Abstract
At the Center for High-Throughput Minimally Invasive Radiation Biodosimetry we have developed a Rapid Automated Biodosimetry Tool (RABiT); this is a completely automated, ultra-high throughput robotically-based biodosimetry workstation designed for use following a large scale radiological event, to perform radiation biodosimetry measurements based on a fingerstick blood sample. High throughput is achieved through purpose built robotics, sample handling in filter-bottomed multi-well plates and innovations in high speed imaging and analysis. Currently, we are adapting the RABiT technologies for use in laboratory settings, for applications in epidemiological and clinical studies. Our overall goal is to extend the RABiT system to directly measure the kinetics of DNA repair proteins. The design of the kinetic/time dependent studies is based on repeated, automated sampling of lymphocytes from a central reservoir of cells housed in the RABiT incubator as a function of time after the irradiation challenge. In the present study, we have characterized the DNA repair kinetics of the following repair proteins: γ-H2AX, 53-BP1, ATM kinase, MDC1 at multiple times (0.5, 2, 4, 7, 24 hours) after irradiation with 4 Gy γ rays. In order to provide a consistent dose exposure at time zero, we have developed an automated capillary irradiator to introduce DNA DSBs into fingerstick-size blood samples within the RABiT. To demonstrate the scalability of the laboratory-based RABiT system, we have initiated a population study using γ-H2AX as a biomarker.
Keywords: ionizing radiation, DNA repair kinetics, human lymphocytes, high throughput, radiation sensitivity
Introduction
The Columbia University Rapid Automated Biodosimetry Tool (RABiT) developed at the Center for High-Throughput Minimally Invasive Radiation Biodosimetry (CHTMIRB), is a fully-automated, ultra-high throughput robotically-based biodosimetry workstation designed for use after small or large scale radiological event to quickly determine individual dose estimates (Chen et al. 2009; Chen et al. 2010; Garty et al. 2010). Analysis is based on a single fingerstick of blood. The RABiT has been designed to automate two well-established manually-based radiation biodosimetry assays, the gamma H2AX assay (Nakamura et al. 2006; Turner et al. 2011) and the micronucleus assay (Fenech et al. 2003; Fenech 2010; Lyulko et al. 2013). High-throughput is achieved through purpose built robotics, sample handling of isolated lymphocytes in filter-bottomed multi-well plates and innovations in high speed imaging (Garty et al. 2010; Garty et al. 2011; Turner et al. 2011). Recent system upgrades include a new robotics system for the parallel processing of four capillaries at once, hardware and laser upgrades which serve to increase the throughput of the system (Garty et al. 2011; Chen et al. 2012) as well as an additional module for measuring lymphocyte yields (Xu et al 2013).
The γ-H2AX assay is a measure of DNA double-strand breaks (DSB) induced by ionizing radiation, and it has a highly linear relationship with dose (Rothkamm and Lobrich 2003; Turner et al. 2011). It quantifies, through immune-staining, the phosphorylated H2AX histone which localizes to DSBs. The yield of phosphorylated γ-H2AX can be quantified by measuring the integrated fluorescent intensity per nucleus (Turner et al. 2011). However, the current RABiT system can assay only one DNA repair protein (typically, γ-H2AX), and at only one time point, as dictated by sample processing prior to loading into the RABiT. The focus of the present work is to extend the application of the RABiT system to immunoassay multiple DNA repair proteins at specific time-points after irradiation challenge to directly measure DNA repair kinetics from a single fingerstick blood sample. This is motivated both by clinical as well as academic desires to study inter-human variation in DNA repair (Collins and Azqueta 2012; Ivashkevich et al. 2012).
To adapt the RABiT system to perform multiple immunohistochemical measurements as a function of time, we have introduced a 96-well culture plate into the assay protocol to serve as a central reservoir for the incubation of the blood lymphocyte samples. At specific time points after irradiation exposure, equal volume aliquots of cell suspension are transferred into a filter-bottomed 96-well plate for immunohistochemical analysis. In previous work, we have developed a Quantitative Light Absorption Analysis (QLAA) method (Xu et al. 2013), to rapidly estimate the number of lymphocytes from small volumes of blood (~10 to 30 μl). In that study, we determined that the lymphocyte counts in finger stick samples collected from 17 healthy volunteers, was in the range 2000-6000 lymphocytes per μl. Thus, a 30 μl blood sample will contain more than enough lymphocytes (~60,000-180,000) for repeat sampling following irradiation exposure.
To provide rapid, automated irradiation within the RABiT, we have built a compact capillary irradiator which will allow the introduction of double strand breaks (DSBs) into the capillary blood sample. The capillary irradiator prototype is based on commercial 90Sr sealed disk sources and has been tested using LiF:Mg,Ti thermoluminescent detector (TLD) rods.
In order to facilitate assay development without the need to reconfigure the RABiT for each experimental study, we have duplicated the major components of the RABiT workstation in the CHTMIRB laboratory. This allows for repeat sampling, in order to characterize double strand break (DSB) DNA repair kinetics of different DNA repair biomarkers, while maintaining high throughput. The four DSB proteins that were studied are: γ-H2AX, ATM kinase (phosphor ser1981), 53BP1 (p-53 binding protein 1), and MDC1 (mediator of DNA damage checkpoint 1) (Rogakou et al. 1998; Schultz et al 2000; Wilson et al. 2010; Jungmichel and Stucki 2010). These DNA repair proteins are known to respond rapidly to ionizing radiation, starting within ~ 3-15 minutes, accumulating in the vicinity of the break site and forming radiation-induced foci (Costes et al. 2010; Hable et al. 2012). Typically, maximum levels of foci are observed within ~30 min of irradiation (Sedelnikova et al. 2003; Bekker-Jensen et al. 2000; Schultz et al. 2000; Andegeko et al. 2001). After the maximum, the number of irradiation-induced foci decline rapidly (characteristic time ~ 1 hour), followed by a slower loss and decay rate over hours or days (Ugenskiene et al. 2009; Kuhne et al. 2004; Rothkamm and Horn 2009). In some scenarios the yield returns essentially to background levels, whilst in others it remains elevated for several days (Banath et al. 2004; Redon et al 2009). Under appropriate conditions, these disappearance kinetics can reflect the efficiency of DSB repair (Kuhne et al. 2004; Löbrich et al. 2005; Bouquet et al. 2006; Markova et al. 2007; Rube et al. 2008), though there is probably not a precise one-toone correlation between DSB yields and expression of any of these repair proteins (Kinner et al. 2008).
In the present work, fingerstick samples were collected using a heparinized capillary tube from multiple healthy volunteers (of no specific gender or age) and irradiated to 4 Gy with γ rays using an external Cs-137 source. Post-irradiation yields of the γ-H2AX, ATM [ser1981], 53BP1, and MDC1 radiation-induced foci were measured at 0.5, 2, 4, 7 and 24 hours using immunohistochemical assays. To validate the laboratory-based RABiT system for high-throughput measurements of DSB repair capacity, we have initiated a population study using γ-H2AX as the repair biomarker. Variations in the γ-H2AX decay rate and repair efficiency up to 24 hours post-irradiation are presented.
Materials and methods
Automated Capillary Irradiator
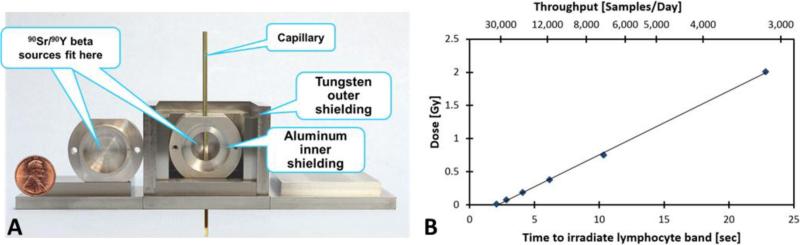
In order to adapt the RABiT to perform DNA repair kinetics one of the first challenges was to design and build a compact capillary irradiator that can deliver a repeatable dose to each capillary blood sample, inducing a known number of DSBs per lymphocyte. Because we wish to irradiate only a short section of each capillary tube (2 mm diameter x 75 mm length of which the lymphocyte band is ~ 1-3 mm thick), on a sequential basis, the irradiator can be made very compact. The tight geometry allows the use of a relatively weak β-emitter, which in turn minimizes the required shielding. Figure 1A, shows the compact capillary irradiator prototype based on two 100 mCi 90Sr/90Y 1” sealed disk sources (Eckert & Zeigler Isotope Products Inc., Valencia, CA). The sources are placed opposite each other with a 1 cm gap providing a uniform dose field, such that the capillary can pass between the sources and is irradiated from both sides. The shielding consists of three concentric housings, with the inner housing made of aluminum, the intermediate housing made of interlocking tungsten plates and the outer housing made of aluminum to lock the tungsten plates in place. This design is ideal for shielding β sources, which require a low Z shielding to minimize the formation of Bremsstrahlung X-rays, surrounded by an external high Z shielding to absorb those X-rays that are formed.
Figure 1.
Compact capillary irradiator. The capillary tube is robotically inserted into the small shielded cavity containing two 100 mCi 90Sr/90Y 1” disk radioactive sources, such that the lymphocyte band will be in centered between the sources (Panel A). The capillary will stay at a position where the estimated dose rate is ~ 6 Gy/min to receive the desired dose. Dose measurements are presented as a function of total time to irradiate 0.5” of the capillary. The upper axis denotes estimated throughput, based on 20 hours of operation per day (Panel B).
Initial dosimetry was performed with LiF:Mg,Ti thermo luminescent detector rods (TLD100, #SNO10106; Thermo Fisher Scientific, Pittsburgh, PA). Four 1 mm diameter x 6 mm long rods were loaded into each capillary and passed through the irradiation system at various speeds. Figure 1B denotes the doses experienced by the average of four rods as a function of irradiation time (the time to load a capillary such that the lymphocyte band is centered on the source, irradiate a 0.5” segment to the prescribed dose and unload the capillary). The actuator used to push the capillary through the irradiator (Digit Linear Actuator, Ultra Motion Inc., Cutchogue, NY) requires a minimum of 2 sec to load and unload each capillary. A secondary axis translates this speed to irradiation throughput, assuming 20 h of operation per day. The response of the TLD100 rods was calibrated using 250kVp X-rays.
Blood Collection and Sample Preparation
Fingerstick blood samples were collected in heparin coated capillaries (Safe-T-Fill capillaries; RAM Scientific Inc., Yonkers, NY) from multiple healthy individuals (males and females aged 19 to 50) using a finger stick lancet (BD Microtainer® Contact-Activated Lancet, #366594, Franklin Lakes, NJ). Informed consent was obtained from all volunteers according to IRB protocol AAAF3516. Following blood collection, the blood-filled capillaries were sealed using Hemato-SealTM tube sealing compound (Fisher Scientific) and placed into a 50 ml conical centrifuge tube and ThermoSafeTM Blood transporter (Fisher Scientific) for transport from the Radiation Oncology clinic to the CHTMIRB laboratory (transit time typically ~ 5-10 minutes). For sample preparation, each donor blood sample (30 μl) was pipetted into heparin-coated PVC capillaries containing 50 μl of lymphocyte separation medium (Histopaque-1083; Invitrogen, Eugene, OR) and sealed using Hemato-Seal™ tube sealing compound. Each blood-filled capillary was irradiated with γ rays to a dose of 4 Gy using a Gammacell 40 Cesium unit (Atomic Energy of Canada, Ltd.). After irradiation, the blood samples are spun at 3750 RPM for 5 min to form a distinct lymphocyte band at the interface between the blood plasma and separation medium. At the Class II Biological Safety Cabinet, the capillaries were cut 5-6 mm below the isolated band and the lymphocyte cells released into a sterile 96-well Eppendorf® Microplate (MTP 96/U-bottom; #951040081, Eppendorf, Hauppauge, NY) containing 170 μl pre-warmed RPMI 1640 culture medium supplemented with 10% FBS and 2% Pen/Strep (all reagents from Invitrogen). The multi-well culture plate was then placed into the incubator (37°C, 5% CO2 at 95% humidity). For each donor sample, equal volume aliquots (~ 20 μl) of lymphocyte cell suspension were transferred from the multi-well plate to a 96-well filter-bottomed multi-well plate (HTS Solubility Filter Plates with polycarbonate filters; Millipore, Billerica, MA) making up the volume to 100 μl with RPMI 1640 culture medium. Lymphocytes were transferred at 0.5 hr, 2 hr, 4 hr, 7 hr and 24 hours post irradiation using an Eppendorf® 12-channel pipetter.
Liquid handling
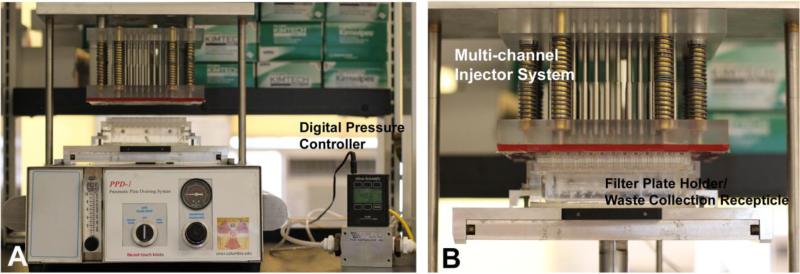
Key to duplicating the RABiT workstation modules for the laboratory was the development of our custom-designed bench-top 96-well plate Pneumatic Plate Draining (PPD) system (Figure 2) that is able to simulate the RABiT’s ability to filter fluid through the 96-well polycarbonate filter plates. The PPD uses a multi-channel injector system, designed to maintain highly uniform positive pressure and uniform air volume dispensed into each individual well of a filter bottom 96 well plate. A sealing gasket is mounted on the bottom of the injector device, serving as an airtight seal between the injector device and the plate. The 96-well plate holder and waste collection receptacle is composed of an acrylic box that can accommodate a standard 96 well plate and brackets used to press the injector device to 96 well plates. The pressure input manifold (not shown) consists of a compressed gas cylinder, regulator, pressure gauge and filtration system gas valve. A digital pressure controller (PCR-100PSIG-D, Alicat Scientific, Tucson, AZ), is used to precisely measure the gas flow into the PPD system as well as calibrate the system (Panel A).
Figure 2.
Pneumatic Plate Draining (PPD). The PPD system (Panel A) comprises of 2 main parts: a) multi-channel injector system and b) plate holder/waste collection receptacle (Magnified in Panel B). The digital pressure controller precisely measures the gas flow into the PPD System.
PPD Calibration and Plate Preparation
Using the digital pressure controller, the PPD system is calibrated for pressure and gas flow with a dry, empty 96 multi-well filter plate. When the empty 96 filter plate is in contact with the sealing gasket (Figure 2, Panel B), the pressure range on the digital pressure controller should read 22 ± 3 PSI. This is the pre-defined range which has been determined to filter 80-100 μl total media volume in each of the micro-wells without compromising the quality of the loaded cells. Prior to cell transfer, each filter-bottomed plate is pre-conditioned by loading and draining 100 μl of 25% methanol. This aids filtering through the polycarbonate membranes as well as serving as confirmation that all 96-channel injectors and micro-wells are functional.
Immunolabeling
Following the transfer of the cells to the filter plate, the cells are washed two times with PBS (100 μl) and fixed with ice-cold methanol for 10 minutes (100 μl) at −20°C. The lymphocyte cells are blocked with 3% bovine serum albumin (BSA; 100 μl) and exposed to one of the following DNA repair protein antibodies for 1 h at room temperature: γ-H2AX (mouse monoclonal, dilution 1:750 #ab18311; Abcam Inc., Cambridge, MA), ATMphospho Ser 1981(mouse monoclonal, dilution 1:500 #ab36810; Abcam Inc.), 53BP1 (rabbit polyclonal, dilution 1:250 #NB100-904, Novus Biologicals, Littleton, CO) or MDC1 (mouse monoclonal, dilution 1:300 #M2444, Sigma-Aldrich Corp. St. Louis, MO). Next, the cells were washed five times with PBS. To visualize the formed foci, the lymphocytes were exposed to a goat anti-mouse or anti-rabbit Alexa Fluor 555 secondary antibodies (dilution 1:1000; Invitrogen) for 50 min, after which time, the cells were subsequently washed five times with PBS. After the final PBS wash, the removable plate underdrain on the 96-well plate is peeled off to expose the polycarbonate membranes. Once the polycarbonate membranes are dry, the polycarbonate membranes are transferred to a transparent, low-fluorescence adhesive film (Clear View™ long lasting packaging tape; Staples, Framingham, MA). This is done by applying the tape to the bottom of the exposed filters and gently/forcefully detached from the plate. Individually attached polycarbonate membranes were mounted onto Fisherbrand Superfrost® Plus slides, counterstained using Vectashield® mounting medium with DAPI (Vector Laboratories, Burlingame, CA) and sealed with a Fisherfinest Premium cover glass slide.
Automated Image Capture
Fluorescent images of DAPI labeled nuclei and AF555-labeled γ-H2AX foci were captured separately for each dose using a 60x oil immersion objective and stored as 16-bit grayscale tiff files using an automated imaging system: Olympus BX-43 Fluorescent Microscope (Olympus, Center Valley, PA) with a Lumen Dynamics X-Cite 120Q Fluorescent Lamp (Lumen Dynamics, Mississauga, Ontario), Prior Proscan III Controller with automated H101A stage and HF110 filter wheels (Prior Scientific, Rockland, MA), Olympus DAPI (U-3N49000) filters, Olympus Cy3 (U-3N49004) filters, and Chroma DAPI/FITC/CY3 (69000BS) Trichroic Mirrors (Chroma, Bellows Falls, VT). Images were captured using a 12 bit Hamamatsu ORCA camera (#C4742-12ERG; Hamamatsu Photonics, Hamamatsu City, Japan)
The automated imaging system is controlled by ImageJ (Rasband 1997-2012) software with the μManager Plugin (Edelstein et al. 2010). Automation was achieved using the μManager Slide Explorer to scan a large portion of a single polycarbonate well for DAPI-stained nuclei. The “Point” tool was used to select a list of multiple points of interest (PoI). The list of PoIs is then exported to the multidimensional acquisition (MDA) to define the acquisition setup for dual channel capture; where the first channel, DAPI was set for 10 ms exposure, and the second channel Cy3 was set for 50 ms exposure with autofocus (AF) performed for each DAPI frame. Thus, the final automated acquisition process at each PoI for each pair of images is: AF DAPI stained nuclei, capture DAPI image, switch to Cy3 filter, and capture image of Cy3 labeled foci. For each PoI, the paired DAPI and Cy3 images were analyzed with our custom-designed software (FluorQuant, Turner et al. 2011) to measure the total fluorescence yields within each cell nucleus. For data acquisition, an average number of 250 paired lymphocyte images were captured and analyzed per data point.
Results
High-Speed Image Analysis
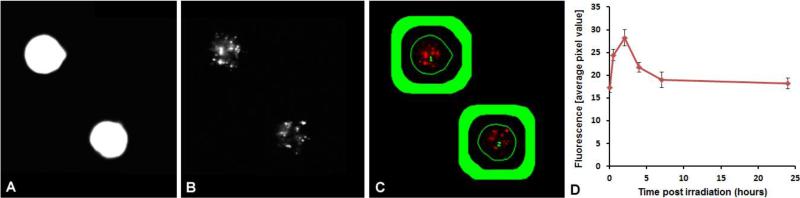
Figure 3 shows a representation of γ-H2AX fluorescence analysis using our FluorQuant software for quantitative fluorescence. The automated imaging system captures paired images of DAPI-labeled (Panel A) and immunofluorescent foci (Panel B), in this case, using a 60x oil immersion objective. Using the FluorQuant software, each image of the nuclei is filtered, binarized, and the boundaries of each cell nucleus identified. Panel C shows an overlay image where the thin green line is the outline of the nucleus shown in Panel A. Background brightness is evaluated from an area surrounding the nucleus, but far enough from it to exclude nonspecific staining at the nuclear boundary (Green annulus in panel C). This annulus was empirically selected with a thickness and gap from the nuclear boundaries of 20 pixels (~ 2 μm) each. For each nucleus matching appropriate morphological criteria (area, elongation, compactness etc’), the fluorescent intensity within each nuclear boundary is integrated and the background is subtracted. The data are saved in Excel format and are plotted as average total fluorescence pixel values against the time post-irradiation in hours (Panel D). Presented here are the mean γ-H2AX responses of DNA repair kinetics in finger stick samples collected from 30 donors (8 male and 22 female) and exposed to 4 Gy γ rays. The data show that radiation-induced γ-H2AX foci rapidly increased within 30 minutes and reached a maximum by ~2 h, after which time there was fast decline in γ-H2AX total fluorescence by 7 hours, followed by a much slower rate of disappearance up to 24 hours. The error bars represent the standard error mean (SEM) based on inter-individual variation among the donors.
Figure 3. Automated Imaging and Scoring of γ-H2AX Fluorescence.
Paired images of DAPI-labeled nuclei (Panel A) and γ-H2AX foci (Panel B) captured using a 60x oil immersion objective integrated over the nuclear area (thin line) with the area covered by the thick line used for background subtraction (Panel C). Averaged plot from 30 donors of γ-H2AX fluorescence after 4 Gy of γ rays (Panel D). Error bars show ± SEM.
DNA repair kinetics
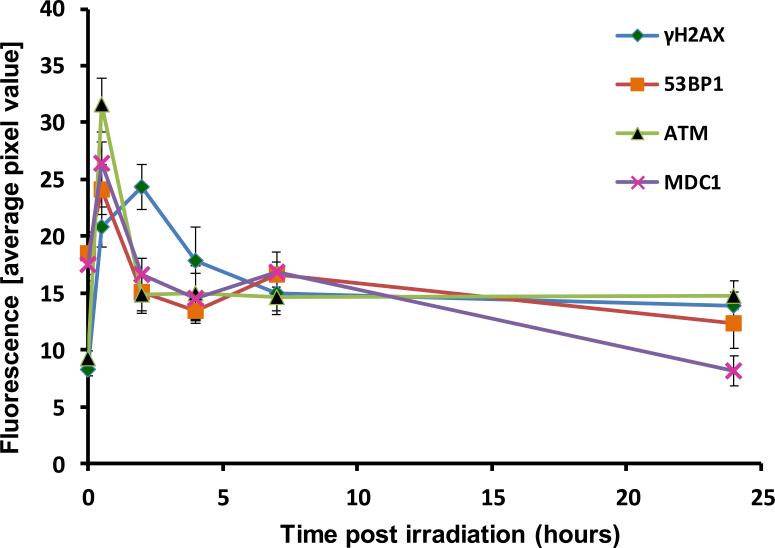
The DNA repair profiles for the four DNA repair proteins, γ-H2AX, 53BP1, MDC1 and ATM[ser1981] are shown in Figure 4. The results show a rapid assembly/recruitment of irradiation-induced foci for all 4 repair proteins at 30 minutes after irradiation with γ-H2AX yields again, peaking by 2 hours. The dissociation kinetics for all 4 biomarkers indicated a fast and slow rate of repair up to 24 h post-irradiation exposure. Twenty four hours post exposure, residual ATM [ser1981] and γ-H2AX foci are apparent that are above background, preirradiation levels, whereas 53BP1 and MDC1 yields have appeared to return to pre-irradiation levels.
Figure 4.
DNA decay kinetics of the four biomarkers γ-H2AX, 53BP1, MDC1 and ATM[ser1981] following in vitro irradiation with 4 Gy γ rays. Presented is the mean response ± SEM for each repair curve derived from an individual donor.
Discussion
The focus of the present study is to extend the application of the RABiT system to directly measure DNA repair kinetics using the same immunofluorescence techniques currently used in the RABiT for γ-H2AX-based biodosimetry (Turner et al. 2011). To do this, we duplicated the major components of the RABiT workstation at the CHTMIRB laboratory and developed protocols for the repeated sampling of multiple DNA repair biomarkers at specific time points up to 24 hours after irradiation. The formation of radiation-induced foci and subsequent dissociation kinetics are presented for: γ-H2AX, 53BP1, ATM and MDC1 (Figure 4). The results show the rapid induction of radiation-induced immunofluorescent foci within 30 minutes of exposure to 4Gy γ rays, followed by fast and slow repair kinetics over the next 24 hours. Residual levels of phosphorylated ATM and γ-H2AX, higher than background levels, were measured 24 hours post-exposure, whereas the yields of 53BP1 and MDC1 have returned to pre-irradiation baseline levels. The fact that the H2AX histone and ATM kinase undergo DSB-induced protein modification whereas 53BP1 and MDC1 undergo DSB-induced redistribution may contribute to the difference in residual total protein levels measured in our system. According to the literature, in general all the common types of DNA damage, including DSB show quite similar repair kinetics (Frankenberg-Schwager 1989), often exhibiting biphasic exponential decay (Metzger and IIiakis 1991; Ang et al. 1992; Van den Aardweg et al. 1996). Although in most studies, these can be reasonably approximated with a single exponential (Ugenskiene et al. 2009). In the broadest of terms, these biphasic shapes indicate different damage types or different (fast/slow) repair processes.
The γ-H2AX repair protein is a useful and reliable biomarker for radiation exposure (Rothkamm and Horn 2009). Previous studies have correlated the rate of loss γ-H2AX foci and the persistence of γ-H2AX residual foci with cellular radiosensitivity (MacPhail et al. 2003; Taneja et al. 2004; Bhogal et al. 2010). More recently, Martin et al. 201l also showed utility in using the kinetic profile of radiation-induced γ-H2AX foci as a surrogate assay to the gold standard colony survival assay to assess radiosensitivity and Ivashkevich et al. 2012 have demonstrated its use to monitor the clinical response to DNA targeted therapies. To demonstrate the scalability of the laboratory-based RABiT system, we have initiated a population study using γ-H2AX as a biomarker. The advantage of using this repair protein for high throughput biodosimetry studies is that γ-H2AX levels are a) largely absent in pre- non irradiated samples (as opposed to 53BP1 and MDC1 proteins which are homogenously expressed and recruited into DSB sites post-irradiation) and b) highly linear with radiation dose and, c) sensitive to a range of radiation doses. To date we have collected finger stick samples from 30 healthy volunteer donors (8 male and 22 female) and plotted the average yields of γ-H2AX fluorescence up to 24 hours following irradiation with 4 Gy γ rays (Figure 3, Panel D). Paired images of immunofluorescent foci counterstained with DAPI were rapidly captured using our automated imaging system and analyzed using our custom-designed FluorQuant software to measure the total fluorescence yields within each cell nucleus (Figure 3, Panels A-C). Our overall goal is to identify outliers in terms of the fitted repair parameters. This work will be presented in a future manuscript.
The compact capillary irradiator that we have developed can be incorporated into the RABiT as part of the lymphocyte harvest module (Garty et al. 2012). This module processes capillaries sequentially, identifying the location of the lymphocyte band prior to cutting the capillary and dispensing the band into a multi-well plate. It would be relatively straightforward to add an irradiation step to this module. Here the capillary and isolated lymphocyte band should be rapidly inserted in between the two centered sources, paused for a few seconds depending on the dose, and then rapidly retracted. The capillary can then be cut and the irradiated lymphocytes dispensed into the multi-well plate. Alternatively, capillary irradiation can be performed prior to loading the capillaries into the RABiT. In this case the entire length of the capillary must be irradiated, decreasing throughput. Since the half-life of 90Sr is sufficiently long (28.8 years), this irradiator will provide a long useful life, once it is incorporated into the RABiT workstation. Once the calibrations have been fully completed for this compact irradiator, we will begin irradiations to compare with the γ ray irradiations presented in this study. In conjunction with integration of the irradiator into the RABiT, future work will automate the planned protocols for the repair kinetic assay protocols within the RABiT system.
Conclusion
In the present study we have 1) extended RABiT technology to directly measure DNA repair protein kinetics, 2) quantified the temporal kinetics for the DNA DSB repair protein biomarkers γ-H2AX, ATM, 53BP1, MDC1 up to 24 hours post-irradiation exposure and 3) initiated a population study to examine global DNA repair kinetics in healthy individuals using γ-H2AX as a biomarker. The modification of our RABiT system for high throughput measurements of radiation-induced foci can potentially provide a practical, high-throughput, and inexpensive tool for assessing global DSB repair capacity on an individual-by-individual basis; thus, paving the way for new individualized cancer therapy approaches and new large-scale epidemiological studies, with the long-term goal of predicting individual disease sensitivity.
Acknowledgments
This work was supported by grant numbers U19-AI067773 and 1R21-ES019494, for the Center for High-Throughput Minimally Invasive Radiation Biodosimetry, from the National Institute of Allergy and Infectious Diseases, National Institute of Environmental Health Sciences and National Institutes of Health. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases, National Institute of Environmental Health Sciences or the National Institutes of Health.
References
- Anderson L, Henderson C, Adachi Y. Phosphorylation and rapid relocalization of 53BP1 to nuclear foci upon DNA damage. Mol Cell Biol. 2001;21:1719–1729. doi: 10.1128/MCB.21.5.1719-1729.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang KK, Jiang GL, Guttenberger R, Thames HD, Stephens LC, Smith CD, Feng Y. Impact of spinal cord repair kinetics on the practice of altered fractionation schedules. Radiother Oncol. 1992;25:287–294. doi: 10.1016/0167-8140(92)90249-t. [DOI] [PubMed] [Google Scholar]
- Banath JP, Macphail SH, Olive PL. Radiation sensitivity, H2AX phosphorylation, and kinetics of repair of DNA strand breaks in irradiated cervical cancer cell lines. Cancer Res. 2004;64:7144–7149. doi: 10.1158/0008-5472.CAN-04-1433. [DOI] [PubMed] [Google Scholar]
- Bekker-Jensen S, Lukas C, Melander F, Bartek J, Lukas J. Dynamic assembly and sustained retention of 53BP1 at the sites of DNA damage are controlled by Mdc1/NFBD1. J Cell Biol. 2005;170:201–211. doi: 10.1083/jcb.200503043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhogal P, Kaspler F, Jalali O, Hyrien R, Chen RP, Hill, Bristow RG. Late residual gamma-H2AX foci in murine skin are dose responsive and predict radiosensitivity in vivo. Radiat. Res. 2010;173:1–9. doi: 10.1667/RR1851.1. [DOI] [PubMed] [Google Scholar]
- Bouquet F, Muller C, Salles B. The loss of gammaH2AX signal is a marker of DNA double strand breaks repair only at low levels of DNA damage. Cell Cycle. 2006;5:1116–1122. doi: 10.4161/cc.5.10.2799. [DOI] [PubMed] [Google Scholar]
- Chen Y, Wang H, Garty G, Xu Y, Lyulko OV, Turner HC, Randers-Pehrson G, Simaan N, Yao YL, Brenner DJ. Design and preliminary validation of a rapid automated biosodimetry tool for high throughput radiological triage. Proc ASME Des Eng Tech Conf. 2009;3:61–67. doi: 10.1115/DETC2009-86425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Wang H, Garty G, Xu Y, Lyulko OV, Turner HC, Randers-Pehrson G, Simaan N, Yao YL, Brenner DJ. Development of a Robotically-based Automated Biodosimetry Tool for High-throughput Radiological Triage. Int J Biomech Biomed Rob. 2010;1:115–125. [Google Scholar]
- Chen Y, Wang H, Zhang J, Garty G, Simaan N, Yao YL, Brenner DJ. Automated Recognition of Robotic Manipulation Failures in High-throughput Biodosimetry Tool. Expert Syst Appl. 2012;39:9602–9611. doi: 10.1016/j.eswa.2012.02.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins AR, Azqueta A. DNA repair as a biomarker in human biomonitoring studies; further applications of the comet assay. Mutat Res. 2012;736:122–129. doi: 10.1016/j.mrfmmm.2011.03.005. [DOI] [PubMed] [Google Scholar]
- Costes SV, Chiolo I, Pluth JM, Barcellos-Hoff MH, Jakob B. Spatiotemporal characterization of ionizing radiation induced DNA damage foci and their relation to chromatin organization. Mutat Res. 2010;704:78–87. doi: 10.1016/j.mrrev.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelstein A, Amodaj N, Hoover K, Vale R, Stuurman N. Computer Control of Microscopes Using μManager. Current Protocols in Mol Biol. 2010:14.20.1–14.20.17. doi: 10.1002/0471142727.mb1420s92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenech M, Chang WP, Kirsch-Volders M, Holland N, Bonassi S, Zeiger E. HUMN project: detailed description of the scoring criteria for the cytokinesis-block micronucleus assay using isolated human lymphocyte cultures. Mutat Res. 2003;534:65–75. doi: 10.1016/s1383-5718(02)00249-8. [DOI] [PubMed] [Google Scholar]
- Fenech M. The lymphocyte cytokinesis-block micronucleus cytome assay and its application in radiation biodosimetry. Health Phys. 2010;98:234–243. doi: 10.1097/HP.0b013e3181b85044. [DOI] [PubMed] [Google Scholar]
- Frankenberg-Schwager M. Review of repair kinetics for DNA damage induced in eukaryotic cells in vitro by ionizing radiation. Radiother Oncol. 1989;14:307–320. doi: 10.1016/0167-8140(89)90143-6. [DOI] [PubMed] [Google Scholar]
- Garty G, Chen Y, Salerno A, Turner HC, Zhang J, Lyulko O, Bertucci A, Xu Y, Wang H, Simaan N, Randers-Pehrson G, Yao YL, Amundson SA, Brenner DJ. The RABiT: A Rapid Automated Biodosimetry Tool for Radiological Triage. Health Phys. 2010;68:209–217. doi: 10.1097/HP.0b013e3181ab3cb6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garty G, Chen Y, Turner HC, Zhang J, Lyulko O, Bertucci A, Xu Y, Wang H, Simaan N, Randers-Pehrson G, Yao YL, Brenner DJ. The RABIT: A Rapid Automated BIodosimetry Tool For Radiological Triage. II. Technological Developments. Int J Radiat Biol. 2011;87:776–790. doi: 10.3109/09553002.2011.573612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hable V, Drexler GA, Brüning T, Burgdorf C, Greubel C, Derer A, Seel J, Strickfaden H, Cremer T, Friedl AA, Dollinger G. Recruitment kinetics of DNA repair proteins Mdc1 and Rad52 but not 53BP1 depend on damage complexity. PLoS One. 2012;7:1–11. doi: 10.1371/journal.pone.0041943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivashkevich A, Redon CE, Nakamura AJ, Martin RF, Martin OA. Use of the γ-H2AX assay to monitor DNA damage and repair in translational cancer research. Cancer Lett. 2012;327:123–133. doi: 10.1016/j.canlet.2011.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungmichel S, Stucki M. MDC1: The art of keeping things in focus. Chromosoma. 2010;119:337–349. doi: 10.1007/s00412-010-0266-9. [DOI] [PubMed] [Google Scholar]
- Kinner A, Wu W, Staudt C, Iliakis G. Gamma-H2AX in recognition and signaling of DNA double-strand breaks in the context of chromatin. Nucleic Acids Res. 2008;36:5678–5694. doi: 10.1093/nar/gkn550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhne M, Riballo E, Rief N, Rothkamm K, Jeggo PA, Lobrich M. A double-strand break repair defect in ATM-deficient cells contributes to radiosensitivity. Cancer Res. 2004;64:500–508. doi: 10.1158/0008-5472.can-03-2384. [DOI] [PubMed] [Google Scholar]
- Löbrich M, Rief N, Kuhne M, Heckmann M, Fleckenstein J, Rube C, Uder M. In vivo formation and repair of DNA double-strand breaks after computed tomography examinations. Proc Natl Acad Sci U S A. 2005;102:8984–8989. doi: 10.1073/pnas.0501895102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyulko OV, Garty G, Randers-Pehrson G, Turner HC, Brenner DJ. Fast image analysis for the micronucleus assay in a fully automated high throughput biodosimetry system. Radiat Res. In review. 2013 doi: 10.1667/RR13441.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacPhail SH, Banath JP, Yu TY, et al. Expression of phosphorylated histone H2AX in cultured cell lines following exposure to X-rays. Int J Radiat Biol. 2003;9:351–358. doi: 10.1080/0955300032000093128. [DOI] [PubMed] [Google Scholar]
- Markova E, Schultz N, Belyaev IY. Kinetics and dose-response of residual 53BP1/gamma-H2AX foci: co-localization, relationship with DSB repair and clonogenic survival. Int J Radiat Biol. 2007;83:319–329. doi: 10.1080/09553000601170469. [DOI] [PubMed] [Google Scholar]
- Martin NT, Nahas SA, Tunuguntla R, Fike F, Gatti RA. Assessing 'radiosensitivity' with kinetic profiles of γ-H2AX, 53BP1 and BRCA1 foci. Radiother Oncol. 2011;101:35–8. doi: 10.1016/j.radonc.2011.05.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger L, Iliakis G. Kinetics of DNA double-strand break repair throughout the cell cycle as assayed by pulsed field gel electrophoresis in CHO cells. Int J Radiat Biol. 1991;59:1325–1339. doi: 10.1080/09553009114551201. [DOI] [PubMed] [Google Scholar]
- Nakamura A, Sedelnikova OA, Redon C, Pilch DR, Sinogeeva NI, Shroff R, Lichten M, Bonner WM. Techniques for gamma-H2AX detection. Methods Enzymol. 2006;409:236–250. doi: 10.1016/S0076-6879(05)09014-2. [DOI] [PubMed] [Google Scholar]
- Rasband WS. ImageJ, U. S. National Institutes of Health, Bethesda, Maryland, USA. 1997-2012 http://imagej.nih.gov/ij/
- Redon CE, Dickey JS, Bonner WM, Sedelnikova OA. gamma-H2AX as a biomarker of DNA damage induced by ionizing radiation in human peripheral blood lymphocytes and artificial skin. Advan Space Res. 2009;43:1171–1178. doi: 10.1016/j.asr.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou EP, Pilch PR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- Rothkamm K, Löbrich M. Evidence for a lack of DNA double-strand break repair in human cells exposed to very low x-ray doses. Proc Natl Acad Sci U S A. 2003;100:5057–5062. doi: 10.1073/pnas.0830918100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothkamm K, Horn S. gamma-H2AX as protein biomarker for radiation exposure. Ann Ist Super Sanita. 2009;45:265–271. [PubMed] [Google Scholar]
- Rube CE, Grudzenski S, Kuhne M, Dong X, Rief N, Lobrich M, Rube C. DNA double-strand break repair of blood lymphocytes and normal tissues analysed in a preclinical mouse model: implications for radiosensitivity testing. Clin Cancer Res. 2008;14:6546–6555. doi: 10.1158/1078-0432.CCR-07-5147. [DOI] [PubMed] [Google Scholar]
- Schultz LB, Chehab NH, Malikzay A, Halazonetis TD. p53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J Cell Biol. 2000;151:1381–1390. doi: 10.1083/jcb.151.7.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedelnikova OA, Pilch DR, Redon C, Bonner WM. Histone H2AX in DNA damage and repair. Cancer Biol Ther. 2003;2:233–235. doi: 10.4161/cbt.2.3.373. [DOI] [PubMed] [Google Scholar]
- Turner HC, Brenner DJ, Chen Y, Bertucci A, Zhang J, Wang H, Lyulko OV, Xu Y, Shuryak I, Schaefer J, Simaan N, Randers-Pehrson G, Yao YL, Amundson SA, Garty G. Adapting the γ-H2AX assay for automated processing in human lymphocytes. 1. Technological aspects. Radiat Res. 2011;175:282–290. doi: 10.1667/RR2125.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taneja N, Davis M, Choy JS, et al. Histone H2AX phosphorylation as a predictor of radiosensitivity and target for radiotherapy. J Biol Chem. 2004;279:2273–2280. doi: 10.1074/jbc.M310030200. [DOI] [PubMed] [Google Scholar]
- Ugenskiene R, Prise K, Folkard M, Lekki J, Stachura Z, Zazula M, Stachura J. Dose response and kinetics of foci disappearance following exposure to high- and low-LET ionizing radiation. Int J Radiat Biol. 2009;85:872–882. [PubMed] [Google Scholar]
- Van den Aardweg GJ, Hopewell JW, Guttenberger R. The kinetics of repair of sublethal radiation-induced damage in pig skin: studies with multiple interfraction intervals. Radiat Res. 1996;145:586–594. [PubMed] [Google Scholar]
- Wilson PF, Nham PB, Urbin SS, Hinz JM, Jones IM, Thompson LH. Inter-individual variation in DNA double-strand break repair in human fibroblasts before and after exposure to low doses of ionizing radiation. Mutat Res. 2010;683:91–97. doi: 10.1016/j.mrfmmm.2009.10.013. [DOI] [PubMed] [Google Scholar]
- Xu Y, Turner HC, Garty G, Brenner DJ. A Rapid, Quantitative Method to Characterize The Human Lymphocyte Concentration for Automated High-Throughput Radiation Biodosimetry. Biomed Eng Res. 2013;2:16–19. doi: 10.5963/ber0201002. [DOI] [PMC free article] [PubMed] [Google Scholar]