Abstract
The desmosomal cadherin desmoglein-1 (DSG1) is an essential intercellular adhesion molecule that is altered in various human cutaneous disorders; however, its regulation and function in allergic disease remains unexplored. Herein, we demonstrate a specific reduction in DSG1 in esophageal biopsies from patients with eosinophilic esophagitis (EoE), an emerging allergic disorder characterized by chronic inflammation within the esophageal mucosa. Further, we show that DSG1 gene silencing weakens esophageal epithelial integrity, and induces cell separation and impaired barrier function (IBF) despite high levels of desmoglein-3 (DSG3). Moreover, DSG1 deficiency induces transcriptional changes that partially overlap with the transcriptome of inflamed esophageal mucosa; notably, periostin, a multipotent pro-inflammatory extracellular matrix molecule, is the top induced overlapping gene. We further demonstrate that IBF is a pathological feature in EoE, which can be partially induced through the downregulation of DSG1 by interleukin-13 (IL-13). Taken together, these data identify a functional role for DSG1 and its dysregulation by IL-13 in the pathophysiology of EoE and suggest that the loss of DSG1 may potentiate allergic inflammation through the induction of pro-inflammatory mediators such as periostin.
Introduction
Eosinophilic esophagitis (EoE) is a chronic inflammatory disease that has emerged over that last decade on a worldwide scale1. Although symptomatically resembling gastroesophageal reflux disease (GERD), EoE is characterized by immune sensitization to a variety of foods and marked Th2-associated allergic inflammation in the esophageal mucosa that is largely refractory to acid-suppressive therapy2. During active disease, the histopathological changes within the inflamed esophageal mucosa include the dense accumulation of activated immune cells, including eosinophils, mast cells, and T and B lymphocytes3. Moreover, evidence of dilated intercellular spaces (DIS) and abnormal epithelial cell proliferation suggest that impaired barrier function (IBF) of the esophageal epithelium may potentially contribute to the pathophysiology of EoE4–6. While dietary modification (i.e., complete or targeted food antigen avoidance) and swallowed glucocorticoids alleviate much of the disease pathology, EoE still has one of the lowest quality-of-life indexes among other chronic pediatric diseases, including inflammatory bowel disease7–9.
Early efforts aimed at the molecular dissection of EoE pathogenesis included gene expression profiling of esophageal mucosal biopsies from patients with active EoE, which identified a striking disease-associated transcript signature that was highly conserved across patients with EoE and largely normalized during glucocorticoid-induced disease remission10, 11. Several pro-inflammatory mediators, such as the chemokine (C-C-motif) ligand 26 (CCL26), periostin (POSTN), and tumor necrosis factor, alpha-induced protein 6 (TNFAIP6), were dramatically elevated in the inflamed esophageal mucosa in EoE10 and in primary esophageal epithelial cells treated with the Th2 cytokine interleukin-13 (IL-13)11. A marked reduction in genes involved in epithelial homeostasis was also observed. In particular, the desmosomal cadherin desmoglein-1 (DSG1) exhibited an approximate 8-fold decrease in patients with active EoE, which only partially normalized upon disease remission11.
DSG1 is a intercellular adhesion molecule belonging to the desmosomal cadherin family, which includes desmogleins (DSG1-4) and desmocollins (DSC1-3)12. Desmogleins and desmocollins are involved in maintaining epithelial homeostasis where they display spatially distinct expression patterns at various levels among different stratified epithelia12, 13. In particular, DSG1 is highly expressed in the epidermis and localized primarily within the suprabasal epithelial layers, where it regulates cell adhesion and supports epithelial cell differentiation14. In contrast, DSG3 is localized to undifferentiated, basal epithelial cells and has been suggested to promote cell proliferation15–17.
While the function of DSG1 has been well characterized in the epidermis, its role in the esophageal epithelium under both homeostatic and inflammatory conditions remains largely unknown. DSG1 mediates intercellular adhesion by forming calcium-dependent heterotypic and homotypic interactions between adjacent epithelial cells via its amino-terminal ectodomain18. However, it is becoming increasingly appreciated that DSG1 has additional roles beyond that of cell adhesion19. Indeed, DSG1 promotes epithelial differentiation through negatively regulating the activation of extracellular signal-regulated kinase (ERK) by epidermal growth factor receptor (EGFR) signaling14. DSG1 can also promote epithelial differentiation through its interactions with ERBIN, which sequesters SHOC2 to attenuate RAS-mediated ERK activation20.
In the present study, we sought to determine the functional consequences of DSG1 dysregulation in EoE pathogenesis as DSG1 was one of the most down-regulated genes in the esophageal mucosa of EoE patients11. We hypothesized that the downregulation of DSG1 contributes to the pathological features of the esophageal epithelium in EoE. We demonstrate a marked reduction in the gene and protein levels of DSG1 but not DSG3 in the esophageal mucosa of patients with EoE. Suppression of DSG1 by shRNA or IL-13 treatment induced IBF in differentiated esophageal epithelial cells grown at the air-liquid interface (ALI); notably, IBF was also observed ex vivo in biopsy samples from patients with EoE. Lastly, knockdown of DSG1 was sufficient to induce a gene expression profile, which included periostin as the top induced gene, that substantially overlaps with the transcriptome of the inflamed esophageal mucosa of patients with EoE. These data suggest that the negative regulation of DSG1 by IL-13 can exacerbate inflammation of the esophageal mucosa in EoE by enhancing IBF and initiating a pro-inflammatory gene expression cascade.
Results
Specific dysregulation of DSG1 in EoE
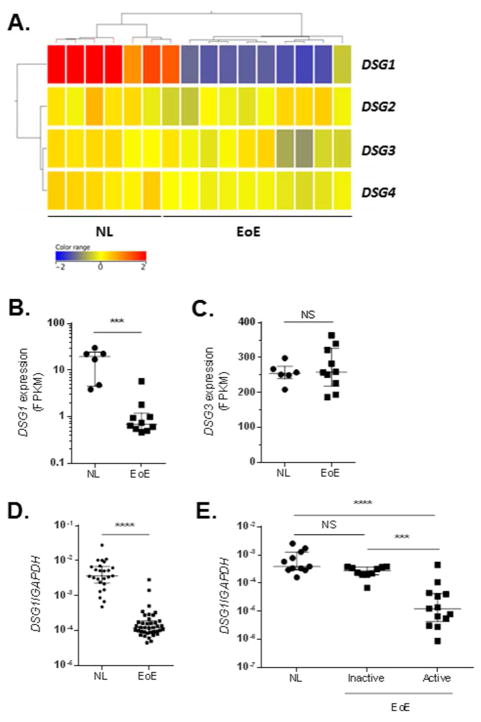
We sought to establish the relative levels of all desmoglein genes (DSG1-4) in the esophagus and to determine whether the downregulation of DSG1 was specific among other DSG family members. Whole-transcriptome RNA sequencing of esophageal biopsies from healthy controls (NL) (n = 6) and patients with active EoE (n = 10) showed a specific and dramatic downregulation of DSG1 in this cohort of patients (Fig. 1A). Indeed, DSG1 exhibited a 12.7 fold reduction in patients with active EoE (FPKM [median + interquartile range] = 19.6 + (4.6 – 24.4) in NL and 0.70 + (0.5 – 1.2) in EoE; p = 1 × 10−3). Notably, the most abundant DSG expressed in the esophageal mucosa was DSG3, which did not display differential expression in EoE (FPKM [median + interquartile range] = 254.4 + (239.0 – 279.5) in NL and 258.2 + (218.6 – 326.1) in EoE; p = 0.75) (Fig. 1C). We examined the downregulation of DSG1 in a larger cohort of NL and patients with active EoE disease (n = 25 and 39, respectively) by quantitative PCR (qPCR) and detected a 22.1-fold reduction (p = 1 × 10−4) in the esophageal expression of DSG1 in active EoE (Fig. 1D). Lastly, we assessed esophageal DSG1 levels in patients with inactive EoE (n = 10) following swallowed glucocorticoid therapy (Fig. 1E). During disease remission, expression of DSG1 normalized to similar levels that were observed in NL (n = 11) yet was significantly different than in patients with active disease (n = 13) (p = 3 × 10−4).
Figure 1. Specific reduction in desmoglein-1 (DSG1) gene expression in EoE.
Heatmap depicting expression levels of desmogleins 1–4 (DSG1–4) (A) and individual FPKM values for DSG1 and DSG3 (B-C) from RNA sequencing of esophageal biopsies from 10 patients with active EoE versus 6 healthy controls (NL). Quantitative PCR (qPCR) analysis of DSG1 expression in esophageal biopsies from NL (n = 25) and patients with active EoE (n = 39) (D). qPCR analysis of DSG1 expression in esophageal biopsies from NL (n = 11) and patients with inactive (n = 10) or active (n = 13) EoE (E). Data are represented as the median + interquartile range: NS (not significant), *** p < 5 × 10−4, and **** p < 1 × 10−4.
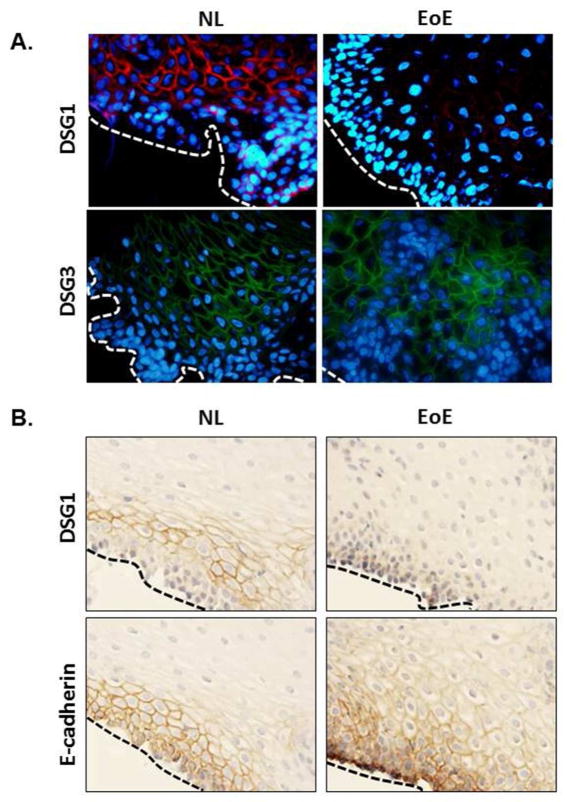
We next performed immunofluorescent and/or immunohistochemical staining for DSG1, DSG3 and E-cadherin in the esophageal mucosa of NL and patients with active EoE to characterize protein expression and localization. Consistent with previous reports21, expression of DSG1 localized to the cell surface and was restricted to the suprabasal esophageal epithelium in NL, while DSG1 staining was remarkably absent in patients with active EoE (Fig. 2A, upper panel). Conversely, DSG3 was abundantly expressed throughout most of the esophageal epithelium in both NL and EoE, with more concentrated staining within the basal epithelial cell layers (Fig. 2A, lower panel). Immunohistochemical staining for DSG1 revealed a similar downregulation in EoE whereas expression of E-cadherin, a ubiquitously expressed cadherin molecule that regulates epithelial homeostasis and barrier formation22, was unchanged between NL and patients with active EoE (Fig. 2B); these findings were supported at the gene level by qPCR analysis of esophageal biopsies from NL and active EoE patients (data not shown). These cumulative data indicate a specific downregulation of the DSG1 gene and protein in EoE, and that this is unique from the expression patterns of other desmogleins and E-cadherin.
Figure 2. Loss of DSG1 protein expression in EoE.
Immunofluorescence (A) or immunohistochemical staining (B) of esophageal biopsy sections from controls (NL) and patients with active EoE. In (A), DSG1 (upper panel, in red) and DSG3 (lower panel, in green) are shown. Nuclei are stained with DAPI (blue). In (B), DSG1 (upper panel, in brown) and E-cadherin (lower panel, in brown) are shown. Dashed lines in (A) and (B) indicate the basal epithelial layer. Images in are representative of 4 patients per group.
Loss of DSG1 regulates esophageal epithelial cell integrity in vitro
In order to investigate the functional consequences of DSG1 dysregulation on the esophageal epithelium, we developed a modified ALI culture system to induce a stratified esophageal epithelium in vitro. Similar ALI models have been used to study differentiated epithelium of both the epidermis and esophagus23, 24. Confluent monolayers of immortalized esophageal epithelial cells (EPC2), which have been previously shown to form stratified esophageal epithelium25–27, were exposed to the ALI in the presence of high Ca2+ (1.8 mM). Following 5–7 days of differentiation at the ALI, the EPC2 cells formed a stratified epithelium as indicated by the differential H&E staining when compared to submerged cells (Fig. 3A). Gene expression analyses after ALI exposure showed significant induction of DSG1 and keratin 10 (KRT10) (411 fold, p < 1 × 10−3 and 6,240 fold, p < 1 × 10−2, respectively), both of which are expressed specifically in differentiated esophageal epithelial cells21, 28, 29 (Fig. 3B–C).
Figure 3. Differentiation of esophageal epithelial cells at the air-liquid interface (ALI).
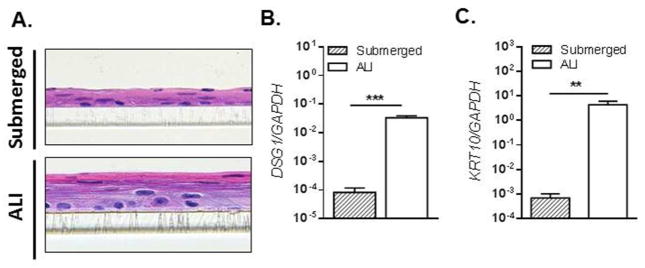
H&E-stained sections of EPC2 cells grown in submerged cultures or differentiated at the ALI (A). qPCR analysis of desmoglein-1 (DSG1) (B) and keratin 10 (KRT10) (C) expression in submerged or ALI-differentiated EPC2 cells. Data are representative of 4 experiments performed in duplicate and are represented as the mean + SEM: ** p < 5 × 10−3 and *** p < 5 × 10−4.
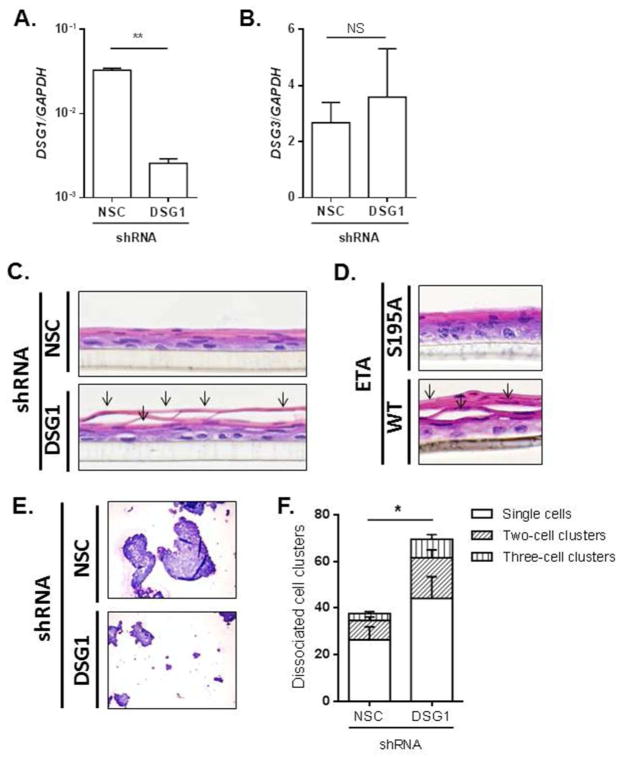
We next utilized lentiviral shRNA gene silencing to directly examine the impact of DSG1 dysregulation on esophageal epithelial cell adhesion. EPC2 cells that were stably transduced with a DSG1 shRNA exhibited a 92% reduction in DSG1 expression compared to cells transduced with a non-silencing control (NSC) shRNA (p < 5 × 10−3) (Fig. 4A), whereas DSG3 levels remained unaffected (Fig. 4B). Histological analyses of the NSC shRNA-transduced cells grown at the ALI showed normal stratification with a differentiated, intact esophageal epithelium (Fig. 4C, upper panel). However, while the differentiation appeared normal in the DSG1 shRNA-transduced cells, prominent cellular separation was evident in the suprabasal layers (arrows, Fig. 4C, lower panel). Importantly, this cellular separation was consistently observed only in the DSG1-deficient cells (and not the NSC controls cells) using multiple DSG1 shRNA clones, indicating that the observed phenotype was not due to sample sectioning (data not shown). A similar phenotype was observed when ALI-differentiated EPC2 cells were treated directly with recombinant wild type (WT) exfoliative toxin A (ETA), a DSG1-specific protease produced by S. aureus and the causative agent of epidermal blistering in staphylococcal scalded-skin syndrome (SSSS)30 (Fig. 4D, lower panel). Notably, the inactive mutant form of ETA (S195A) did not induce cellular separation (Fig. 4D, upper panel). To directly measure whether loss of DSG1 reduced esophageal epithelial cell adhesion, NSC or DSG1 shRNA-transduced cells were subjected to a dispase adhesion assay. Following mechanical disruption, a significantly greater amount of cell dissociation was observed in cells deficient in DSG1 than in control cells (p < 5 × 10−2) (Fig. 4E–F).
Figure 4. Loss of DSG1 reduces esophageal epithelial cell adhesion.
qPCR analysis of DSG1 (A) and DSG3 (B) in ALI-differentiated EPC2 cells stably transduced with non-silencing control (NSC) or DSG1 shRNA. H&E-stained sections from stably transduced cells differentiated at the ALI (C). H&E-stained sections from EPC2 cells exposed to the air interface and treated with 10 μg/mL ETA (WT) or the S195A inactive mutant for 24 h (D). Arrows (C–D) indicate cell separation within the suprabasal epithelium. Cytospins from NSC or DSG1 shRNA-transduced EPC2 cells following dispase adhesion assays (E) and quantification of dissociated cell clusters are shown (F). Images in (C–E) are representative of 4–5 experiments performed in duplicate. Data in (A–B) and (E) are from 3 experiments performed in duplicate and are represented as the mean + SEM: NS (not significant), * p < 5 × 10−2 and ** p < 5 × 10−3.
IBF in EoE and in DSG1-deficient esophageal epithelial cells in vitro
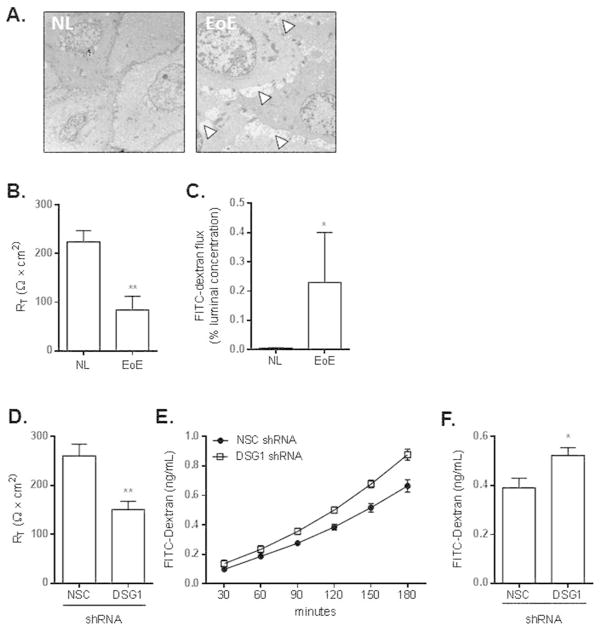
To assess the integrity of the esophageal epithelium in EoE at the ultra-structural level, we performed electron microscopy on NL and EoE patient biopsies. While the esophageal epithelium of NL patients was composed of cohesive, intact epithelia, prominent DIS were evident throughout the esophageal epithelium of patients with EoE (arrowheads, Fig. 5A). These findings, which have been noted in previous studies of both EoE and GERD31, 32, were consistent across multiple sections from the same patient and among different patients. We next assessed whether IBF is indeed a pathological defect in EoE by measuring transepithelial electrical resistance (TER) ex vivo in esophageal biopsies from NL and patients with EoE (Fig. 5B). We found that transcellular permeability was reduced by approximately 62% in patients with EoE compared to in NL (RT = 84 ± 28 vs. 224 ± 23 ohms × cm2, respectively; p < 5 × 10−3). We also analyzed paracellular permeability in EoE by measuring macromolecular flux in esophageal biopsies and observed an approximate 46-fold increase in those from patients with EoE compared to NL (p < 5 × 10−2). Notably, esophageal expression of occludin (OCLN) and genes in the claudin (CLDN) and tight junction protein (TJP) families was not reduced in EoE, suggesting the IBF in EoE occurs in the absence of marked alteration to tight junctions (Supplementary Fig. 1A–C).
Figure 5. Impaired barrier function (IBF) in EoE can be replicated in DSG1 deficient esophageal epithelial cells.
Representative electron micrographs of esophageal biopsies from healthy (NL) controls (n = 3) and patients with active EoE (n = 3). Arrowheads indicate the presence of dilated intercellular spaces (DIS) in EoE (A). TER (RT) measurements from esophageal biopsies from healthy (NL) control and patients with active EoE (n = 6 and 9, respectively) (B). FITC-dextran flux assays from NL and active EoE esophageal biopsies (n = 6 and 4, respectively) (C). TER (RT) measurements from NSC and DSG1 shRNA-transduced EPC2 cells following ALI differentiation (D). Kinetic analysis of FITC-dextran flux was also performed (E). Total FITC-dextran flux following 180 minutes are depicted in (F). Data in (D–F) are from two independent experiments performed in quadruplicate. All data are represented as the mean + SEM: *, p < 5 × 10−2 ; **, p < 5 × 10−3.
We hypothesized that the cellular separation that occurs within the esophageal epithelium following the loss of DSG1 (Fig. 4C) may contribute to the IBF observed in EoE (Fig. 5A–C). To test this possibility, we measured TER and paracellular permeability in the ALI-differentiated NSC or DSG1 shRNA-transduced EPC2 cells. Reduced DSG1 expression resulted in impaired TER (p < 5 × 10−3, Fig. 5D) and increased FITC-dextran flux (p < 5 × 10−2, Fig. 5E–F) by approximately 42% and 33%, respectively. These data identify a novel mechanism whereby loss of DSG1 negatively affects esophageal epithelial integrity and is sufficient to induce IBF.
IL-13 regulates DSG1 and promotes IBF in differentiated esophageal epithelial cells
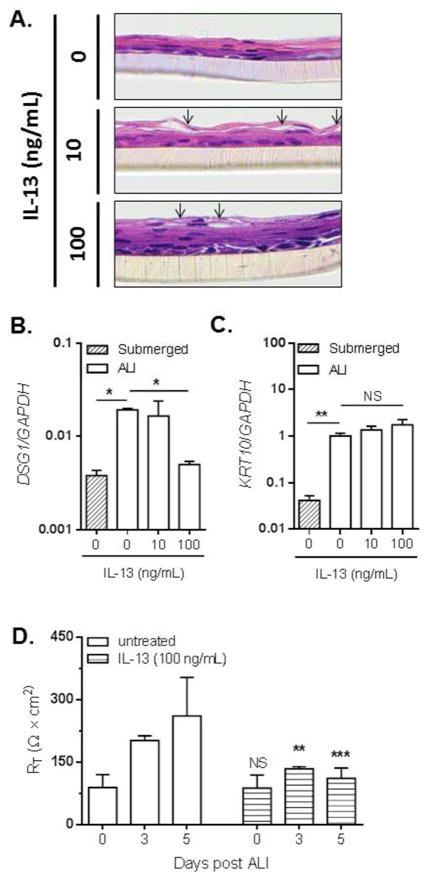
IL-13 is a critical Th2 cytokine capable of eliciting some of the transcriptional changes within the inflamed esophageal epithelium that are associated with EoE, including the downregulation of multiple epithelial cell differentiation genes11, 33. Therefore, we assessed the effects of IL-13 on the integrity and barrier formation of ALI-differentiated esophageal epithelial cells. Esophageal epithelial cells were left untreated or treated with 10 or 100 ng/mL IL-13 continuously throughout the ALI differentiation process. Both concentrations of IL-13 induced partial cellular separation within the suprabasal epithelial layers (arrows, Fig. 6A, middle and lower panels).
Figure 6. IL-13 downregulates DSG1 and promotes IBF in esophageal epithelial cells.
H&E-stained sections of EPC2 cells differentiated at the ALI in the presence of 0 (untreated), 10, or 100 ng/mL IL-13 (A). Arrows indicate a cell separation within to the suprabasal epithelium. Images are representative of 3 experiments performed in duplicate. Expression levels of DSG1 (B) and KRT10 (C) were measured by qPCR in submerged or ALI-differentiated EPC2 cells in the absence (0 ng/mL) or presence of IL-13 (10 or 100 ng/mL). TER (RT) measurements on EPC2 cells at 0, 3, and 5 days following differentiation at the ALI in the absence (untreated) or presence of IL-13 (100 ng/mL) (D). Data are from 3 experiments performed in duplicate and are represented as the mean + SEM: NS (not significant), ** p < 5 × 10−3 and *** p < 1 ×10−4 as compared to the untreated cells at the same days post-ALI differentiation.
As the IL-13-induced suprabasal cell separation partially reflected the phenotype of DSG1-deficient cells (Fig. 4C), we next investigated the ability of IL-13 to regulate DSG1 expression. IL-13 treatment suppressed the induction of DSG1 in ALI-differentiated esophageal epithelial cells (Fig. 6B), whereas induction of KRT10 (Fig. 6C) and DSG3 (data not shown) were unaffected. Importantly, IL-13 also promoted IBF, as a significant reduction in TER was observed at both 3 and 5 days post treatment with IL-13 (100 ng/mL) compared to untreated cells (Fig. 6D).
We also tested whether IL-13 could attenuate DSG1 expression in vivo using a transgenic murine model of EoE. Doxycycline (Dox)-inducible expression of IL-13 in the lung (using the Clara cell-specific promoter CC10) has been shown to induce an EoE phenotype in mice34. Elevated IL-13 levels in the BAL and induced esophageal eosinophilia were detected in the Dox-treated animals (Supplementary Fig. 1A–B). Overexpression of IL-13 in treated (+ Dox) mice reduced DSG1 mRNA and protein levels in the esophageal mucosa as compared to untreated (-Dox) mice (Supplementary Fig. 1C–E).
Loss of DSG1 primes for the innate inflammatory transcript signature
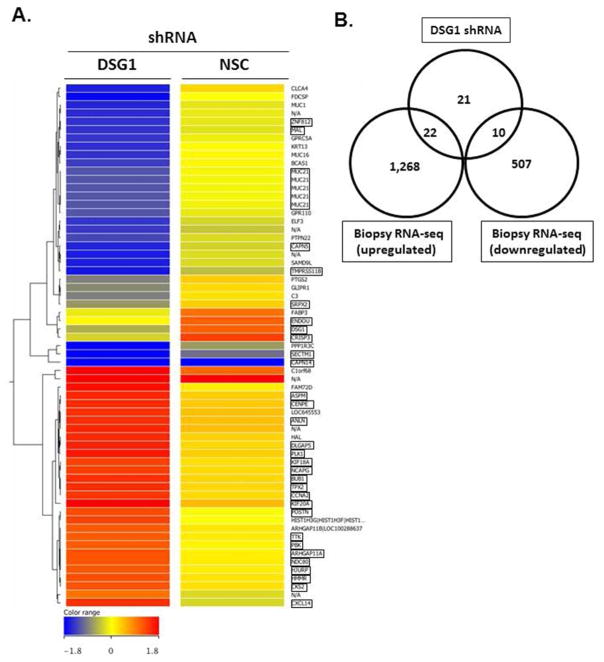
DSG1 has been shown to counter-regulate EGFR signaling, and more recently RAS-mediated signaling, through mechanisms that are independent of its full-length, adhesive N-terminal domain14, 20. Therefore, we hypothesized that DSG1 deficiency may also regulate downstream signaling responses associated with the Th2 inflammation in EoE. To test this possibility, we performed microarray analyses on NSC and DSG1 shRNA-transduced EPC2 cells following ALI-differentiation. Interestingly, 63 transcripts coding for 53 unique genes were differentially expressed in the DSG1-deficient cells compared to in control cells (p < 5 × 10−2, fold change > 2.0) (Fig. 7A). Notably, there was a substantial overlap between this transcript profile and the EoE transcriptome identified in patient esophageal biopsies by RNA sequencing (RNA-seq); 60% of the unique DSG1-regulated transcripts were also dysregulated in esophageal biopsies of patients with EoE (Fig. 7B and Supplementary Table 1). In particular, periostin (POSTN) was the most highly induced gene upon DSG1 knockdown in esophageal epithelial cells (2.5 fold) and was dramatically elevated in esophageal tissue of patients with EoE (384 fold) (Supplementary Table 1).
Figure 7. Loss of DSG1 promotes epithelial pro-inflammatory transcriptional responses.
Heatmap of 63 transcripts with differential expression (p < 5 × 10−2, fold change > 2.0) in DSG1-deficient EPC2 cells compared to non-silencing control (NSC) cells following ALI-differentiation (A). Venn diagram depicting the number of genes (n = 32) (boxed transcripts in [A]) dysregulated following DSG1 knockdown that overlap with differentially expressed genes in esophageal mucosa of EoE patients (n = 10) compared to healthy control patients (NL) (n = 6) identified by RNA sequencing (B).
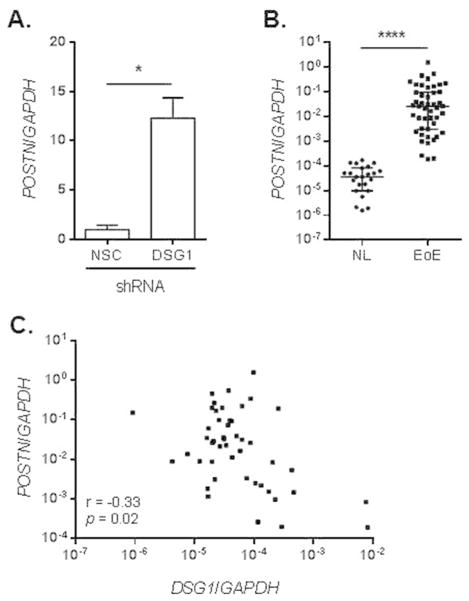
We next validated the significant increase in POSTN levels in DSG1-deficient esophageal epithelial cells by qPCR and observed a 17-fold increase in POSTN in the DSG1 versus NSC shRNA-transduced cells (p < 5 × 10−2) (Fig. 8A). Furthermore, qPCR analysis demonstrated a significant and dramatic increase in POSTN expression in the esophageal mucosa of patients with active EoE compared to NL (p < 1 × 10−4, fold change = 2,047) (Fig. 8B). Notably, POSTN levels in EoE showed a significant inverse correlation with DSG1 expression (same cohort as in Fig. 1D) (Spearman r = −0.33, p = 0.02) (Fig. 8C). Together, these data indicate that IL-13-induced loss of DSG1 augments the allergic inflammatory response by promoting IBF and likely elevates pro-inflammatory gene expression in EoE (Fig. 9).
Figure 8. DSG1 deficiency increases periostin (POSTN) expression.
qPCR analysis of POSTN expression in non-silencing control (NSC) and DSG1 shRNA-transduced EPC2 cells following ALI differentiation (A) and in patient biopsies (same cohort as in Fig. 1D) (B). Spearman correlation between esophageal expression of POSTN and DSG1 (from Fig. 1D) in patients with active EoE (C). Data in (A) are representative of three independent experiments performed in duplicate and represented as the mean + SEM: * p < 5 × 10−2 and **** p < 1 × 10−4.
Figure 9. Model of DSG1 dysregulation in EoE pathogenesis.
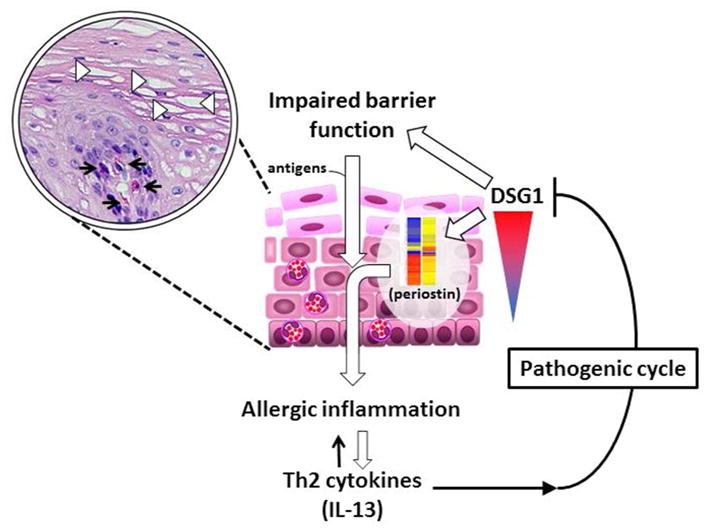
Downregulation of DSG1 by select Th2 cytokines (e.g. IL-13) results in impaired barrier function (IBF) and increased antigen exposure as well as the expression of pro-allergic mediators including periostin (POSTN), forming a pathogenic cycle to further exacerbate allergic inflammation. Inset shows H&E staining of inflamed esophageal mucosa with dilated intercellular spaces (DIS) (arrowheads) and eosinophilic infiltration (arrows).
Discussion
The data presented herein characterize the pathological impact of DSG1 dysregulation in EoE and define a non-redundant role for DSG1 in the regulation of the esophageal epithelial barrier function and homeostasis. In particular, we demonstrated a specific and marked downregulation of DSG1 in the esophageal epithelium of patients with EoE that was largely reversible with patient disease status. This loss in DSG1 expression was sufficient to induce cell separation and IBF in the esophageal epithelium. Moreover, DSG1 deficiency led to an esophageal epithelial transcript signature that included increased gene expression of the pro-inflammatory extracellular matrix molecule periostin. These findings are particularly notable in view of the relatively high abundance of DSG3 expression in EoE and in our in vitro model, demonstrating the lack of redundancy for DSG1’s regulation of esophageal responses, particularly during allergic inflammation. Interestingly, in GERD, in which DIS and IBF have been attributed to acid exposure of the esophageal epithelium, esophageal expression of DSG1 (and DSG3 to a lesser extent) is increased35, further suggesting a unique, non-redundant and essential role for DSG1 in regulating disease processes specifically linked with EoE pathology.
Our data identify IL-13 as a potent regulator of DSG1 expression in human esophageal epithelial cells cultured at the ALI and in mouse esophageal epithelial cells in a murine model of EoE. This latter finding is quite striking given the physiological differences between the human and mouse esophagus (not keratinized versus keratinized, respectively) and the existence of multiple murine Dsg1 isoforms (α, β, and γ)36, which were universally detected in our qPCR analysis and immunofluorescent staining (see Supplementary Materials and Methods). Previous work has demonstrated a pronounced effect of IL-13 on global gene expression in primary esophageal epithelial cells, and in particular, genes involved in epithelial differentiation11, 33. DSG1 was not shown to be regulated by IL-13 in these data, which, on the basis of our data in the ALI system (Fig. 3), we hypothesize was due to the low levels of DSG1 expression in esophageal epithelial cells grown in the submerged culture conditions used in these studies. Interestingly, in skin keratinocytes, which express high baseline levels of DSG1 even in submerged cultures, IL-4 has been shown to downregulate DSG1 expression in vitro37. Notably, both IL-13 and IL-4 are significantly increased in the peripheral blood and esophageal mucosa of patients with active EoE38, 39 and preliminary data indicate that IL-4 is also capable of negatively regulating DSG1 expression and inducing IBF in ALI-differentiated esophageal epithelial cells in vitro (data not shown). These data, together with our findings that esophageal expression of DSG1 normalizes in EoE patients during disease remission, indicate DSG1 is negatively regulated by Th2 cytokines during allergic inflammation.
Within the epithelial barrier of the epidermis and intestine, the selective ability to discriminate the uptake of molecules based on size and charge is attributed to the tight junction complex40. For example, several tight junction genes are expressed within the epidermis including CLDN1, CLDN4, TJP1, and OCLN41. While only CLDN1 has been demonstrated to have an essential role in epidermal barrier function in vivo42, a recent study has shown CLDN4, TJP1, and OCLN can also independently regulate epidermal permeability in vitro43. Although the esophageal epithelial barrier has been studied primarily in the context of GERD, tight junction proteins have been shown to have no correlation with GERD pathogenesis44. Moreover, our data (Supplementary Fig. 1) demonstrate no decrease in genes encoding claudins, tight junction proteins, or occludin in EoE; none of these genes were dramatically altered in DSG1-deficient cells following shRNA transduction or IL-13 treatment (Supplementary Table 3). Together, these data suggest physiologically distinct mechanisms regulate the epithelial barrier properties of the esophagus and the epidermis.
Despite our evidence substantiating IBF as a bona fide pathological feature in EoE, little is known regarding the role of the esophageal barrier in regulating allergic inflammation. Within the epidermis, Langerhans cells continually sample external antigens by inducing transient reorganization of tight junctions within the stratum corneum without affecting barrier integrity; this homeostatic surveillance is further amplified in activated Langerhans cells45. These data suggest a potential mechanism by which IBF could lead to increased antigen sensitization, which is dependent on tissue resident antigen presenting cells. It is important to note that Langerhans cells46, 47 and high baseline expression of the high affinity IgE receptor FcεRI48 have been observed in both healthy and inflamed esophageal epithelium. Moreover, data suggesting esophageal epithelial cells can present antigen and MHC class II expression is increased in EoE49 further suggest that the esophagus is not a unifunctional, static organ involved solely in food transport, but rather has an active role in immunosurveillance during allergic inflammation.
Our finding of elevated periostin expression in DSG1-deficient cells is particularly notable as periostin has been implicated in multiple allergic inflammatory disease including asthma, atopic dermatitis, and EoE50–52. Periostin can directly enhance eosinophil adhesion50, 53, as well as increase keratinocyte production of thymic stromal lymphopoetin (TSLP)52, a potent Th2-skewing cytokine that has been genetically linked to EoE susceptibility54, 55. Although periostin is expressed primarily in fibroblasts, treatment of both bronchial epithelial cells56 and esophageal epithelial cells50 with IL-13 induces periostin expression in these cell types. Interestingly, periostin has been shown to enhance signaling through EGFR and integrin αvβ5 to induce epithelial-mesenchymal transition (EMT)57, a process associated with the loss of epithelial cell markers as epithelial cells adopt a fibroblast-like phenotype and increased migratory properties (e.g., loss of cell adhesion)58. While a previous report demonstrated that DSG1 supports epithelial differentiation and reduces proliferative capacity through negative regulation of EGFR signaling14, a similar suppressive effect of DSG1 on EMT induction has not been addressed. As it has been recently proposed that EMT is actively occurring in EoE59, 60, perhaps the loss of DSG1-dependent adhesion and the upregulation of periostin synergistically disrupt the homeostatic interactions between epithelial cells and fibroblasts during allergic inflammation.
The clinical importance of DSG1 and its role in maintaining epithelial integrity has been reported in several cutaneous diseases of autoimmune, infectious, and genetic origin30. In pemphigus foliaceous, autoantibodies against the ectodomain of DSG1 induce severe epidermal acantholysis30; interestingly, heightened Th2 inflammation, including elevated Th2 cytokine expression61 and the presence of activated eosinophilic infiltrates62, are observed in some instances. In Netherton syndrome, an epidermal inflammatory disease involving IBF with marked eosinophilia and elevated IgE levels63, the primary etiology has been attributed to the genetic loss of the epithelial-derived serine protease inhibitor, Kazal type, 5 (SPINK5)64. In this disease, the uncontrolled activity of endogenous trypsin-like proteases leads to aberrant cleavage of epithelial barrier proteins within the stratum corneum (including DSG1). Mice deficient in Spink5 display increased DSG1 degradation and significant epidermal pathology, including epidermal acantholysis and IBF65. However, it has been unclear whether the loss of DSG1 alone is sufficient to induce IBF as several proteases (e.g., kallikriens 5, 7, and 14 and elastase 2) and stratum corneum proteins are involved in barrier formation (e.g., filaggrin, involucrin, and loricrin) and are also altered in Netherton syndrome65–67.
Recently, genetic variants in DSG1 were identified in a new clinical syndrome manifesting with severe atopic dermatitis, multiple allergies, and metabolic wasting (SAM syndrome) in two consanguineous families68. Homozygous loss-of-function variants led to reduced expression of DSG1 and loss of cell adhesion68. Notably, one of three patients with SAM syndrome was reported to have EoE as well as elevated keratinocyte expression of IL5 and TSLP68. Interestingly, in a genome-wide association study, we identified a non-coding variant in intron 1 of DSG1 with suggestive association with EoE risk (meta p = 6.57 × 10−6)54, the function of which has yet to be determined. Nonetheless, these supportive findings in SAM coincide with our data as they provide additional evidence that links DSG1 deficiency as an initiating factor to IBF in allergic inflammation.
In summary, the observations presented herein support a pathophysiological role for DSG1 dysregulation in EoE. We propose a pathogenic cycle in which the localized increase in select Th2 cytokines (e.g. IL-13) within the inflamed esophagus decreases expression of DSG1, which is sufficient to weaken esophageal epithelial integrity and induce IBF. This loss of DSG1 initiates a pro-allergic transcriptional response (e.g., increased periostin expression) that potentiates the inflammatory response and may have far-reaching implications beyond the initial IBF insult, such as promoting EMT (Fig. 9).
Methods
Human subjects
NL (healthy control patients) were defined as having no history of EoE diagnosis with 0 eosinophils per high-power field (HPF) and no evidence of esophagitis within distal esophageal biopsies obtained during the same endoscopy procedure as the analyzed samples. Patients with EoE had clinician-diagnosed EoE and as having active disease in concomitant distal esophageal biopsies with greater than 15 eosinophils per HPF. EoE patients in disease remission (inactive) had clinician-diagnosed EoE and distal esophageal biopsies with less than 15 eosinophils per HPF (range = 0–1) following swallowed glucocorticoid therapy.
RNA sequencing (RNA-seq) and bioinformatic analyses
RNA isolated from esophageal biopsies of 6 NL and 10 patients with active EoE (mean eosinophils/HPF = 164 ± 29 SEM) was subjected to RNA sequencing at the CCHMC Gene Discovery and Genetic Variation Core as previously described69. The paired-end sequencing reads were aligned against the GRCh37 genome model using TopHat 2.04 with Bowtie 2.0370, 71. The separate alignments were then merged using Cuffmerge with RefSeq gene models as a reference. The aligned reads were then quantified for differential expression analysis using Cuffdiff72. Statistical significance was determined using a t-test with a threshold of p < 5 × 10−2 and a 2.0-fold cut-off filter in GeneSpringR 11.5 (Agilent Technologies Incorporated, Clara, CA, USA).
qPCR analysis
RNA samples were prepared as previously described34. Briefly, total RNA (250–500 ng) was DNAase treated, and cDNA was generated using the iScriptTM cDNA synthesis kit (Bio-Rad Laboratories, Hercules, CA, USA). qPCR was performed, and SYBRR Green incorporation was analyzed using iQ5 software (Bio-Rad Laboratories, Hercules, CA, USA). Specific primer sequences are listed in Supplementary Table 2.
Immunohistochemical and immunofluorescent staining. For immunohistochemical staining, formalin-fixed, paraffin-embedded distal esophageal biopsies were serially sectioned and de-paraffinized using xylene followed by graded ethanol washes. Heat-induced epitope retrieval in sodium citrate buffer (10 mM sodium citrate, 0.05% Tween 20, p.H. 6.0) was used, and endogenous peroxidase activity was quenched in 2% H2O2. Slides were blocked in PBS with 3% goat serum for 1 hr followed by overnight incubation at 4°C in the following primary antibodies (2 μg/mL): anti-DSG1 (sc-2011) (Santa Cruz Biotechnology, Santa Cruz, CA, USA) or anti-E-cadherin (#3195, Cell Signaling Technology, Inc., Danvers, MA, USA). Slides were then washed, incubated for 1 hr at room temperature in biotin-conjugated anti-rabbit IgG (1:250), and developed using the Vectastain ABC System according to manufacturer’s protocol (Vector Laboratories, Burlingame, CA, USA). Lastly, developed slides were counterstained with Harris hematoxylin. Immunofluorescent staining was performed as previously described50 using the following primary antibodies (2 μg/mL): anti-DSG1 (sc-2011) or anti-DSG3 (sc-23912) (Santa Cruz Biotechnology, Santa Cruz, CA, USA). Nuclei were stained with DAPI. Patients with active EoE that were analyzed by staining had eosinophil levels ranging from 98 to 265 eosinophils per HPF.
ALI culture system
The esophageal epithelial cell line (human telomerase reverse transcriptase [hTERT]-immortalized EPC2 cell line) was a kind gift from Dr. Anil Rustgi (University of Pennsylvania, Philadelphia, PA, USA) and has been extensively characterized in previous studies25, 26, 73. For the ALI culture system, EPC2 cells were grown to confluence while fully submerged in low-calcium ([Ca2+] = 0.09 μM) keratinocyte serum-free media (K-SFM) (Life Technologies, Grand Island, NY, USA) on 0.4-μm pore-size permeable supports (Corning Incorporated, Corning, NY, USA). Confluent monolayers were then switched to high-calcium ([Ca2+] = 1.8 μM) K-SFM for an additional 3–5 days. To induce epithelial stratification and differentiation, the culture medium was removed from the inner chamber of the permeable support in order to expose the cell monolayer to the air interface. Differentiated esophageal epithelial equivalents were analyzed 5–7 days post exposure.
DSG1 knockdown
EPC2 cells were transduced with shRNA targeting the last exon of DSG1 or a NSC shRNA using the GIPZ lentiviral system (Thermo Fisher Scientific, Rockford, IL, USA). Lentiviral particles were prepared at the CCHMC Viral Vector Core facility. Forty-eight hours post transduction, cells were selected for stable integration using puromycin (1 μg/mL), which was maintained throughout all subsequent experiments. Transduction efficiency was assessed by GFP fluorescence, and knockdown efficiency as compared to NSC shRNA-transduced cells was assessed by qPCR and immunofluorescence staining as described herein.
Expression and purification of recombinant ETA
The plasmids encoding WT ETA or the inactive ETA mutant S195A74 with a 5X histidine epitope tag75 were kindly provided by Dr. John Stanley (University of Pennsylvania, Philadelphia, PA, USA). The cDNAs were subcloned into the pT7-7 vector, validated by Sanger sequencing, and purified from isopropyl β-D-1-thiogalactopyranoside (IPTG)-induced BL21 (DE3) pLysS E. coli cell lysates using Ni+-NTA agarose column chromatography (QIAGEN Incorporated, Germantown, MD, USA). Purified ETA WT and S195A proteins were analyzed by SDS-PAGE and Coomassie staining or western blot using anti-histidine antibodies; in transiently transfected HEK293T cells, cleavage of DSG1 by WT ETA protein and non-cleavage of DSG1 by S195A ETA protein were confirmed by western blot (data not shown). Final protein concentrations were determined by bicinchoninic acid (BCA) assay (Thermo Fisher Scientific, Rockford, IL, USA).
Dispase adhesion assays
Dispase adhesion assays were performed as previously described76. Briefly, confluent monolayers of NSC or DSG1 shRNA-transduced EPC2 cells were grown in 1.8 mM Ca2+ for 24 h, washed twice with PBS, and then detached from tissue culture wells by incubation in 2.4 U/mL dispase (Life Technologies Corporation, Grand Island, NY, USA) for 15–20 min at 37°C. Detached monolayers were then subjected to mechanical stress by pipetting with a 1-mL pipet 5 times. Aliquots were then cyto-centrifuged and stained with Hema 3R (Thermo Fisher Scientific, Rockford, IL, USA). One-, two-, and three-cell clusters were counted from the entire field under 10X magnification.
Microarray analyses
RNA was isolated using the miRNeasy kit (QIAGEN Incorporated, Germantown, MD) according to the manufacturer’s protocol. RNA quality assessment, library preparation, hybridization to the GeneChipR Human Gene 2.0 ST exon array (Affymetrix, Santa Clara, CA, USA), and analysis were performed at the CCHMC Gene Expression Microarray Core. Expression profiles were analyzed using GeneSpringR 11.5 (Agilent Technologies Incorporated, Clara, CA, USA), and statistical significance was determined using a t-test with a threshold of p < 5 × 10−2 and a 2.0-fold cut-off filter.
Electron microscopy
Transmission electron microscopy was performed at the Pathology Research Core at Cincinnati Children’s Hospital Medical Center (CCHMC). Biopsy specimens used in this analysis were from NL (n = 3) and from patients with active EoE (n = 3) who had a previous EoE diagnosis and an esophageal biopsy with greater than 15 eosinophils per HPF (range = 60–100 eosinophils/HPF). The representative images shown were taken at 10,000X magnification.
TER and paracellular flux assays
For ex vivo studies, esophageal biopsies from NL or patients with active EoE were mounted into mini-Ussing chambers, and TER measurements and paracellular flux assays using FITC-dextran (average molecular weight = 4 kDa) were performed as previously described77. A total of 6 NL biopsies were analyzed for TER and paracellular permeability. A total of 9 and 4 biopsies from patients with EoE were assessed for TER and paracellular permeability, respectively. To account for potential differences in biopsy thickness across the different patient groups, an arbitrary cut-off of 2% FITC-dextran flux was used, resulting in 5 of the 9 biopsies from patients with EoE being excluded. The data shown represent the mean ± SEM. In vitro measurements for TER following IL-13 treatment were assessed using an EVOM (World Precision Instruments, Inc., Sarasota, FL, USA), whereas TER and paracellular flux assays in DSG1-deficient cells were performed as previously described78.
Statistical analyses
Statistical significance was determined using a t-test (two-tailed). Non-normally distributed data from patient biopsy samples were analyzed using a Mann-Whitney test, and the Spearman correlation was used to test for correlated gene expression. All statistical analyses were performed using GraphPad PrismR (GraphPad Software Incorporated, La Jolla, CA, USA).
Study approvals
For human subjects, written informed consent was obtained prior to a patient’s enrollment in the studies, and all human studies were approved by the CCHMC Institutional Review Board (IRB protocol 2008-0090). All experiments involving mice were approved by the CCHMC IACUC.
Supplementary Material
Acknowledgments
This work was supported in part by NIH U19 AI070235, NIH R01 DK076893, the PHS Grant P30 DK0789392, Food Allergy Research & Education (FARE, formerly Food Allergy Project [FAP] / Food Allergy Initiative [FAI] and Food Allergy & Anaphylaxis Network [FAAN]), the Buckeye Foundation, the Campaign Urging Research for Eosinophilic Diseases (CURED) Foundation, and the Angels for Eosinophilic Research Foundation. J.D.S. was supported by a T32 NIH training grant (HL091805) and the Thrasher Research Fund Early Career Award (NR-0171). We would like to Dr. John Stanley (University of Pennsylvania) for the ETA constructs and Dr. Anil Rustgi (University of Pennsylvania) for the EPC2-hTERT cell line. We would also like to thank Shawna Hottinger for editorial assistance, all of the participating families and the Cincinnati Center for Eosinophilic Disorders, and members of the Division of Allergy and Immunology for improving the health of children with allergic and immune conditions through innovative research, outstanding clinical care, and education of the current and next generation of leaders in healthcare and research.
Footnotes
Disclosures
R.C.O. has research funding from Takeda Pharmaceuticals and Astra Zeneca. S.P.H. is a consultant with Immune Pharmaceuticals. M.E.R. has an equity interest in reslizumab (Teva Pharmaceuticals), is a consultant for Immune Pharmaceuticals, is the inventor of several eosinophilic esophagitis (EoE)-related patents owned by Cincinnati Children’s Hospital Medical Center (CCHMC), is on the American Partnership for Eosinophilic Disorders (APFED) Medical Advisory Board, and is on the Steering Committees of the International Eosinophil Society (IES) and The International Gastrointestinal Eosinophil Researchers (TIGERS). The remaining authors have no disclosures.
References
- 1.Mulder DJ, Justinich CJ. Understanding eosinophilic esophagitis: the cellular and molecular mechanisms of an emerging disease. Mucosal Immunol. 2011;4(2):139–147. doi: 10.1038/mi.2010.88. [DOI] [PubMed] [Google Scholar]
- 2.Liacouras CA, Furuta GT, Hirano I, Atkins D, Attwood SE, Bonis PA, et al. Eosinophilic esophagitis: updated consensus recommendations for children and adults. J Allergy Clin Immunol. 2011;128(1):3–20. e26. doi: 10.1016/j.jaci.2011.02.040. quiz 21–22. [DOI] [PubMed] [Google Scholar]
- 3.Abonia JP, Rothenberg ME. Eosinophilic esophagitis: rapidly advancing insights. Annual review of medicine. 2012;63:421–434. doi: 10.1146/annurev-med-041610-134138. [DOI] [PubMed] [Google Scholar]
- 4.Parfitt JR, Gregor JC, Suskin NG, Jawa HA, Driman DK. Eosinophilic esophagitis in adults: distinguishing features from gastroesophageal reflux disease: a study of 41 patients. Mod Pathol. 2006;19(1):90–96. doi: 10.1038/modpathol.3800498. [DOI] [PubMed] [Google Scholar]
- 5.Ravelli AM, Villanacci V, Ruzzenenti N, Grigolato P, Tobanelli P, Klersy C, et al. Dilated intercellular spaces: a major morphological feature of esophagitis. Journal of pediatric gastroenterology and nutrition. 2006;42(5):510–515. doi: 10.1097/01.mpg.0000215312.78664.b9. [DOI] [PubMed] [Google Scholar]
- 6.Mueller S, Neureiter D, Aigner T, Stolte M. Comparison of histological parameters for the diagnosis of eosinophilic oesophagitis versus gastro-oesophageal reflux disease on oesophageal biopsy material. Histopathology. 2008;53(6):676–684. doi: 10.1111/j.1365-2559.2008.03187.x. [DOI] [PubMed] [Google Scholar]
- 7.Ingerski LM, Modi AC, Hood KK, Pai AL, Zeller M, Piazza-Waggoner C, et al. Health-related quality of life across pediatric chronic conditions. J Pediatr. 2010;156(4):639–644. doi: 10.1016/j.jpeds.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Menard-Katcher P, Marks KL, Liacouras CA, Spergel JM, Yang YX, Falk GW. The natural history of eosinophilic oesophagitis in the transition from childhood to adulthood. Alimentary pharmacology & therapeutics. 2013;37(1):114–121. doi: 10.1111/apt.12119. [DOI] [PubMed] [Google Scholar]
- 9.Harris RF, Menard-Katcher C, Atkins D, Furuta GT, Klinnert MD. Psychosocial Dysfunction in Children and Adolescents with Eosinophilic Esophagitis. J Pediatr Gastroenterol Nutr. 2013 doi: 10.1097/MPG.0b013e31829ce5ad. [DOI] [PubMed] [Google Scholar]
- 10.Blanchard C, Wang N, Stringer KF, Mishra A, Fulkerson PC, Abonia JP, et al. Eotaxin-3 and a uniquely conserved gene-expression profile in eosinophilic esophagitis. J Clin Invest. 2006;116(2):536–547. doi: 10.1172/JCI26679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blanchard C, Mingler MK, Vicario M, Abonia JP, Wu YY, Lu TX, et al. IL-13 involvement in eosinophilic esophagitis: transcriptome analysis and reversibility with glucocorticoids. J Allergy Clin Immunol. 2007;120(6):1292–1300. doi: 10.1016/j.jaci.2007.10.024. [DOI] [PubMed] [Google Scholar]
- 12.Simpson CL, Patel DM, Green KJ. Deconstructing the skin: cytoarchitectural determinants of epidermal morphogenesis. Nature reviews Molecular cell biology. 2011;12(9):565–580. doi: 10.1038/nrm3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanakawa Y, Amagai M, Shirakata Y, Yahata Y, Tokumaru S, Yamasaki K, et al. Differential effects of desmoglein 1 and desmoglein 3 on desmosome formation. J Invest Dermatol. 2002;119(6):1231–1236. doi: 10.1046/j.1523-1747.2002.19648.x. [DOI] [PubMed] [Google Scholar]
- 14.Getsios S, Simpson CL, Kojima S, Harmon R, Sheu LJ, Dusek RL, et al. Desmoglein 1-dependent suppression of EGFR signaling promotes epidermal differentiation and morphogenesis. J Cell Biol. 2009;185(7):1243–1258. doi: 10.1083/jcb.200809044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen YJ, Lee LY, Chao YK, Chang JT, Lu YC, Li HF, et al. DSG3 Facilitates Cancer Cell Growth and Invasion through the DSG3-Plakoglobin-TCF/LEF-Myc/Cyclin D1/MMP Signaling Pathway. PLoS One. 2013;8(5):e64088. doi: 10.1371/journal.pone.0064088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown L, Waseem A, Cruz IN, Szary J, Gunic E, Mannan T, et al. Desmoglein 3 promotes cancer cell migration and invasion by regulating activator protein 1 and protein kinase C-dependent-Ezrin activation. Oncogene. 2013 doi: 10.1038/onc.2013.186. [DOI] [PubMed] [Google Scholar]
- 17.Mannan T, Jing S, Foroushania SH, Fortune F, Wan H. RNAi-mediated inhibition of the desmosomal cadherin (desmoglein 3) impairs epithelial cell proliferation. Cell proliferation. 2011;44(4):301–310. doi: 10.1111/j.1365-2184.2011.00765.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Green KJ, Simpson CL. Desmosomes: new perspectives on a classic. J Invest Dermatol. 2007;127(11):2499–2515. doi: 10.1038/sj.jid.5701015. [DOI] [PubMed] [Google Scholar]
- 19.Green KJ, Gaudry CA. Are desmosomes more than tethers for intermediate filaments? Nature reviews Molecular cell biology. 2000;1(3):208–216. doi: 10.1038/35043032. [DOI] [PubMed] [Google Scholar]
- 20.Harmon RM, Simpson CL, Johnson JL, Koetsier JL, Dubash AD, Najor NA, et al. Desmoglein-1/Erbin interaction suppresses ERK activation to support epidermal differentiation. The Journal of clinical investigation. 2013;123(4):1556–1570. doi: 10.1172/JCI65220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shirakata Y, Amagai M, Hanakawa Y, Nishikawa T, Hashimoto K. Lack of mucosal involvement in pemphigus foliaceus may be due to low expression of desmoglein 1. J Invest Dermatol. 1998;110(1):76–78. doi: 10.1046/j.1523-1747.1998.00085.x. [DOI] [PubMed] [Google Scholar]
- 22.Tunggal JA, Helfrich I, Schmitz A, Schwarz H, Gunzel D, Fromm M, et al. E-cadherin is essential for in vivo epidermal barrier function by regulating tight junctions. EMBO J. 2005;24(6):1146–1156. doi: 10.1038/sj.emboj.7600605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kalabis J, Wong GS, Vega ME, Natsuizaka M, Robertson ES, Herlyn M, et al. Isolation and characterization of mouse and human esophageal epithelial cells in 3D organotypic culture. Nature protocols. 2012;7(2):235–246. doi: 10.1038/nprot.2011.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frankart A, Malaisse J, De Vuyst E, Minner F, de Rouvroit CL, Poumay Y. Epidermal morphogenesis during progressive in vitro 3D reconstruction at the air-liquid interface. Exp Dermatol. 2012;21(11):871–875. doi: 10.1111/exd.12020. [DOI] [PubMed] [Google Scholar]
- 25.Andl CD, Mizushima T, Nakagawa H, Oyama K, Harada H, Chruma K, et al. Epidermal growth factor receptor mediates increased cell proliferation, migration, and aggregation in esophageal keratinocytes in vitro and in vivo. J Biol Chem. 2003;278(3):1824–1830. doi: 10.1074/jbc.M209148200. [DOI] [PubMed] [Google Scholar]
- 26.Oyama K, Okawa T, Nakagawa H, Takaoka M, Andl CD, Kim SH, et al. AKT induces senescence in primary esophageal epithelial cells but is permissive for differentiation as revealed in organotypic culture. Oncogene. 2007;26(16):2353–2364. doi: 10.1038/sj.onc.1210025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Michaylira CZ, Wong GS, Miller CG, Gutierrez CM, Nakagawa H, Hammond R, et al. Periostin, a cell adhesion molecule, facilitates invasion in the tumor microenvironment and annotates a novel tumor-invasive signature in esophageal cancer. Cancer research. 2010;70(13):5281–5292. doi: 10.1158/0008-5472.CAN-10-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shirakawa Y, Naomoto Y, Kimura M, Kawashima R, Yamatsuji T, Tamaki T, et al. Topological analysis of p21WAF1/CIP1 expression in esophageal squamous dysplasia. Clinical cancer research : an official journal of the American Association for Cancer Research. 2000;6(2):541–550. [PubMed] [Google Scholar]
- 29.Ohkawa T, Naomoto Y, Takaoka M, Nobuhisa T, Noma K, Motoki T, et al. Localization of heparanase in esophageal cancer cells: respective roles in prognosis and differentiation. Laboratory investigation; a journal of technical methods and pathology. 2004;84(10):1289–1304. doi: 10.1038/labinvest.3700159. [DOI] [PubMed] [Google Scholar]
- 30.Amagai M, Matsuyoshi N, Wang ZH, Andl C, Stanley JR. Toxin in bullous impetigo and staphylococcal scalded-skin syndrome targets desmoglein 1. Nat Med. 2000;6(11):1275–1277. doi: 10.1038/81385. [DOI] [PubMed] [Google Scholar]
- 31.Rodrigo S, Abboud G, Oh D, DeMeester SR, Hagen J, Lipham J, et al. High intraepithelial eosinophil counts in esophageal squamous epithelium are not specific for eosinophilic esophagitis in adults. Am J Gastroenterol. 2008;103(2):435–442. doi: 10.1111/j.1572-0241.2007.01594.x. [DOI] [PubMed] [Google Scholar]
- 32.Orlando LA, Orlando RC. Dilated intercellular spaces as a marker of GERD. Curr Gastroenterol Rep. 2009;11(3):190–194. doi: 10.1007/s11894-009-0030-6. [DOI] [PubMed] [Google Scholar]
- 33.Blanchard C, Stucke EM, Burwinkel K, Caldwell JM, Collins MH, Ahrens A, et al. Coordinate interaction between IL-13 and epithelial differentiation cluster genes in eosinophilic esophagitis. J Immunol. 2010;184(7):4033–4041. doi: 10.4049/jimmunol.0903069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zuo L, Fulkerson PC, Finkelman FD, Mingler M, Fischetti CA, Blanchard C, et al. IL-13 induces esophageal remodeling and gene expression by an eosinophil-independent, IL-13R alpha 2-inhibited pathway. J Immunol. 2010;185(1):660–669. doi: 10.4049/jimmunol.1000471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wex T, Monkemuller K, Stahr A, Kuester D, Fry LC, Volkel S, et al. Gastro-oesophageal reflux disease is associated with up-regulation of desmosomal components in oesophageal mucosa. Histopathology. 2012;60(3):405–415. doi: 10.1111/j.1365-2559.2011.04123.x. [DOI] [PubMed] [Google Scholar]
- 36.Brennan D, Hu Y, Kljuic A, Choi Y, Joubeh S, Bashkin M, et al. Differential structural properties and expression patterns suggest functional significance for multiple mouse desmoglein 1 isoforms. Differentiation; research in biological diversity. 2004;72(8):434–449. doi: 10.1111/j.1432-0436.2004.07208009.x. [DOI] [PubMed] [Google Scholar]
- 37.Hatano Y, Adachi Y, Elias PM, Crumrine D, Sakai T, Kurahashi R, et al. The Th2 cytokine, interleukin-4, abrogates the cohesion of normal stratum corneum in mice: implications for pathogenesis of atopic dermatitis. Exp Dermatol. 2013;22(1):30–35. doi: 10.1111/exd.12047. [DOI] [PubMed] [Google Scholar]
- 38.Blanchard C, Stucke EM, Rodriguez-Jimenez B, Burwinkel K, Collins MH, Ahrens A, et al. A striking local esophageal cytokine expression profile in eosinophilic esophagitis. J Allergy Clin Immunol. 2011;127(1):208–217. 217 e201–207. doi: 10.1016/j.jaci.2010.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vicario M, Blanchard C, Stringer KF, Collins MH, Mingler MK, Ahrens A, et al. Local B cells and IgE production in the oesophageal mucosa in eosinophilic oesophagitis. Gut. 2010;59(1):12–20. doi: 10.1136/gut.2009.178020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsukita S, Furuse M, Itoh M. Multifunctional strands in tight junctions. Nat Rev Mol Cell Biol. 2001;2(4):285–293. doi: 10.1038/35067088. [DOI] [PubMed] [Google Scholar]
- 41.Yoshida K, Yokouchi M, Nagao K, Ishii K, Amagai M, Kubo A. Functional tight junction barrier localizes in the second layer of the stratum granulosum of human epidermis. Journal of dermatological science. 2013;71(2):89–99. doi: 10.1016/j.jdermsci.2013.04.021. [DOI] [PubMed] [Google Scholar]
- 42.Furuse M, Hata M, Furuse K, Yoshida Y, Haratake A, Sugitani Y, et al. Claudin-based tight junctions are crucial for the mammalian epidermal barrier: a lesson from claudin-1-deficient mice. The Journal of cell biology. 2002;156(6):1099–1111. doi: 10.1083/jcb.200110122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kirschner N, Rosenthal R, Furuse M, Moll I, Fromm M, Brandner JM. Contribution of tight junction proteins to ion, macromolecule, and water barrier in keratinocytes. The Journal of investigative dermatology. 2013;133(5):1161–1169. doi: 10.1038/jid.2012.507. [DOI] [PubMed] [Google Scholar]
- 44.Monkemuller K, Wex T, Kuester D, Fry LC, Kandulski A, Kropf S, et al. Role of tight junction proteins in gastroesophageal reflux disease. BMC gastroenterology. 2012;12:128. doi: 10.1186/1471-230X-12-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kubo A, Nagao K, Yokouchi M, Sasaki H, Amagai M. External antigen uptake by Langerhans cells with reorganization of epidermal tight junction barriers. The Journal of experimental medicine. 2009;206(13):2937–2946. doi: 10.1084/jem.20091527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lucendo AJ, Navarro M, Comas C, Pascual JM, Burgos E, Santamaria L, et al. Immunophenotypic characterization and quantification of the epithelial inflammatory infiltrate in eosinophilic esophagitis through stereology: an analysis of the cellular mechanisms of the disease and the immunologic capacity of the esophagus. The American journal of surgical pathology. 2007;31(4):598–606. doi: 10.1097/01.pas.0000213392.49698.8c. [DOI] [PubMed] [Google Scholar]
- 47.Yen EH, Hornick JL, Dehlink E, Dokter M, Baker A, Fiebiger E, et al. Comparative analysis of Fcepsilon RI expression patterns in patients with eosinophilic and reflux esophagitis. J Pediatr Gastroenterol Nutr. 2010;51(5):584–592. doi: 10.1097/MPG.0b013e3181de7685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bannert C, Bidmon-Fliegenschnee B, Stary G, Hotzy F, Stift J, Nurko S, et al. Fc-epsilon-RI, the high affinity IgE-receptor, is robustly expressed in the upper gastrointestinal tract and modulated by mucosal inflammation. PLoS One. 2012;7(7):e42066. doi: 10.1371/journal.pone.0042066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mulder DJ, Pooni A, Mak N, Hurlbut DJ, Basta S, Justinich CJ. Antigen presentation and MHC class II expression by human esophageal epithelial cells: role in eosinophilic esophagitis. Am J Pathol. 2011;178(2):744–753. doi: 10.1016/j.ajpath.2010.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Blanchard C, Mingler MK, McBride M, Putnam PE, Collins MH, Chang G, et al. Periostin facilitates eosinophil tissue infiltration in allergic lung and esophageal responses. Mucosal Immunol. 2008;1(4):289–296. doi: 10.1038/mi.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jia G, Erickson RW, Choy DF, Mosesova S, Wu LC, Solberg OD, et al. Periostin is a systemic biomarker of eosinophilic airway inflammation in asthmatic patients. The Journal of allergy and clinical immunology. 2012;130(3):647–654. e610. doi: 10.1016/j.jaci.2012.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Masuoka M, Shiraishi H, Ohta S, Suzuki S, Arima K, Aoki S, et al. Periostin promotes chronic allergic inflammation in response to Th2 cytokines. J Clin Invest. 2012;122(7):2590–2600. doi: 10.1172/JCI58978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Johansson MW, Annis DS, Mosher DF. alpha(M)beta(2) integrin-mediated adhesion and motility of IL-5-stimulated eosinophils on periostin. American journal of respiratory cell and molecular biology. 2013;48(4):503–510. doi: 10.1165/rcmb.2012-0150OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rothenberg ME, Spergel JM, Sherrill JD, Annaiah K, Martin LJ, Cianferoni A, et al. Common variants at 5q22 associate with pediatric eosinophilic esophagitis. Nat Genet. 2010;42(4):289–291. doi: 10.1038/ng.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sherrill JD, Gao PS, Stucke EM, Blanchard C, Collins MH, Putnam PE, et al. Variants of thymic stromal lymphopoietin and its receptor associate with eosinophilic esophagitis. J Allergy Clin Immunol. 2010;126(1):160–165. e163. doi: 10.1016/j.jaci.2010.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Takayama G, Arima K, Kanaji T, Toda S, Tanaka H, Shoji S, et al. Periostin: a novel component of subepithelial fibrosis of bronchial asthma downstream of IL-4 and IL-13 signals. The Journal of allergy and clinical immunology. 2006;118(1):98–104. doi: 10.1016/j.jaci.2006.02.046. [DOI] [PubMed] [Google Scholar]
- 57.Yan W, Shao R. Transduction of a mesenchyme-specific gene periostin into 293T cells induces cell invasive activity through epithelial-mesenchymal transformation. The Journal of biological chemistry. 2006;281(28):19700–19708. doi: 10.1074/jbc.M601856200. [DOI] [PubMed] [Google Scholar]
- 58.Kalluri R, Neilson EG. Epithelial-mesenchymal transition and its implications for fibrosis. The Journal of clinical investigation. 2003;112(12):1776–1784. doi: 10.1172/JCI20530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kagalwalla AF, Akhtar N, Woodruff SA, Rea BA, Masterson JC, Mukkada V, et al. Eosinophilic esophagitis: epithelial mesenchymal transition contributes to esophageal remodeling and reverses with treatment. The Journal of allergy and clinical immunology. 2012;129(5):1387–1396. e1387. doi: 10.1016/j.jaci.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Muir AB, Lim DM, Benitez AJ, Modayur Chandramouleeswaran P, Lee AJ, Ruchelli ED, et al. Esophageal epithelial and mesenchymal cross-talk leads to features of epithelial to mesenchymal transition in vitro. Experimental cell research. 2013;319(6):850–859. doi: 10.1016/j.yexcr.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lin MS, Fu CL, Aoki V, Hans-Filho G, Rivitti EA, Moraes JR, et al. Desmoglein-1-specific T lymphocytes from patients with endemic pemphigus foliaceus (fogo selvagem) J Clin Invest. 2000;105(2):207–213. doi: 10.1172/JCI8075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Simon D, Hoesli S, Roth N, Staedler S, Yousefi S, Simon HU. Eosinophil extracellular DNA traps in skin diseases. J Allergy Clin Immunol. 2011;127(1):194–199. doi: 10.1016/j.jaci.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 63.Smith DL, Smith JG, Wong SW, deShazo RD. Netherton’s syndrome: a syndrome of elevated IgE and characteristic skin and hair findings. The Journal of allergy and clinical immunology. 1995;95(1 Pt 1):116–123. doi: 10.1016/s0091-6749(95)70159-1. [DOI] [PubMed] [Google Scholar]
- 64.Chavanas S, Bodemer C, Rochat A, Hamel-Teillac D, Ali M, Irvine AD, et al. Mutations in SPINK5, encoding a serine protease inhibitor, cause Netherton syndrome. Nature genetics. 2000;25(2):141–142. doi: 10.1038/75977. [DOI] [PubMed] [Google Scholar]
- 65.Descargues P, Deraison C, Bonnart C, Kreft M, Kishibe M, Ishida-Yamamoto A, et al. Spink5-deficient mice mimic Netherton syndrome through degradation of desmoglein 1 by epidermal protease hyperactivity. Nat Genet. 2005;37(1):56–65. doi: 10.1038/ng1493. [DOI] [PubMed] [Google Scholar]
- 66.Deraison C, Bonnart C, Lopez F, Besson C, Robinson R, Jayakumar A, et al. LEKTI fragments specifically inhibit KLK5, KLK7, and KLK14 and control desquamation through a pH-dependent interaction. Molecular biology of the cell. 2007;18(9):3607–3619. doi: 10.1091/mbc.E07-02-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bonnart C, Deraison C, Lacroix M, Uchida Y, Besson C, Robin A, et al. Elastase 2 is expressed in human and mouse epidermis and impairs skin barrier function in Netherton syndrome through filaggrin and lipid misprocessing. The Journal of clinical investigation. 2010;120(3):871–882. doi: 10.1172/JCI41440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Samuelov L, Sarig O, Harmon RM, Rapaport D, Ishida-Yamamoto A, Isakov O, et al. Desmoglein 1 deficiency results in severe dermatitis, multiple allergies and metabolic wasting. Nature genetics. 2013 doi: 10.1038/ng.2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lu TX, Sherrill JD, Wen T, Plassard AJ, Besse JA, Abonia JP, et al. MicroRNA signature in patients with eosinophilic esophagitis, reversibility with glucocorticoids, and assessment as disease biomarkers. J Allergy Clin Immunol. 2012;129(4):1064–1075. e1069. doi: 10.1016/j.jaci.2012.01.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nature biotechnology. 2013;31(1):46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Garber M, Grabherr MG, Guttman M, Trapnell C. Computational methods for transcriptome annotation and quantification using RNA-seq. Nature methods. 2011;8(6):469–477. doi: 10.1038/nmeth.1613. [DOI] [PubMed] [Google Scholar]
- 73.Harada H, Nakagawa H, Oyama K, Takaoka M, Andl CD, Jacobmeier B, et al. Telomerase induces immortalization of human esophageal keratinocytes without p16INK4a inactivation. Mol Cancer Res. 2003;1(10):729–738. [PubMed] [Google Scholar]
- 74.Vath GM, Earhart CA, Rago JV, Kim MH, Bohach GA, Schlievert PM, et al. The structure of the superantigen exfoliative toxin A suggests a novel regulation as a serine protease. Biochemistry. 1997;36(7):1559–1566. doi: 10.1021/bi962614f. [DOI] [PubMed] [Google Scholar]
- 75.Hanakawa Y, Schechter NM, Lin C, Garza L, Li H, Yamaguchi T, et al. Molecular mechanisms of blister formation in bullous impetigo and staphylococcal scalded skin syndrome. J Clin Invest. 2002;110(1):53–60. doi: 10.1172/JCI15766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ishii K, Harada R, Matsuo I, Shirakata Y, Hashimoto K, Amagai M. In vitro keratinocyte dissociation assay for evaluation of the pathogenicity of anti-desmoglein 3 IgG autoantibodies in pemphigus vulgaris. J Invest Dermatol. 2005;124(5):939–946. doi: 10.1111/j.0022-202X.2005.23714.x. [DOI] [PubMed] [Google Scholar]
- 77.Jovov B, Que J, Tobey NA, Djukic Z, Hogan BL, Orlando RC. Role of E-cadherin in the pathogenesis of gastroesophageal reflux disease. Am J Gastroenterol. 2011;106(6):1039–1047. doi: 10.1038/ajg.2011.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wu D, Ahrens R, Osterfeld H, Noah TK, Groschwitz K, Foster PS, et al. Interleukin-13 (IL-13)/IL-13 receptor alpha1 (IL-13Ralpha1) signaling regulates intestinal epithelial cystic fibrosis transmembrane conductance regulator channel-dependent Cl-secretion. J Biol Chem. 2011;286(15):13357–13369. doi: 10.1074/jbc.M110.214965. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.