. doi: 10.1093/schbul/sbi043. [DOI] [PubMed] [Google Scholar]
- Uht RM, Amos S, Martin PM, Riggan AE, Hussaini IM. The protein kinase C-eta isoform induces proliferation in glioblastoma cell lines through an ERK/Elk-1 pathway. Oncogene. 2007;26 (20):2885–2893. doi: 10.1038/sj.onc.1210090. [DOI] [PubMed] [Google Scholar]
- Velanac V, Unterbarnscheidt T, Hinrichs W, Gummert MN, Fischer TM, Rossner MJ, Trimarco A, Brivio V, Taveggia C, Willem M, Haass C, Mobius W, Nave KA, Schwab MH. Bace1 processing of NRG1 type III produces a myelin-inducing signal but is not essential for the stimulation of myelination. Glia. 2012;60 (2):203–217. doi: 10.1002/glia.21255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt Weisenhorn DM, Roback LJ, Kwon JH, Wainer BH. Coupling of cAMP/PKA and MAPK signaling in neuronal cells is dependent on developmental stage. Experimental neurology. 2001;169 (1):44–55. doi: 10.1006/exnr.2001.7651. [DOI] [PubMed] [Google Scholar]
- Walss-Bass C, Liu W, Lew DF, Villegas R, Montero P, Dassori A, Leach RJ, Almasy L, Escamilla M, Raventos H. A novel missense mutation in the transmembrane domain of neuregulin 1 is associated with schizophrenia. Biological psychiatry. 2006;60 (6):548–553. doi: 10.1016/j.biopsych.2006.03.017. [DOI] [PubMed] [Google Scholar]
- Weickert CS, Tiwari Y, Schofield PR, Mowry BJ, Fullerton JM. Schizophrenia-associated HapICE haplotype is associated with increased NRG1 type III expression and high nucleotide diversity. Translational Psychiatry. 2012;2:e104. doi: 10.1038/tp.2012.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Pan Y, Zhu Q, Gong S, Tao J, Xu GY, Jiang X. Arcuate Src activation-induced phosphorylation of NR2B NMDA subunit contributes to inflammatory pain in rats. Journal of neurophysiology. 2012;108 (11):3024–3033. doi: 10.1152/jn.01047.2011. [DOI] [PubMed] [Google Scholar]
- Yao H, York RD, Misra-Press A, Carr DW, Stork PJ. The cyclic adenosine monophosphate-dependent protein kinase (PKA) is required for the sustained activation of mitogen-activated kinases and gene expression by nerve growth factor. The Journal of biological chemistry. 1998;273 (14):8240–8247. doi: 10.1074/jbc.273.14.8240. [DOI] [PubMed] [Google Scholar]
- Zhao H, Cao X, Wu G, Loh HH, Law PY. Neurite outgrowth is dependent on the association of c-Src and lipid rafts. Neurochemical research. 2009;34 (12):2197–2205. doi: 10.1007/s11064-009-0016-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file Table S1 – Peptides differentially phosphorylated in Val/Val and Val/Leu cell lines