Abstract
Co-translational protein targeting by the Signal Recognition Particle (SRP) is an essential cellular pathway that couples the synthesis of nascent proteins to their proper cellular localization. The bacterial SRP, which contains the minimal ribonucleoprotein core of this universally conserved targeting machine, has served as a paradigm for understanding the molecular basis of protein localization in all cells. In this review, we highlight recent biochemical and structural insights into the molecular mechanisms by which fundamental challenges faced by protein targeting machineries are met in the SRP pathway. Collectively, these studies elucidate how an essential SRP RNA and two regulatory GTPases in the SRP and SRP receptor (SR) enable this targeting machinery to recognize, sense and respond to its biological effectors, i.e. the cargo protein, the target membrane and the translocation machinery, thus driving efficient and faithful co-translational protein targeting.
Keywords: protein targeting, SecYEG, GTPases, molecular recognition and regulation, ribosome
1. Overview of protein targeting in bacteria
A major challenge for all cells is to correctly transport newly synthesized proteins from the cytosol, where they are initially synthesized, to their final cellular destination. In the 1970s, Günter Blobel postulated that newly synthesized proteins carry intrinsic signals, termed signal sequences, that encode information about their cellular location [1]. This finding spawned a new era in cell biology. In subsequent years, the signal sequences for various organelles including the endoplasmic reticulum (ER), nucleus, mitochondria and chloroplasts were identified. Targeting factors were also identified that recognize these distinct signal sequences and mediate the delivery of the substrate proteins to their respective target membranes [2].
Despite the lack of sub-cellular organelles, bacterial cells also contain distinct sites to which newly synthesized proteins must be correctly localized, including the plasma membrane and the extracellular space. Additional destinations in Gram-negative bacteria include the periplasmic space and the outer membrane. Across all bacterial species, the major protein trafficking route involves the transport of newly synthesized membrane and secretory proteins from the cytosol to the plasma membrane. As often occurs in microorganisms, bacteria have evolved multiple pathways for the targeted delivery of these proteins (Fig. 1) [3, 4].
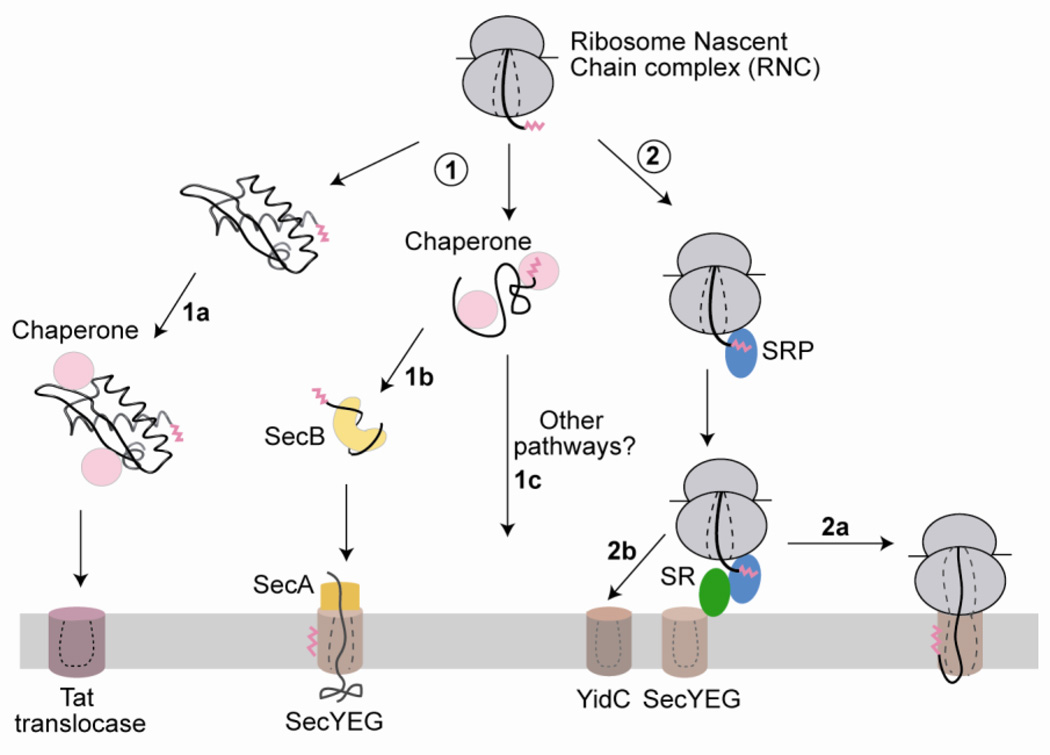
Fig. 1.
A schematic depiction of various targeting pathways for delivering proteins to the bacterial inner membrane. Newly synthesized proteins with N-terminal targeting sequences (magenta) can be targeted either post-translationally (route 1) or co-translationally (route 2). Post-translational targeting (route 1) involves targeting of the nascent protein either in a fully folded state via the Tat pathway (1a) or in an unfolded state via the chaperone SecB and the ATPase SecA (1b). Both pathways may also involve general chaperones (pink) that maintain the proteins in a translocation-competent state. The co-translational targeting pathway (route 2), which primarily handles inner membrane proteins in bacteria, is mediated by the signal recognition particle (SRP, blue) and its receptor (SR, green) (2a). Both SecA (yellow) and SRP deliver proteins to the SecYEG protein-conducting channel and may co-operate in the translocation of membrane proteins with large periplasmic domains. Translating ribosomes may also be directly delivered to the YidC translocase (2b), which may either act independently or in conjunction with SecYEG. Whether additional pathways exist for the targeting of substrates, such as tail-anchored proteins, remains to be determined (1c). The same color scheme is maintained throughout the paper.
Protein targeting in bacteria can be divided into two major routes: (a) Post-translational pathways, in which the nascent protein is completely synthesized and released from the ribosome prior to targeting (Fig. 1, route 1); (b) the co-translational pathway, in which the targeting and translocation of the nascent cargo protein is coupled to its ongoing synthesis by the ribosome (Fig. 1, route 2). Co-translational targeting is preserved throughout evolution and is the major pathway for targeting all secretory and membrane proteins to the endoplasmic reticulum in higher eukaryotes. In contrast, most secretory proteins in bacteria are targeted to the plasma membrane via post-translational mechanisms (Fig. 1, route 1). Why bacteria have evolved these different mechanisms remains unclear. It has been suggested that since protein translation is slower than translocation, it is beneficial to uncouple these pathways in rapidly growing organisms, like bacteria and yeast, to fully utilize the limited number of SecYEG translocation channels, a major translocon in the bacterial inner membrane [5]. Additional targeting mechanisms may have also evolved to accommodate specific substrates unable to use the Sec translocon (e.g. Tat pathway, see below).
Co-translational targeting is carried out by a universally conserved ribonucleoprotein complex, the Signal Recognition Particle (SRP) (Fig. 1, route 2a), which primarily mediates the targeted delivery of ribosomes translating integral membrane proteins and some periplasmic proteins to the Sec translocon (SecYEG in bacteria, Sec61p in eukaryotes) at the plasma membrane [6]. Here, a continuous channel is formed from the ribosome exit tunnel to the SecYEG translocation pore, allowing the nascent protein to be directly released into the membrane. The co-translational mode of targeting ensures that proteins containing highly hydrophobic transmembrane domains are sequestered from the aqueous environment of the cytosol and thus protected from misfolding or aggregation.
While SecYEG is the main site for protein insertion, other translocation machineries are often found to participate in membrane protein insertion in bacteria. The most notable of these is the non-homologous YidC translocon [7], which is essential in bacteria and is also found in organelles derived from them. In vivo, YidC appears to exist in two pools: one that is tightly associated with SecYEG and assists in the integration of polytopic membrane proteins [8–11], and another that acts independently of SecYEG to mediate the integration of several multi-spanning membrane proteins [12–14]. Targeting to YidC (Fig. 1, route 2b) is thought to occur via the SRP pathway, although SRP-independent mechanisms may also be involved [15]. Although YidC has been shown to bind translating ribosomes [16, 17], the mechanism by which YidC mediates insertion of its substrates is not well understood [14, 18].
Post-translational targeting of many periplasmic, outer membrane, and secretory proteins to SecYEG is carried out by the chaperone SecB, which captures newly synthesized substrate proteins in a translocation-competent state and delivers them to the ATPase SecA. SecA tightly associates with SecYEG and inserts the unfolded substrate protein across it using ATP-driven conformational changes (Fig. 1, route 1b) [3, 4, 19]. Other general chaperones, such as trigger factor (TF), may also be involved in maintaining the nascent polypeptides in a translocation-competent unfolded state. Recent reports suggest that SecA can also associate with ribosomes bearing the SecM nascent chain, raising the intriguing possibility that post-translational targeting machineries could also exert some of their actions co-translationally [20].
In an alternative targeting route, a subset of secretory proteins may be translocated in a completely folded state. This may be essential for substrate proteins that fold quickly, require cytosolic co-factors for maturation, or are multi-protein complexes in which only one subunit has a signal sequence. Substrates for this pathway have a twin arginine motif in their signal sequence and are translocated via the Tat translocon, composed of TatA, TatB and TatC subunits (Fig. 1, route 1a) [21]. How the substrate proteins, which presumably fold in the cytosol, are targeted to and translocated across the membrane by this pathway remains a mystery [22].
In addition to these pathways, there may be other mechanisms for targeting proteins to the bacterial membrane (Fig. 1, route 1c). For example, bacteria contain several proteins with putative C-terminal transmembrane domains (called tail-anchored proteins) that lack an N-terminal targeting sequence [23]. The mechanism by which these proteins are targeted to the membrane is not known. In a radically distinct mechanism, targeting could also precede translation and may instead rely on cis-acting elements in the TM-encoding regions of the mRNA [24]. The detailed mechanisms for targeting of these substrates have not been elucidated.
Despite the diversity of trafficking pathways, protein targeting can be divided into three key steps that are common to all pathways: recognition of substrates in the cytosol, their delivery to the target membrane, and passage through the membrane. The SRP pathway embodies these general principles and has served as a paradigm for understanding the molecular basis of protein localization in all cells. In this review, we focus on key events in the bacterial SRP pathway and highlight recent advances in our understanding of co-translational protein targeting.
2. SRP-mediated co-translational targeting
Although the size and composition of SRP varies significantly across species, the bacterial SRP contains the essential ribonucleoprotein core of SRP that can replace its more complex eukaryotic homologues to carry out efficient protein targeting to the endoplasmic reticulum [25, 26], highlighting the remarkable evolutionary conservation of this pathway. As such, the much simpler bacterial SRP has served as a model system to understand the fundamental molecular mechanisms and energetic principles of this targeting machine in both prokaryotic and eukaryotic cells.
Bacterial SRP is comprised of the protein Ffh (a homologue of SRP54, the only evolutionarily conserved protein component of eukaryotic SRP) bound to a 4.5S SRP RNA [25, 26]. Ffh has two functional domains connected by a flexible linker: a C-terminal M-domain, which contains the binding site for the SRP RNA and the signal peptide [27–30]; and an NG-domain composed of an N-terminal N-domain packed tightly against a central G-domain. The helical N-domain binds the ribosomal protein L23 at the ribosomal tunnel exit site, while the G-domain harbors the GTPase activity of Ffh and interacts with the SRP receptor.
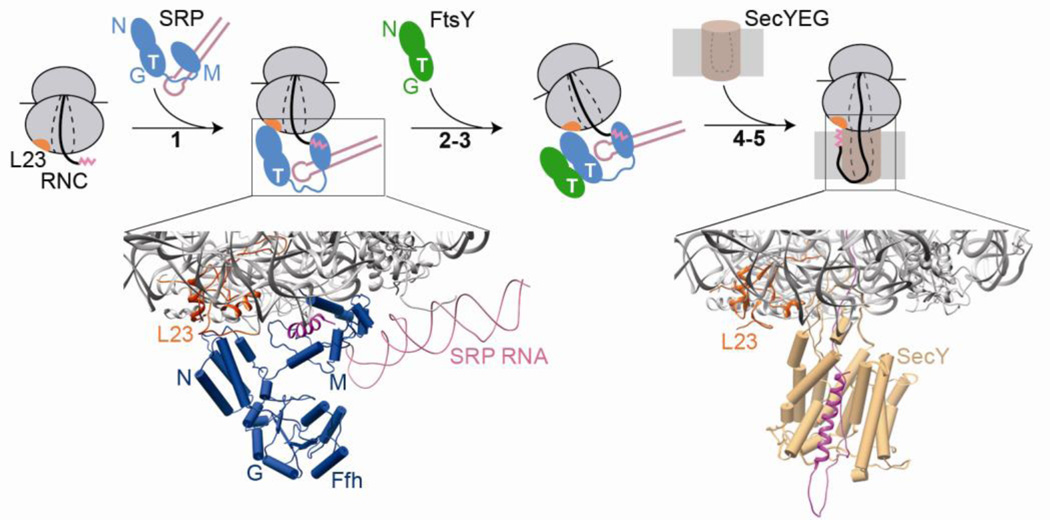
The SRP targeting cycle begins when SRP recognizes N-terminal signal sequences displayed on proteins destined for the plasma membrane as they emerge from the translating ribosome (Fig 2, step 1). The ribosome nascent chain complex (RNC or cargo) is delivered to the target membrane via the interaction of SRP with its receptor SR, which associates peripherally with the membrane (Fig 2, steps 2–3). At the membrane, the cargo is transferred to the SecYEG translocation channel (Fig 2, steps 4–5). Here, the nascent protein is either integrated into or translocated across the membrane. Meanwhile, SRP and SR dissociate and begin another round of targeting [6, 31]. Below we discuss each of these steps in greater detail.
Fig. 2.
An overview of co-translational protein targeting by the bacterial SRP. Step 1: a ribosome-nascent chain complex (RNC) displaying an SRP signal sequence (magenta) is recognized by SRP, primarily via interactions of the SRP N-domain with the ribosomal protein L23 (orange), and the SRP M-domain with the signal sequence. The lower panel shows a molecular model of the RNC-SRP complex, derived from docking the individual crystal structures of the ribosome (grey) and SRP into a cryo-electron microscopy reconstruction of the complex (PDB ID: 2j28). For clarity, only the region near the ribosome exit site (boxed in the cartoon) is shown. Steps 2–3: binding of cargo-loaded SRP to the SRP receptor (FtsY), via their homologous NG domains, localizes this complex to the membrane. Steps 4–5: the translating ribosome is transferred to the SecYEG protein-conducting channel (brown) at the membrane, which binds to the same sites on the RNC as the SRP. The lower panel shows a molecular model of RNC bound to SecYEG derived from docking the individual crystal structures of the ribosome and a homology model of SecYEG into a cryo-electron microscopy reconstruction of the complex (PDB ID: 3j00/3j01). The steps are numbered to be consistent with Figs 3 and 4.
(a) Cargo Recognition
SRP-dependent signal sequences are characterized by a stretch of hydrophobic amino acids that are minimally 8–12 residues long and preferentially adopt an α-helical structure. Thus the first transmembrane helix of an integral membrane protein can often serve as a signal sequence for SRP. These signals are highly divergent in sequence, length and amino acid composition, and lack any known consensus motifs [5, 32]. How does SRP recognize such diverse signal sequences? Early cross-linking and sequence analyses identified the M-domain of SRP as the signal sequence-binding site [33–36]. This was supported by the notion that the methionine rich M-domain of SRP provides a hydrophobic environment with sufficient plasticity to accommodate a variety of signal sequences. The crystal structures of Ffh [30] and SRP54-signal peptide fusions [28, 29] showed that the signal sequence binds into a deep, hydrophobic groove in the M-domain. Interestingly, two crystal structures solved to-date for the SRP54-signal-peptide fusions [28, 29] show different docking modes of the signal peptide, highlighting the flexibility of the signal sequence-M domain interaction.
The SRP M-domain also contains a flexible finger loop, which lines the signal-sequence binding groove of SRP [30]. The fingerloop was proposed to be important in signal sequence binding based on structural studies and the finding that mutations in this conserved region abolish the ability of SRP RNA to stimulate SRP•SR complex assembly, a process normally triggered by signal sequences or their mimics (see section 3 below) [30, 37, 38]. However, recent biochemical studies that directly measured the contribution of this interaction to cargo-SRP binding suggest that the role of fingerloop in signal sequence recognition is small [39]. Rather, it plays a crucial role in mediating communication between the two functional domains of SRP by conveying information about binding of the signal sequence in the M-domain to its NG-domain.
Nevertheless, the binding of isolated signal peptides to SRP is weak, with a dissociation constant in the micromolar range [40]. In contrast, vacant ribosomes bind SRP with an affinity of 80–100 nM [41–43]. Thus the interaction with the ribosome makes a significant contribution to the RNC-SRP binding energy and provides an important driving force for SRP recruitment to RNCs. The site of the SRP-ribosome interaction was identified from cross-linking analysis [44, 45] and cryo-EM reconstructions [46, 47] of the RNC• SRP complex. Together, these studies showed that the primary interaction occurs between the SRP N-domain and the ribosomal protein L23 adjacent to the ribosomal tunnel exit (Fig 2, step 1 and lower left panel). Additional contacts are observed between the N-domain and the ribosomal protein L29 near the ribosome exit site, and between the M-domain and the 23S ribosomal RNA and the ribosomal protein L22, although the contribution of these contacts to SRP-RNC binding remain to be determined. These multi-dentate interactions allow the SRP to bind RNCs with low to sub-nanomolar affinity [41–43, 48, 49].
(b) Interaction of SRP with SRP receptor
The membrane localization of cargo-bound SRP is mediated via interaction between the NG domains of SRP and SR. The SRP receptor, called FtsY in bacteria, is a peripheral membrane protein with an NG-domain that is highly homologous to the NG-domain of Ffh [50, 51]. As described in section (c) below, FtsY associates with membrane dynamically [52–54] and its membrane binding is enhanced by its GTP-dependent interaction with SRP [55], suggesting that the bacterial receptor likely cycles between the membrane and the cytosol.
The FtsY NG-domain is preceded by an acidic A-domain, which is thought to anchor the targeting complex to the membrane [56, 57] and mediate interactions with the SecYEG translocon [58]. The GTPase G-domains of SRP and SR share the classic Ras GTPase fold and contain the four conserved sequence motifs (GI-GIV) of the GTPase superfamily [59, 60]. Unique to the SRP family of GTPases are two additional features: (i) an insertion box domain (IBD) comprised of a β-α-β-α motif, which contains multiple catalytic residues required for GTP hydrolysis; (ii) the N-domain, which is a four-helix bundle that packs tightly against the G-domain to form a structural and functional unit (the NG domain) and plays crucial roles in SRP function (see below).
Free Ffh and FtsY have low nucleotide binding affinity and display open nucleotide-binding pockets in their crystal structures allowing free exchange of nucleotides [60–62]. In this state they exhibit low basal GTPase activity, as the IBD loops are not correctly aligned for GTP hydrolysis [63]. Ffh and FtsY also do not exhibit significant conformational differences in the apo, GDP-, or GTP- bound states. Thus unlike the canonical GTPases, they do not require GTPase activating proteins (GAPs) or guanine nucleotide exchange factors (GEFs) to regulate their GTPase cycle [64]. Instead, the GTPase cycle of SRP and SR is controlled by nucleotide-dependent dimerization, which leads to their GTPase activation (see next paragraph). Other members of this novel family of dimerization-activated GTPases include FlhF, MinD, MnmE, the dynamins, Toc proteins and septins [64–67].
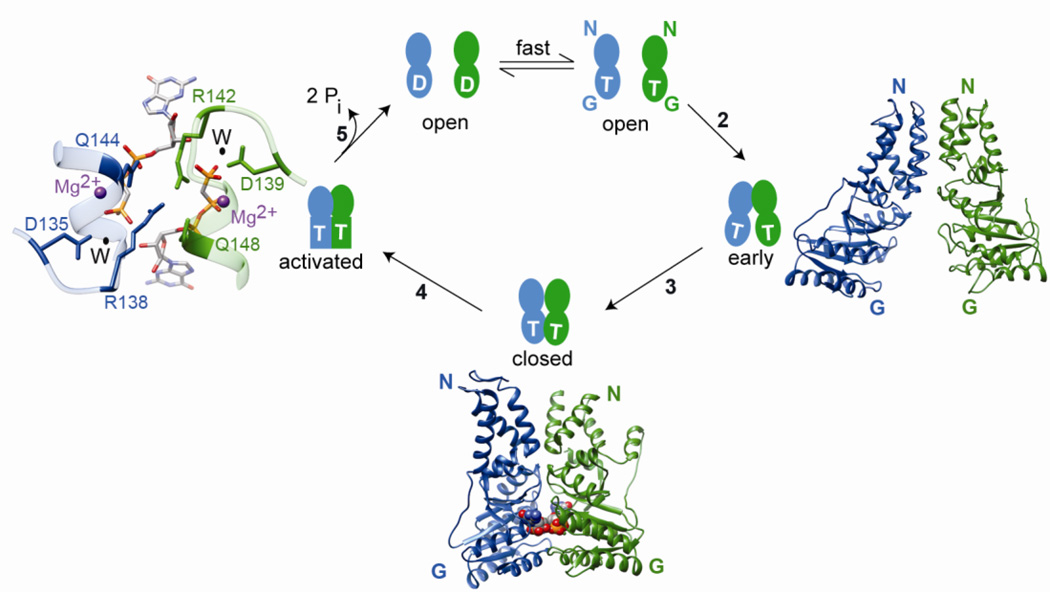
A series of discrete conformational changes occur during the dimerization of the SRP and FtsY NG-domains, which culminate in reciprocal GTPase activation in both proteins (Fig 3). SRP and FtsY can initially associate to form a transient ‘early’ intermediate independently of GTP (Fig 3, step 2) [68]. This intermediate is unstable (Kd ~ 4–10 µM) and involves electrostatic contacts between the N-domains of SRP and FtsY (Fig 3, right panel) [69–71]. The presence of bound GTP in both proteins induces a conformational change involving adjustments of the NG-interface [50, 51, 72, 73] and removal of an inhibitory N-terminal helix of FtsY [55, 74–76] (Fig 3, step 3). This results in a stable ‘closed’ complex with extensive interfacial interactions between the two G domains (Fig 3, bottom panel). The two GTP molecules also interact with each other across the dimer interface via hydrogen bonds between the 3′-OH of one GTP and the γ-phosphoryl oxygen of the other, which contributes to the enhanced stability of the closed complex and its specificity for GTP [50, 51]. The final rearrangement in the GTPase cycle involves repositioning of the catalytic residues in the IBD loop at the active site, so that the GTPases are ‘activated’ to trigger efficient GTP hydrolysis (Fig 3, step 4). Three catalytic residues in each IBD loop (Asp 135, Arg138 and Gln144 in Ffh and Asp 139, Arg142 and Gln148 in FtsY) coordinate the nucleophilic water, the active site magnesium and the γ-phosphoryl oxygen, respectively, forming a symmetric composite active site at the heterodimer interface primed for GTPase activation (Fig 3, left panel) [50, 51, 72]. Hydrolysis of GTP results in loss of stabilizing contacts mediated by the γ-phosphate at the heterodimer interface, driving the irreversible dissociation and recycling of SRP and SR (Fig 3, step 5) [63, 77].
Fig. 3.
SRP and SR are multi-state regulatory GTPases that undergo a series of conformational changes during their GTPase cycle. For clarity, only the NG-domains of SRP and SR are shown. T and D represent GTP and GDP, respectively. Under cellular conditions, nucleotide exchange on free SRP and SR is rapid and the proteins exist predominantly in the GTP-bound state. Step 2: SRP and SR GTPases first associate to form an early intermediate, which primarily involves interactions between the two N-domains. The right panel shows a molecular model for the early complex (PDB ID: 2xkv). Step 3: the G-domains of both proteins gain closer approach to one another, forming a closed complex with an extensive binding interface. The bottom panel shows a co-crystal structure of the SRP-FtsY NG domain complex (PDB ID: 1rj9) in the closed/activated conformation. The non-hydrolyzable GTP analog GMPPCP is shown in space filling model. Step 4: rearrangement of the IBD loops optimizes the position of catalytic residues relative to GTP, generating the activated conformation for efficient GTP hydrolysis. The left panel shows a magnification of the composite active site at the dimer interface for GTPase activation. The active site Mg2+ is in magenta, nucleophilic water (W) is in black and the catalytic residues of SRP (blue) and SR (green) are indicated. Step 5: GTP hydrolysis drives the disassembly and recycling of SRP and SR. The steps are numbered to be consistent with Figures 2 and 4.
Importantly, the GTPase cycle of SRP and SR is tightly coupled to their biological function. Every conformational step in this cycle is regulated by its respective effector in the targeting pathway, including the cargo protein, anionic phospholipids, and the SecYEG translocon (Fig 4), thus allowing the recognition of cargo to be effectively coupled to its efficient delivery at the membrane. For example, in the absence of biological cues, stable complex formation between SRP and SR is too slow (kon ~ 102–103 M−1s−1) [40, 63] to sustain the protein targeting reaction. An SRP-dependent substrate can strongly stabilize the otherwise labile early complex (Fig 4, step 2), thereby accelerating the stable SRP-FtsY complex assembly 1000-fold [70]. Likewise, anionic phospholipids can accelerate complex formation 160-fold by preorganizing FtsY into the closed conformation (Fig 4, step 3) [55, 78–80]. These effects ensure rapid delivery of cargo to the membrane and prevent futile cycles of GTP binding and hydrolysis.
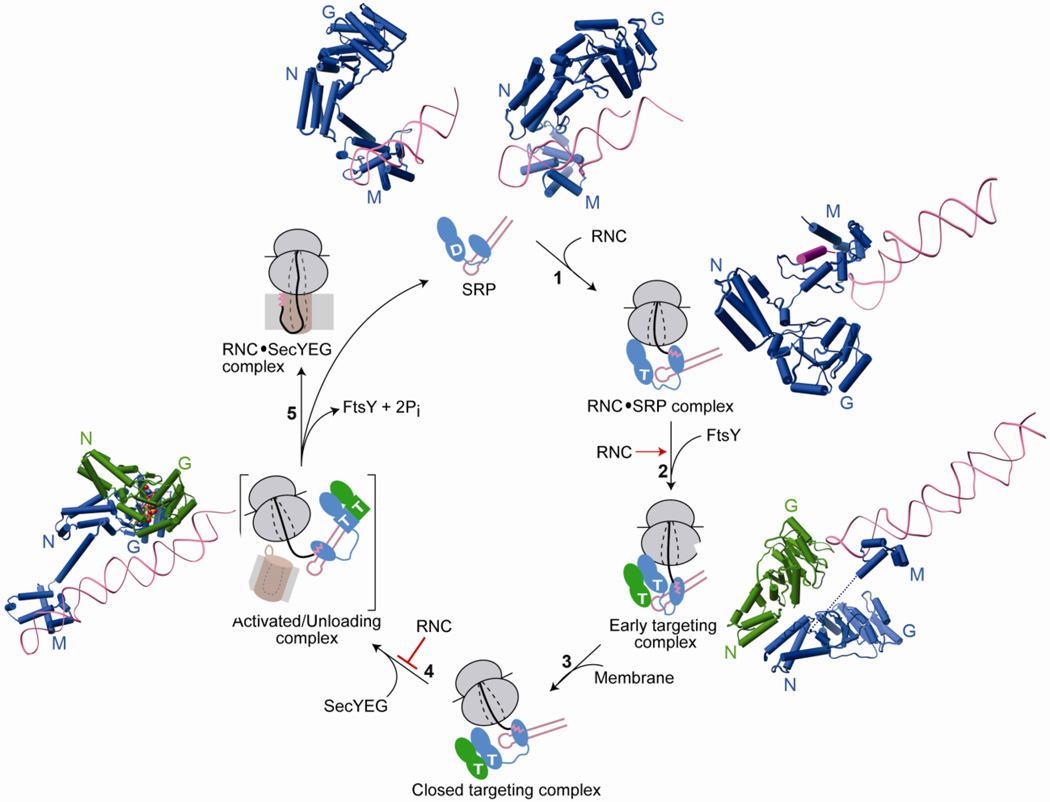
Fig. 4.
Conformational changes in SRP and SR GTPases are coupled to global reorganization of the SRP particle and are regulated by biological effectors for the pathway. Free SRP exists in a number of conformations in which the NG-domain of Ffh is oriented differently with respect to the M-domain and the SRP RNA. The top panel shows structures of SRP from S. solfataricus (PDB ID: 1qzw, left) and M. jannaschii (PDB ID: 2v3c, right) highlighting its conformational flexibility. The binding of RNC to SRP favors an SRP conformation in which the tetraloop of the SRP RNA is poised to interact with the G-domain of SR (step 1). This interaction strongly stabilizes the early targeting complex resulting in very efficient assembly of this complex (step 2). Top right (PDB ID: 2j28) and bottom right (PDB ID: 2xkv) panels show molecular models of the interaction of RNC with SRP without or with FtsY. Anionic phospholipids in the membrane strongly accelerate the rearrangement of the early targeting complex to the closed state (step 3). Interaction with SecYEG induces the SRP/SR complex into the activated state (step 4) in which the NG-domain complex relocalizes to the distal end of SRP RNA (left panel, PDB ID: 2xxa). This movement is negatively regulated by the RNC allowing a productive search for the translocon. The activated complex is shown in brackets to indicate that it is a proposed intermediate with transient lifetime, and its precise structure is not known. Hydrolysis of GTP triggers disassembly of the GTPase complex while the cargo is transferred to the translocon (step 5).
Interestingly, the cargo also slows down the rearrangement of the GTPases to the closed state and delays conformational changes that lead to GTPase activation (Fig 4, step 4) [43, 70]. This generates a highly stable RNC•SRP•FtsY complex paused at the early conformational stage, in which a strong cargo is estimated to bind SRP with picomolar affinity. What could be the role of such a ‘pausing’ effect? On the one hand, pausing delays GTP hydrolysis and thus lengthens the lifetime of the targeting complex from <1 s to ~ 8 s [70], likely providing an important time window for the targeting complex to productively search for the membrane and thus preventing abortive targeting cycles. On the other hand, pausing also provides a strategy for the SRP to discriminate against incorrect substrates, as described in section 4 [43].
Although beneficial at the early stages of targeting, continued tight binding of SRP to its cargo will be detrimental for cargo unloading. A partial resolution to this problem is provided by the conformational rearrangement of the GTPases to the closed and activated states, which is predicted to weaken cargo-SRP binding by ~400-fold and thus switch the SRP from a cargo-binding to a cargo-releasing mode [70]. In agreement with this model, cryo-EM [81] and cross-linking experiments [45] with eukaryotic SRP•SR complexes show that the NG-domain of SRP becomes mobile and detaches from its binding site on the ribosomal protein L23. Mutant GTPases that specifically inhibit the rearrangement to the activated state strongly inhibit protein targeting [82], consistent with the importance of the late GTPase rearrangements in cargo unloading. Remarkably, anionic phospholipids strongly favor the rearrangement of the targeting complex to the closed state, thus spatially coupling the delivery of the cargo to its subsequent unloading at the membrane [55, 78]. Finally, it was recently shown that SecYEG partially negates the cargo-induced stabilization of the early state and actively promotes reactivation of GTP hydrolysis [83]. These studies show that SecYEG is not a passive channel, rather it plays an active role in driving the rearrangement of the targeting complex to the activated state in which the cargo can be more readily unloaded from the SRP (Fig 4, step 5) [83]. Collectively, these results provide a coherent model for how the novel GTPase cycles in the SRP and SR provide exquisite spatial and temporal co-ordination of co-translational protein targeting.
(c) Interaction of SR with the membrane and SecYEG
Several lines of evidence including in vivo co-localization [53, 54], cell-fractionation [52] and in vitro liposome binding experiments [55, 56, 84] suggest that the interaction of FtsY with the membrane is weak and dynamic. Although the A-domain of FtsY was thought to mediate its localization at the membrane, recent studies show that a truncated version of FtsY (termed NG+1), containing an additional residue preceding the NG domain (Phe196), is sufficient for lipid binding [56, 85, 86]. This observation can be explained from a comparison of the crystal structures of the FtsY(NG) and FtsY(NG+1) constructs, which show that the presence of Phe196 in FtsY(NG+1) induces the folding of an otherwise unstructured region into an amphipathic α-helix at the N-terminus [56, 60]. This helix, rich in basic residues, is the primary lipid-binding motif of FtsY. Consistent with this finding, FtsY(NG+1) can support co-translational targeting both in vitro and in vivo [85, 86].
In vitro binding studies also show that FtsY preferentially binds anionic phospholipids, phosphatidylglycerol (PG) and cardiolipin [55, 56, 84]. These observations are supported by in vivo growth assays, in which the upregulation of genes responsible for PG and cardiolipin biosynthesis rescue an FtsY mutant defective in lipid binding [80]. Given that SecYEG and SecA also preferentially interact with anionic phospholipids [87, 88], this suggests that regions of bacterial membrane enriched in these phospholipids may act as favored sites for protein targeting and translocation.
Cross-linking and co-purification assays further suggest that FtsY can also interact with SecYEG, which could provide an attractive mechanism to localize the targeting complex to translocation sites on the membrane [89, 90]. Mutagenesis and cross-linking experiments have identified residues in the A-domain of FtsY that interact with loops connecting TMs 6–7 and TMs 8–9 (called loops c4 and c5 in bacteria) [57, 58, 90]. The importance of these interactions for co-translational protein targeting has been difficult to gauge, since the A-domain is poorly conserved and is dispensable in vivo. Further, the same residues in loops c4 and c5 of SecYEG also interact with the ribosome [58], suggesting that their interaction with FtsY is transient and needs to be broken for stable binding of SecYEG to the translating ribosome. The precise nature of these interactions and their roles in co-translational targeting remain to be determined.
(d) Cargo-SecYEG interaction
In the last step of co-translational protein targeting, the ribosome must be transferred to an essential and highly conserved protein-conducting channel, a heterotrimeric complex composed of the SecY, E, and G subunits [91]. The mechanism by which SecYEG mediates the translocation of secretory proteins across the membrane, or the integration of membrane proteins into the lipid bilayer, has been studied extensively through biochemical, genetic, and cross-linking experiments (for reviews see ref [92–95]). These studies were corroborated by the crystal structure of M. jannaschii Sec YEβ, an archeal SecYEG homolog [96], which showed that ten transmembrane helices of the SecY subunit of the translocon form an hourglass-shaped channel that provides the passageway for translocated proteins across the cell membrane. A lateral gate formed by two transmembrane helices (TM2b and TM7) serves as a binding site for signal and signal anchor sequences, and allows membrane proteins to exit the translocon laterally into the lipid bilayer [97–100]. Although SecYEG mediates the translocation of both co- and post-translationally targeted proteins, here we focus on the role of SecYEG during co-translational protein targeting.
Biochemical and genetic studies [101, 102], together with cryo-EM reconstructions of the RNC-translocon complex [103–106], showed that highly conserved basic residues in the cytosolic loops c4 and c5 interact with the ribosomal proteins L23 and L29 at the ribosome exit site (Fig 2, lower right panel). Intriguingly, both the SRP and SecYEG bind to overlapping sites on the RNC. Thus the binding of these two factors to RNC is expected to be mutually exclusive, requiring SRP to detach from the RNC to allow its stable engagement with the translocon. This raises puzzling questions as to how abortive loss of cargo is prevented and how cargo is retained at the membrane during transfer. A plausible resolution to this puzzle could involve a concerted mechanism of cargo transfer that proceeds via the formation of a RNC•SRP•FtsY•SecYEG quaternary complex. Support for such a mechanism has come from a kinetic analysis of SRP•FtsY GTPase cycle [83] and from recent studies with the SRP RNA as described in the next section.
3. SRP RNA: An active scaffold to mediate conformational changes of SRP and SR
The SRP RNA is an evolutionary conserved RNA found in all SRPs. Since its serendipitous discovery by Peter Walter in the mammalian system, it was largely thought to be a passive scaffold necessary for the correct assembly of the six mammalian SRP protein subunits. The discovery of bacterial SRP RNA ten years later challenged this view [107]. SRP RNA was found to be essential in bacteria in spite of the fact that bacterial SRP contains only one protein subunit, implying that this RNA played a more active role in co-translational protein targeting beyond scaffolding [108]. Recent biochemical and structural studies have demonstrated that indeed, the SRP RNA actively mediates the global reorganization of the SRP in response to cargo binding, thus allowing communication between the cargo and SRP/SR GTPases during co-translational protein targeting.
Bacterial SRP, which contains the most conserved domain IV of the SRP RNA, forms a hairpin structure capped by a highly conserved GGAA tetraloop. It binds with picomolar affinity to the M-domain of Ffh via two internal loops, A and B, adjacent to the tetraloop [27]. By itself, the SRP particle can attain multiple conformations in which the orientation of the Ffh NG-domain with respect to the RNA is variable as evidenced from crystal structures and structural mapping experiments (Fig 4, top panel) [30, 37, 109–111]. These results suggest that free SRP is a highly dynamic particle that can undergo substantial structural rearrangements, likely due to the 30-amino acid flexible linker connecting the M- and NG- domains of Ffh.
The binding of RNC induces a global conformational change in SRP (Fig 4, step 1) [46, 47, 112]. The M and NG domains, which bind the signal sequence and the ribosome respectively, are reoriented such that the SRP RNA lies almost perpendicularly to the ribosomal exit tunnel and its tetraloop end is next to the surface of Ffh that interacts with FtsY (Fig 4, top right panel). This is crucial because the RNA tetraloop is required to catalyze the rapid assembly of the SRP•FtsY complex [63, 68, 113, 114]. Based on kinetic and sequence analysis [115], footprinting experiments [116] and cryo-EM data [71], a key electrostatic interaction is made between the SRP RNA tetraloop and conserved basic residues including Lys399 in the G-domain of FtsY (Fig 4, step 2 and lower right panel). By stabilizing the otherwise highly labile early intermediate, this interaction accelerates the assembly of the SRP•FtsY complex by 2–3 orders of magnitude [68, 115]. Consistent with the structural observations, the stimulatory effect of the RNA tetraloop is only observed with RNCs bearing SRP-dependent signal sequences [115, 117] or with signal peptides and their mimics [40]. Together, these studies show that RNCs bearing SRP substrates favor an SRP conformation that is more conducive to rapid recruitment of the receptor, thereby ensuring efficiency and fidelity of targeting.
Although these results confirmed an essential role for the tetraloop end of the SRP RNA in protein targeting, they did not explain why bacteria needed an elongated SRP RNA containing 114 nucleotides that span >100 Å [108]. The answer to this question came from a recent crystal structure, which trapped a closed/activated state of the GTPase complex at the opposite end of the SRP RNA ~100 Å away from the tetraloop end (Fig 4, left panel) [118]. The structure was corroborated by biochemical studies, which showed that mutations at the distal site compromised the GTPase activity of the SRP•FtsY complex. These results suggest a model in which the SRP•FtsY NG domains, after initial assembly at the tetraloop end of the RNA, relocalize to its distal end where GTP hydrolysis is activated (Fig 4, steps 3–4). This movement was directly visualized by single molecule fluorescence microscopy experiments, which also showed that interaction with the RNA distal end further stimulated GTP hydrolysis in the NG-domain complex another 100-fold [119]. Importantly, the movement of the NG dimer to the RNA distal end is negatively regulated by the translating ribosome and restored by the SecYEG complex [83, 119], explaining the molecular basis for the cargo-induced ‘pausing’ of the GTPases and how this pausing effect is relieved by the SecYEG channel.
These findings also provide the first experimental support for a concerted mechanism of cargo handover from the SRP to the SecYEG complex: the movement of the GTPase complex to the SRP RNA distal end vacates the ribosomal protein L23, thereby making it accessible to SecYEG. Consistent with this notion, cross-linking and cryo-EM reconstructions of the closed targeting complex indicated the absence of SRP and FtsY NG-domains from the vicinity of the ribosome exit site [45, 81]. Co-localization and kinetic measurements further provided some evidence that the transfer of cargo happens via the formation of a RNC• SRP• SR• SecYEG quaternary complex [83, 119]. The detailed molecular mechanism of RNC transfer to the translocon and the precise nature of the quaternary intermediate remain to be elucidated. Nevertheless, the movement of the NG-dimer to the RNA distal end switches the SRP to a conformation more conducive to the unloading and transfer of cargo. This movement, which is actively promoted by SecYEG [83], provides an attractive mechanism to couple the unloading of cargo to GTP hydrolysis (Fig 4, step 5), thereby minimizing futile GTPase cycles and abortive targeting reactions.
Thus, the SRP RNA is an active molecular scaffold that can mediate large-scale protein rearrangements and exchange of distinct factors via multiple protein interaction sites, thus allowing effective co-ordination of a complex cellular pathway. Such RNA-mediated movement of proteins has been observed in other ribonucleoprotein complexes including the spliceosome [120], helicases [121] and restriction endonucleases [122]. These studies provide a general framework to facilitate understanding of similar mechanisms in other ribonucleoprotein particles.
4. Fidelity in the SRP pathway
SRP signal sequences are highly divergent in composition and lack a consensus motif [5, 32]. Thus, SRP must be sufficiently flexible to accommodate diverse signal sequences. On the other hand, accurate protein localization within the cell requires the SRP to remain highly faithful to its cognate substrates and effectively reject non-cognate substrates based on small differences in their signal sequences. How SRP meets these challenges and achieves a high fidelity of protein localization was not understood for a long time. It was previously thought that the major discrimination between SRP-dependent and SRP-independent substrates came from the weaker binding of SRP to incorrect substrates (Fig 4, step 1). Recent kinetic analyses indeed show that incorrect substrates are released from SRP significantly faster than correct ones [49]. However, quantitative measurements also show that the SRP can nevertheless bind to incorrect cargos and vacant ribosomes with substantial affinity (Kd ~ 13–100 nM) [41–43]. Thus given the cellular concentration of SRP (~ 400 nM), a significant fraction of these incorrect cargos will still bind SRP and might compromise its fidelity. How are these challenges overcome by SRP?
A quantitative analysis of the bacterial SRP pathway revealed that the conformational rearrangements during the SRP•SR GTPase cycle introduce additional fidelity checkpoints for rejecting incorrect cargos [43]. These include: (a) the efficient formation of an SRP•SR early intermediate, which is strongly stabilized by the correct cargos but not by incorrect cargos (Fig 4, step 2); (b) subsequent rearrangement of the early complex to the closed state, which is ~10 fold faster for correct cargo (Fig 4, step 3); and (c) the pausing of GTP hydrolysis in the SRP•SR complex by the correct, but not the incorrect cargos (Fig 4, step 4). This sets a differential ‘timer’ for the targeting complexes: those that carry the correct cargos have a much longer time window to locate the SecYEG translocon, whereas those carrying the incorrect cargos are aborted through premature GTP hydrolysis. A mathematical analysis of the kinetic and thermodynamic parameters of each step suggests that all of these factors are necessary to reproduce the experimentally observed pattern of substrate selection by the SRP in a reconstituted protein targeting assay [43]. Thus, fidelity in the SRP pathway is achieved via a combination of mechanisms including preferential binding, induced fit, and kinetic proofreading. These characteristics are highly reminiscent of other important biological machines including the DNA and RNA polymerases [123, 124], spliceosome [125], tRNA synthetases [126], and the ribosome [127], and may represent a general mechanism for pathways that need to differentiate between correct and incorrect substrates based on small differences.
A crucial factor that contributes to the fidelity of the SRP is the kinetic competition of the targeting pathway with the elongation of the nascent polypeptide by the ribosome. Multiple lines of evidence suggest that SRP loses its ability to target nascent proteins longer than ~140 amino acids [42, 128]. This effect might be more prominent for the bacterial SRP which, unlike the mammalian SRP, does not cause translation arrest [129, 130]. In vitro and in vivo targeting experiments show that a slower rate of translation elongation can rescue substrate proteins bearing mutant signal sequences that are sub-optimal in co-translational protein targeting under normal conditions [130, 131]. Similar observations were made either when the SRP subunits were depleted or when the kinetics of the SRP-receptor binding was compromised [129–131]. These data suggest a model in which targeting by SRP is in kinetic competition with ongoing translation and provides an important driving force for fidelity of SRP.
Additional in vivo conditions could further modulate the fidelity of the SRP-mediated protein targeting. The ribosome exit site is a crowded environment where various chaperones, modification enzymes and transport factors compete for binding the nascent chain [132, 133]. For example, the nascent chain associated complex (NAC) is a co-translational chaperone in yeast [134], which has overlapping substrate specificity with SRP. It was recently shown that the presence of NAC could modulate the binding of SRP to its substrates and help to enhance the fidelity of SRP [135, 136]. Similar mechanisms are also likely to operate in bacteria further improving the overall fidelity of the SRP-mediated protein-targeting pathway.
5. Conclusions
In summary, the biochemical accessibility of the bacterial SRP pathway has allowed an in-depth mechanistic understanding of the molecular mechanisms that underlie co-translational protein targeting. These studies show that SRP and SR are multi-state regulatory GTPases that directly respond to the biological effectors in the pathway including the cargo protein, anionic phospholipids and the translocon. The SRP RNA plays a critical role in this process by acting as a scaffold that actively drives large-scale conformational rearrangements. A concerted action of these machineries ensures the efficiency and fidelity of protein targeting. The challenges faced by SRP are general to protein targeting machineries, and the lessons learned here may be applicable to other protein targeting pathways.
HIGHLIGHTS.
Diverse pathways mediate protein targeting to the plasma membrane in bacteria
Bacterial SRP has served as a model system to understand protein targeting
GTPase rearrangements in the SRP and SRP receptor drive protein targeting
Cargo protein and the membrane translocon actively regulate SRP/SRP receptor GTPases to ensure efficient and faithful targeting
SRP RNA provides an active scaffold to mediate communications
Acknowledgments
We thank Sandra Schmid, Jennifer Doudna, Peter Walter, Ramanujan Hegde, Douglas Rees, Raymond Deshaies and Bil Clemons for support and insightful discussions over the years, and David Akopian for critical reading of the manuscript. S.S. was supported by NIH grant GM078024 and by career awards from the Henry and Camille Dreyfus foundation, the Arnold and Mabel Beckman foundation, and the David and Lucile Packard foundation. I.S. was supported by a grant from the Betty and Gordon Moore Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Blobel G. Intracellular Protein Topogenesis. Proc. Nat. Acad. Sci. USA. 1980;77:1496–1500. doi: 10.1073/pnas.77.3.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Protein targeting, transport and translocation. London: Academic Press; 2002. [Google Scholar]
- 3.Fekkes P, Driessen AJM. Protein targeting to the bacterial cytoplasmic membrane. Microbiology and Molecular Biology Reviews. 1999;63:161–173. doi: 10.1128/mmbr.63.1.161-173.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cross BCS, Sinning I, Luirink J, High S. Delivering proteins for export from the cytosol. Nat. Rev. Mol. Cell. Biol. 2009;10:255–264. doi: 10.1038/nrm2657. [DOI] [PubMed] [Google Scholar]
- 5.Hegde RS, Bernstein HD. The surprising complexity of signal sequences. Trends Biochem. Sci. 2006;31:563–571. doi: 10.1016/j.tibs.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 6.Keenan RJ, Freymann DM, Stroud RM, Walter P. The signal recognition particle. Ann. Rev. Biochem. 2001;70:755–775. doi: 10.1146/annurev.biochem.70.1.755. [DOI] [PubMed] [Google Scholar]
- 7.Wang P, Dalbey RE. Inserting membrane proteins: The YidC/Oxa1/Alb3 machinery in bacteria, mitochondria, and chloroplasts. Biochim. Biophys. Acta, Biomembr. 2011;1808:866–875. doi: 10.1016/j.bbamem.2010.08.014. [DOI] [PubMed] [Google Scholar]
- 8.Scotti PA, Urbanus ML, Brunner J, de Gier JWL, von Heijne G, van der Does C, Driessen AJM, Oudega B, Luirink J. YidC, the Escherichia coli homologue of mitochondrial Oxa1p, is a component of the Sec translocase. EMBO J. 2000;19:542–549. doi: 10.1093/emboj/19.4.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nouwen N, Driessen AJM. SecDFyajC forms a heterotetrameric complex with YidC. Molecular Microbiology. 2002;44:1397–1405. doi: 10.1046/j.1365-2958.2002.02972.x. [DOI] [PubMed] [Google Scholar]
- 10.du Plessis DJF, Nouwen N, Driessen AJM. Subunit a of cytochrome o oxidase requires both YidC and SecYEG for membrane insertion. J. Biol. Chem. 2006;281:12248–12252. doi: 10.1074/jbc.M600048200. [DOI] [PubMed] [Google Scholar]
- 11.Kol S, Majczak W, Heerlien R, van der Berg JP, Nouwen N, Driessen AJM. Subunit a of the F1F0 ATP Synthase Requires YidC and SecYEG for Membrane Insertion. J. Mol. Biol. 2009;390:893–901. doi: 10.1016/j.jmb.2009.05.074. [DOI] [PubMed] [Google Scholar]
- 12.Samuelson JC, Chen MY, Jiang FL, Moller I, Wiedmann M, Kuhn A, Phillips GJ, Dalbey RE. YidC mediates membrane protein insertion in bacteria. Nature. 2000;406:637–641. doi: 10.1038/35020586. [DOI] [PubMed] [Google Scholar]
- 13.van der Laan M, Bechtluft P, Kol S, Nouwen N, Driessen AJM. F1F0 ATP synthase subunit c is a substrate of the novel YidC pathway for membrane protein biogenesis. J. Cell Biol. 2004;165:213–222. doi: 10.1083/jcb.200402100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van der Laan M, Urbanus ML, ten Hagen-Jongman CM, Nouwen N, Oudega B, Harms N, Driessen AJM, Luirink J. A conserved function of YidC in the biogenesis of respiratory chain complexes. Proc. Nat. Acad. Sci. USA. 2003;100:5801–5806. doi: 10.1073/pnas.0636761100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Welte T, Kudva R, Kuhn P, Sturm L, Braig D, Mueller M, Warscheid B, Drepper F, Koch H-G. Promiscuous targeting of polytopic membrane proteins to SecYEG or YidC by the Escherichia coli signal recognition particle. Mol. Biol. Cell. 2012;23:464–479. doi: 10.1091/mbc.E11-07-0590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kedrov A, M S, de Keyzer J, J CJ, C WZ, Driessen AJM. Elucidating the native architecture of the YidC:Ribosome complex. J. Mol. Biol. 2013 doi: 10.1016/j.jmb.2013.07.042. 10.1016/j.jmb.2013.1007.1042. [DOI] [PubMed] [Google Scholar]
- 17.Kohler R, Boehringer D, Greber B, Bingel-Erienmeyer R, Collinson I, Schaffitzel C, Ban N. YidC and Oxa1 Form Dimeric Insertion Pores on the Translating Ribosome. Mol. Cell. 2009;34:344–353. doi: 10.1016/j.molcel.2009.04.019. [DOI] [PubMed] [Google Scholar]
- 18.van der Laan M, Houben ENG, Nouwen N, Luirink J, Driessen AJM. Reconstitution of Sec-dependent membrane protein insertion: nascent FtsQ interacts with YidC in a SecYEG-dependent manner. EMBO Reports. 2001;2:519–523. doi: 10.1093/embo-reports/kve106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cabelli RJ, Chen LL, Tai PC, Oliver DB. SecA protein is required for secretory protein translocation into Escherichia-coli membrane-vesicles. Cell. 1988;55:683–692. doi: 10.1016/0092-8674(88)90227-9. [DOI] [PubMed] [Google Scholar]
- 20.Huber D, Rajagopalan N, Preissler S, Rocco MA, Merz F, Kramer G, Bukau B. SecA Interacts with Ribosomes in Order to Facilitate Posttranslational Translocation in Bacteria. Mol. Cell. 2011;41:343–353. doi: 10.1016/j.molcel.2010.12.028. [DOI] [PubMed] [Google Scholar]
- 21.Palmer T, Berks BC. The twin-arginine translocation (Tat) protein export pathway. Nat. Rev. Microbiol. 2012;10:483–496. doi: 10.1038/nrmicro2814. [DOI] [PubMed] [Google Scholar]
- 22.Perez-Rodriguez R, Fisher AC, Perlmutter JD, Hicks MG, Chanal A, Santini C-L, Wu L-F, Palmer T, DeLisa MP. An essential role for the DnaK molecular chaperone in stabilizing over-expressed substrate proteins of the bacterial twin-arginine translocation pathway. J. Mol. Biol. 2007;367:715–730. doi: 10.1016/j.jmb.2007.01.027. [DOI] [PubMed] [Google Scholar]
- 23.Borgese N, Righi M. Remote Origins of Tail-Anchored Proteins. Traffic. 2010;11:877–885. doi: 10.1111/j.1600-0854.2010.01068.x. [DOI] [PubMed] [Google Scholar]
- 24.Nevo-Dinur K, Nussbaum-Shochat A, Ben-Yehuda S, Amster-Choder O. Translation-Independent Localization of mRNA in E. coli. Science. 2011;331:1081–1084. doi: 10.1126/science.1195691. [DOI] [PubMed] [Google Scholar]
- 25.Bernstein HD, Zopf D, Freymann DM, Walter P. Functional substitution of the signal recognition particle 54-KDa subunit by its Escherichia-coli homolog. Proc. Nat. Acad. Sci. USA. 1993;90:5229–5233. doi: 10.1073/pnas.90.11.5229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Powers T, Walter P. Co-translational protein targeting catalyzed by the Escherichia coli signal recognition particle and its receptor. EMBO J. 1997;16:4880–4886. doi: 10.1093/emboj/16.16.4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Batey RT, Rambo RP, Lucast L, Rha B, Doudna JA. Crystal structure of the ribonucleoprotein core of the signal recognition particle. Science. 2000;287:1232–1239. doi: 10.1126/science.287.5456.1232. [DOI] [PubMed] [Google Scholar]
- 28.Hainzl T, Huang SH, Merilainen G, Brannstrom K, Sauer-Eriksson AE. Structural basis of signal-sequence recognition by the signal recognition particle. Nat. Struct. Mol. Biol. 2011;18:389–391. doi: 10.1038/nsmb.1994. [DOI] [PubMed] [Google Scholar]
- 29.Janda CY, Li J, Oubridge C, Hernandez H, Robinson CV, Nagai K. Recognition of a signal peptide by the signal recognition particle. Nature. 2010;465:507–510. doi: 10.1038/nature08870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keenan RJ, Freymann DM, Walter P, Stroud RM. Crystal structure of the signal sequence binding subunit of the signal recognition particle. Cell. 1998;94:181–191. doi: 10.1016/s0092-8674(00)81418-x. [DOI] [PubMed] [Google Scholar]
- 31.Akopian D, Shen K, Zhang X, Shan S-O. Signal recognition particle: an essential protein-targeting machine. Ann. Rev. Biochem. 2013;82:693–721. doi: 10.1146/annurev-biochem-072711-164732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng N, Gierasch LM. Signal sequences: The same yet different. Cell. 1996;86:849–852. doi: 10.1016/s0092-8674(00)80159-2. [DOI] [PubMed] [Google Scholar]
- 33.Krieg UC, Walter P, Johnson AE. Photo-Cross-Linking of the Signal Sequence of Nascent Preprolactin to the 54-Kilodalton Polypeptide of the Signal Recognition Particle. Proc. Nat. Acad. Sci. USA. 1986;83:8604–8608. doi: 10.1073/pnas.83.22.8604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kurzchalia TV, Wiedmann M, Girshovich AS, Bochkareva ES, Bielka H, Rapoport TA. The signal sequence of nascent preprolactin interacts with the 54k polypeptide of the signal recognition particle. Nature. 1986;320:634–636. doi: 10.1038/320634a0. [DOI] [PubMed] [Google Scholar]
- 35.Zopf D, Bernstein HD, Johnson AE, Walter P. The Methionine-Rich Domain of the 54 Kd Protein Subunit of the Signal Recognition Particle Contains an Rna-Binding Site and Can Be Cross-Linked to a Signal Sequence. EMBO J. 1990;9:4511–4517. doi: 10.1002/j.1460-2075.1990.tb07902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bernstein HD, Poritz MA, Strub K, Hoben PJ, Brenner S, Walter P. Model for signal sequence recognition from amino-acid-sequence of 54K subunit of signal recognition particle. Nature. 1989;340:482–486. doi: 10.1038/340482a0. [DOI] [PubMed] [Google Scholar]
- 37.Rosendal KR, Wild K, Montoya G, Sinning L. Crystal structure of the complete core of archaeal signal recognition particle and implications for interdomain communication. Proc. Nat. Acad. Sci. USA. 2003;100:14701–14706. doi: 10.1073/pnas.2436132100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bradshaw N, Walter P. The signal recognition particle (SRP) RNA links conformational changes in the SRP to protein targeting. Mol. Biol. Cell. 2007;18:2728–2734. doi: 10.1091/mbc.E07-02-0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ariosa AR, Duncan SS, Saraogi I, Lu X, Brown A, Phillips GJ, Shan S-O. Fingerloop activates cargo delivery and unloading during cotranslational protein targeting. Mol. Biol. Cell. 2013;24:63–73. doi: 10.1091/mbc.E12-06-0434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bradshaw N, Neher SB, Booth DS, Walter P. Signal Sequences Activate the Catalytic Switch of SRP RNA. Science. 2009;323:127–130. doi: 10.1126/science.1165971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bornemann T, Jockel J, Rodnina MV, Wintermeyer W. Signal sequence-independent membrane targeting of ribosomes containing short nascent peptides within the exit tunnel. Nat. Struct. Mol. Biol. 2008;15:494–499. doi: 10.1038/nsmb.1402. [DOI] [PubMed] [Google Scholar]
- 42.Flanagan JJ, Chen JC, Miao YW, Shao YL, Lin JL, Bock PE, Johnson AE. Signal recognition particle binds to ribosome-bound signal sequences with fluorescence-detected subnanomolar affinity that does not diminish as the nascent chain lengthens. J. Biol. Chem. 2003;278:18628–18637. doi: 10.1074/jbc.M300173200. [DOI] [PubMed] [Google Scholar]
- 43.Zhang X, Rashid R, Wang K, Shan SO. Sequential Checkpoints Govern Substrate Selection During Cotranslational Protein Targeting. Science. 2010;328:757–760. doi: 10.1126/science.1186743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gu SQ, Peske F, Wieden HJ, Rodnina MV, Wintermeyer W. The signal recognition particle binds to protein L23 at the peptide exit of the Escherichia coli ribosome. RNA. 2003;9:566–573. doi: 10.1261/rna.2196403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pool MR, Stumm J, Fulga TA, Sinning I, Dobberstein B. Distinct modes of signal recognition particle interaction with the ribosome. Science. 2002;297:1345–1348. doi: 10.1126/science.1072366. [DOI] [PubMed] [Google Scholar]
- 46.Halic M, Blau M, Becker T, Mielke T, Pool MR, Wild K, Sinning I, Beckmann R. Following the signal sequence from ribosomal tunnel exit to signal recognition particle. Nature. 2006;444:507–511. doi: 10.1038/nature05326. [DOI] [PubMed] [Google Scholar]
- 47.Schaffitzel C, Oswald M, Berger I, Ishikawa T, Abrahams JP, Koerten HK, Koning RI, Ban N. Structure of the E-coli signal recognition particle bound to a translating ribosome. Nature. 2006;444:503–506. doi: 10.1038/nature05182. [DOI] [PubMed] [Google Scholar]
- 48.Saraogi I, Zhang D, Chandrasekaran S, Shan S. Site-specific fluorescent labeling of nascent proteins on the translating ribosome. J. Amer. Chem. Soc. 2011;133:14936–14939. doi: 10.1021/ja206626g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Holtkamp W, Lee S, Bornemann T, Senyushkina T, Rodnina MV, Wintermeyer W. Dynamic switch of the signal recognition particle from scanning to targeting. Nat. Struct. Mol. Biol. 2012;19:1332–1337. doi: 10.1038/nsmb.2421. [DOI] [PubMed] [Google Scholar]
- 50.Egea PF, Shan SO, Napetschnig J, Savage DF, Walter P, Stroud RM. Substrate twinning activates the signal recognition particle and its receptor. Nature. 2004;427:215–221. doi: 10.1038/nature02250. [DOI] [PubMed] [Google Scholar]
- 51.Focia PJ, Shepotinovskaya IV, Seidler JA, Freymann DM. Heterodimeric GTPase core of the SRP targeting complex. Science. 2004;303:373–377. doi: 10.1126/science.1090827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Luirink J, Tenhagenjongman CM, Vanderweijden CC, Oudega B, High S, Dobberstein B, Kusters R. An alternative protein targeting pathway in Escherichia-coli -studies on the role of FtsY. EMBO J. 1994;13:2289–2296. doi: 10.1002/j.1460-2075.1994.tb06511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rubio A, Jiang X, Pogliano K. Localization of translocation complex components in Bacillus subtilis: Enrichment of the signal recognition particle receptor at early sporulation septa. J. Bacteriol. 2005;187:5000–5002. doi: 10.1128/JB.187.14.5000-5002.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mircheva M, Boy D, Weiche B, Hucke F, Graumann P, Koch H-G. Predominant membrane localization is an essential feature of the bacterial signal recognition particle receptor. BMC Biology. 2009;7:76. doi: 10.1186/1741-7007-7-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lam VQ, Akopian D, Rome M, Henningsen D, Shan S. Lipid activation of the signal recognition particle receptor provides spatial coordination of protein targeting. J. Cell Biol. 2010;190:623–635. doi: 10.1083/jcb.201004129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Parlitz R, Eitan A, Stjepanovic G, Bahari L, Bange G, Bibi E, Sinning I. Escherichia coli signal recognition particle receptor FtsY contains an essential and autonomous membrane-binding amphipathic helix. J. Biol. Chem. 2007;282:32176–32184. doi: 10.1074/jbc.M705430200. [DOI] [PubMed] [Google Scholar]
- 57.Weiche B, Buerk J, Angelini S, Schiltz E, Thumfart JO, Koch H-G. A cleavable N-terminal membrane anchor is involved in membrane binding of the Escherichia coli SRP receptor. J. Mol. Biol. 2008;377:761–773. doi: 10.1016/j.jmb.2008.01.040. [DOI] [PubMed] [Google Scholar]
- 58.Kuhn P, Weiche B, Sturm L, Sommer E, Drepper F, Warscheid B, Sourjik V, Koch H-G. The Bacterial SRP Receptor, SecA and the Ribosome Use Overlapping Binding Sites on the SecY Translocon. Traffic. 2011;12:563–578. doi: 10.1111/j.1600-0854.2011.01167.x. [DOI] [PubMed] [Google Scholar]
- 59.Freymann D, Keenan R, Stroud R, Walter P. Crystal structure of the 'NG' GTPase of the prokaryotic SRP54 homolog Ffh. FASEB Journal. 1997;11:A1063–A1063. [Google Scholar]
- 60.Montoya G, Svensson C, Luirink J, Sinning I. Crystal structure of the NG domain from the signal-recognition particle receptor FtsY. Nature. 1997;385:365–368. doi: 10.1038/385365a0. [DOI] [PubMed] [Google Scholar]
- 61.Freymann DM, Keenan RJ, Stroud RM, Walter P. Structure of the conserved GTPase domain of the signal recognition particle. Nature. 1997;385:361–364. doi: 10.1038/385361a0. [DOI] [PubMed] [Google Scholar]
- 62.Freymann DM, Keenan RJ, Stroud RM, Walter P. Functional changes in the structure of the SRP GTPase on binding GDP and Mg(2+)GDP. Nat. Struct. Biol. 1999;6:793–801. doi: 10.1038/11572. [DOI] [PubMed] [Google Scholar]
- 63.Peluso P, Shan SO, Nock S, Herschlag D, Walter P. Role of SRP RNA in the GTPase cycles of ffh and FtsY. Biochemistry. 2001;40:15224–15233. doi: 10.1021/bi011639y. [DOI] [PubMed] [Google Scholar]
- 64.Gasper R, Meyer S, Gotthardt K, Sirajuddin M, Wittinghofer A. It takes two to tango: regulation of G proteins by dimerization. Nat. Rev. Mol. Cell. Biol. 2009;10:423–429. doi: 10.1038/nrm2689. [DOI] [PubMed] [Google Scholar]
- 65.Leipe DD, Wolf YI, Koonin EV, Aravind L. Classification and evolution of P-loop GTPases and related ATPases. J. Mol. Biol. 2002;317:41–72. doi: 10.1006/jmbi.2001.5378. [DOI] [PubMed] [Google Scholar]
- 66.Bange G, Kuemmerer N, Grudnik P, Lindner R, Petzold G, Kressler D, Hurt E, Wild K, Sinning I. Structural basis for the molecular evolution of SRP-GTPase activation by protein. Nat. Struct. Mol. Biol. 2011;18:1376–U1390. doi: 10.1038/nsmb.2141. [DOI] [PubMed] [Google Scholar]
- 67.Chappie JS, Acharya S, Leonard M, Schmid SL, Dyda F. G domain dimerization controls dynamin's assembly-stimulated GTPase activity. Nature. 2010;465:U435–U454. doi: 10.1038/nature09032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang X, Kung S, Shan SO. Demonstration of a multistep mechanism for assembly of the SRP.SRP receptor complex: Implications for the catalytic role of SRP RNA. J. Mol. Biol. 2008;381:581–593. doi: 10.1016/j.jmb.2008.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang X, Lam VQ, Mou Y, Kimura T, Chung J, Chandrasekar S, Winkler JR, Mayo SL, Shan S. Direct visualization reveals dynamics of a transient intermediate during protein assembly. Proc. Nat. Acad. Sci. USA. 2011;108:6450–6455. doi: 10.1073/pnas.1019051108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang X, Schaffitzel C, Ban N, Shan SO. Multiple conformational switches in a GTPase complex control co-translational protein targeting. Proc. Nat. Acad. Sci. USA. 2009;106:1754–1759. doi: 10.1073/pnas.0808573106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Estrozi LF, Boehringer D, Shan S, Ban N, Schaffitzel C. Cryo-EM structure of the E. coli translating ribosome in complex with SRP and its receptor. Nat. Struct. Mol. Biol. 2011;18:88–90. doi: 10.1038/nsmb.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shan SO, Stroud RM, Walter P. Mechanism of association and reciprocal activation of two GTPases. PLOS Biol. 2004;2:1572–1581. doi: 10.1371/journal.pbio.0020320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shan SO, Walter P. Induced nucleotide specificity in a GTPase. Proc. Nat. Acad. Sci. USA. 2003;100:4480–4485. doi: 10.1073/pnas.0737693100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Neher SB, Bradshaw N, Floor SN, Gross JD, Walter P. SRP RNA controls a conformational switch regulating the SRP-SRP receptor interaction. Nat. Struct. Mol. Biol. 2008;15:916–923. doi: 10.1038/nsmb.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gawronski-Salerno J, Freymann DM. Structure of the GMPPNP-stabilized NG domain complex of the SRP GTPases Ffh and FtsY. J. Struct. Biol. 2007;158:122–128. doi: 10.1016/j.jsb.2006.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shepotinovskaya IV, Freymann DM. Conformational change of the N-domain on formation of the complex between the GTPase domains of Thermus aquaticus Ffh and FtsY. Biochim. Biophys. Acta, Protein Struct. 2002;1597:107–114. doi: 10.1016/s0167-4838(02)00287-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Connolly T, Rapiejko PJ, Gilmore R. Requirement of GTP hydrolysis for dissociation of the signal recognition particle from its receptor. Science. 1991;252:1171–1173. doi: 10.1126/science.252.5009.1171. [DOI] [PubMed] [Google Scholar]
- 78.Braig D, Mircheva M, Sachelaru I, van der Sluis EO, Sturm L, Beckmann R, Koch H-G. Signal sequence-independent SRP-SR complex formation at the membrane suggests an alternative targeting pathway within the SRP cycle. Mol. Biol. Cell. 2011;22:2309–2323. doi: 10.1091/mbc.E11-02-0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stjepanovic G, Kapp K, Bange G, Graf C, Parlitz R, Wild K, Mayer MP, Sinning I. Lipids Trigger a Conformational Switch That Regulates Signal Recognition Particle (SRP)-mediated Protein Targeting. J. Biol. Chem. 2011;286:23489–23497. doi: 10.1074/jbc.M110.212340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Erez E, Stjepanovic G, Zelazny AM, Brugger B, Sinning I, Bibi E. Genetic Evidence for Functional Interaction of the Escherichia coli Signal Recognition Particle Receptor with Acidic Lipids in Vivo. J. Biol. Chem. 2010;285:40508–40514. doi: 10.1074/jbc.M110.140921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Halic M, Gartmann M, Schlenker O, Mielke T, Pool MR, Sinning I, Beckmann R. Signal recognition particle receptor exposes the ribosomal translocon binding site. Science. 2006;312:745–747. doi: 10.1126/science.1124864. [DOI] [PubMed] [Google Scholar]
- 82.Shan S, Chandrasekar S, Walter P. Conformational changes in the GTPase modules of the signal reception particle and its initiation of protein translocation. J. Cell Biol. 2007;178:611–620. doi: 10.1083/jcb.200702018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Akopian D, Dalal K, Shen K, Duong F, Shan S-o. SecYEG activates GTPases to drive the completion of cotranslational protein targeting. J. Cell Biol. 2013;200:397–405. doi: 10.1083/jcb.201208045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.de Leeuw E, Kaat KT, Moser C, Menestrina G, Demel R, de Kruijff B, Oudega B, Luirink J, Sinning I. Anionic phospholipids are involved in membrane association of FtsY and stimulate its GTPase activity. EMBO J. 2000;19:531–541. doi: 10.1093/emboj/19.4.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bahari L, Parlitz R, Eitan A, Stjepanovic G, Bochkareva ES, Sinning I, Bibi E. Membrane targeting of Ribosomes and their release require distinct and separable functions of ftsy. J. Biol. Chem. 2007;282:32168–32175. doi: 10.1074/jbc.M705429200. [DOI] [PubMed] [Google Scholar]
- 86.Eitan A, Bibi E. The core Escherichia coli signal recognition particle receptor contains only the N and G domains of FtsY. J. Bacteriol. 2004;186:2492–2494. doi: 10.1128/JB.186.8.2492-2494.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gold VAM, Robson A, Bao H, Romantsov T, Duong F, Collinson I. The action of cardiolipin on the bacterial translocon. Proc. Nat. Acad. Sci. USA. 2010;107:10044–10049. doi: 10.1073/pnas.0914680107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lill R, Dowhan W, Wickner W. The ATPase activity of SecA is regulated by acidic phospholipids, SecY, and the leader and mature domains of precursor proteins. Cell. 1990;60:271–280. doi: 10.1016/0092-8674(90)90742-w. [DOI] [PubMed] [Google Scholar]
- 89.Angelini S, Deitermann S, Koch HG. FtsY, the bacterial signal-recognition particle receptor, interacts functionally and physically with the SecYEG translocon. EMBO Reports. 2005;6:476–481. doi: 10.1038/sj.embor.7400385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Angelini S, Boy D, Schiltz E, Koch H-G. Membrane binding of the bacterial signal recognition particle receptor involves two distinct binding sites. J. Cell Biol. 2006;174:715–724. doi: 10.1083/jcb.200606093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Driessen AJM, Nouwen N. Protein translocation across the bacterial cytoplasmic membrane. Ann. Rev. Biochem. 2008;77:643–667. doi: 10.1146/annurev.biochem.77.061606.160747. [DOI] [PubMed] [Google Scholar]
- 92.Du Plessis DJF, Nouwen N, Driessen AJM. The Sec translocase. Biochim. Biophys. Acta, Biomembr. 2011;1808:851–865. doi: 10.1016/j.bbamem.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 93.Rapoport TA. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature. 2007;450:663–669. doi: 10.1038/nature06384. [DOI] [PubMed] [Google Scholar]
- 94.Johnson AE, van Waes MA. The translocon: A dynamic gateway at the ER membrane. Ann. Rev. Cell Dev. Biol. 1999;15:799–842. doi: 10.1146/annurev.cellbio.15.1.799. [DOI] [PubMed] [Google Scholar]
- 95.Mandon EC, Trueman SF, Gilmore R. Translocation of proteins through the Sec61 and SecYEG channels. Current Opinion in Cell Biology. 2009;21:501–507. doi: 10.1016/j.ceb.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.van der Berg B, Clemons WM, Collinson I, Modis Y, Hartmann E, Harrison SC, Rapoport TA. X-ray structure of a protein-conducting channel. Nature. 2004;427:36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- 97.Cannon KS, Or E, Clemons WM, Shibata Y, Rapoport TA. Disulfide bridge formation between SecY and a translocating polypeptide localizes the translocation pore to the center of SecY. J. Cell Biol. 2005;169:219–225. doi: 10.1083/jcb.200412019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Plath K, Wilkinson BM, Stirling CJ, Rapoport TA. Interactions between Sec complex and prepro-alpha-factor during posttranslational protein transport into the endoplasmic reticulum. Mol. Biol. Cell. 2004;15:1–10. doi: 10.1091/mbc.E03-06-0390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mothes W, Jungnickel B, Brunner J, Rapoport TA. Signal sequence recognition in cotranslational translocation by protein components of the endoplasmic reticulum membrane. J. Cell Biol. 1998;142:355–364. doi: 10.1083/jcb.142.2.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Heinrich SU, Mothes W, Brunner J, Rapoport TA. The Sec61p complex mediates the integration of a membrane protein by allowing lipid partitioning of the transmembrane domain. Cell. 2000;102:233–244. doi: 10.1016/s0092-8674(00)00028-3. [DOI] [PubMed] [Google Scholar]
- 101.Cheng ZL, Jiang Y, Mandon EC, Gilmore R. Identification of cytoplasmic residues of Sec61p involved in ribosome binding and cotranslational translocation. J. Cell Biol. 2005;168:67–77. doi: 10.1083/jcb.200408188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Menetret J-F, Schaletzky J, Clemons WM, Jr, Osborne AR, Skanland SS, Denison C, Gygi SP, Kirkpatrick DS, Park E, Ludtke SJ, Rapoport TA, Akey CW. Ribosome binding of a single copy of the SecY complex: Implications for protein translocation. Mol. Cell. 2007;28:1083–1092. doi: 10.1016/j.molcel.2007.10.034. [DOI] [PubMed] [Google Scholar]
- 103.Becker T, Bhushan S, Jarasch A, Armache J-P, Funes S, Jossinet F, Gumbart J, Mielke T, Berninghausen O, Schulten K, Westhof E, Gilmore R, Mandon EC, Beckmann R. Structure of Monomeric Yeast and Mammalian Sec61 Complexes Interacting with the Translating Ribosome. Science. 2009;326:1369–1373. doi: 10.1126/science.1178535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Beckmann R, Spahn CMT, Eswar N, Helmers J, Penczek PA, Sali A, Frank J, Blobel G. Architecture of the protein-conducting channel associated with the translating 80S ribosome. Cell. 2001;107:361–372. doi: 10.1016/s0092-8674(01)00541-4. [DOI] [PubMed] [Google Scholar]
- 105.Frauenfeld J, Gumbart J, van der Sluis EO, Funes S, Gartmann M, Beatrix B, Mielke T, Berninghausen O, Becker T, Schulten K, Beckmann R. Cryo-EM structure of the ribosome-SecYE complex in the membrane environment. Nat. Struct. Mol. Biol. 2011;18:U614–U127. doi: 10.1038/nsmb.2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mitra K, Schaffitzel C, Shaikh T, Tama F, Jenni S, Brooks CL, Ban N, Frank J. Structure of the E-coli protein-conducting channel bound to a translating ribosome. Nature. 2005;438:318–324. doi: 10.1038/nature04133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Poritz MA, Bernstein HD, Strub K, Zopf D, Wilhelm H, Walter P. An Escherichia-coli ribonucleoprotein containing 4.5S RNA resembles mammalian Signal Recognition Particle. Science. 1990;250:1111–1117. doi: 10.1126/science.1701272. [DOI] [PubMed] [Google Scholar]
- 108.Poritz MA, Strub K, Walter P. Human SRP RNA and Escherichia-coli 4.5S RNA contain a highly homologous structural domain. Cell. 1988;55:4–6. doi: 10.1016/0092-8674(88)90003-7. [DOI] [PubMed] [Google Scholar]
- 109.Hainzl T, Huang S, Sauer-Eriksson AE. Interaction of signal-recognition particle 54 GTPase domain and signal-recognition particle RNA in the free signal-recognition particle. Proc. Nat. Acad. Sci. USA. 2007;104:14911–14916. doi: 10.1073/pnas.0702467104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Buskiewicz I, Kubarenko A, Peske F, Rodnina MV, Wintermeyer W. Domain rearrangement of SRP protein Ffh upon binding 4.5S RNA and the SRP receptor FtsY. RNA. 2005;11:947–957. doi: 10.1261/rna.7242305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Buskiewicz I, Peske F, Wieden HJ, Gryczynski I, Rodnina MV, Wintermeyer W. Conformations of the signal recognition particle protein Ffh from Escherichia coli as determined by FRET. J. Mol. Biol. 2005;351:417–430. doi: 10.1016/j.jmb.2005.06.023. [DOI] [PubMed] [Google Scholar]
- 112.Buskiewicz IA, Jockel J, Rodnina MV, Wintermeyer W. Conformation of the signal recognition particle in ribosomal targeting complexes. RNA. 2009;15:44–54. doi: 10.1261/rna.1285609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jagath JR, Matassova NB, De Leeuw E, Warnecke JM, Lentzen G, Rodnina MV, Luirink J, Wintermeyer W. Important role of the tetraloop region of 4.5S RNA in SRP binding to its receptor FtsY. RNA. 2001;7:293–301. doi: 10.1017/s1355838201002205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Siu FY, Spanggord RJ, Doudna JA. SRP RNA provides the physiologically essential GTPase activation function in cotranslational protein targeting. RNA. 2007;13:240–250. doi: 10.1261/rna.135407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Shen K, Shan S. Transient tether between the SRP RNA and SRP receptor ensures efficient cargo delivery during cotranslational protein targeting. Proc. Nat. Acad. Sci. USA. 2010;107:7698–7703. doi: 10.1073/pnas.1002968107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Spanggord RJ, Siu F, Ke AL, Doudna JA. RNA-mediated interaction between the peptide-binding and GTPase domains of the signal recognition particle. Nat. Struct. Mol. Biol. 2005;12:1116–1122. doi: 10.1038/nsmb1025. [DOI] [PubMed] [Google Scholar]
- 117.Shen K, Zhang X, Shan S. Synergistic actions between the SRP RNA and translating ribosome allow efficient delivery of the correct cargos during cotranslational protein targeting. RNA. 2011;17:892–902. doi: 10.1261/rna.2610411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ataide SF, Schmitz N, Shen K, Ke A, Shan S, Doudna JA, Ban N. The Crystal Structure of the Signal Recognition Particle in Complex with Its Receptor. Science. 2011;331:881–886. doi: 10.1126/science.1196473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Shen K, Arslan S, Akopian D, Ha T, Shan S-o. Activated GTPase movement on an RNA scaffold drives co-translational protein targeting. Nature. 2012;492:271–275. doi: 10.1038/nature11726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hoskins AA, Gelles J, Moore MJ. New insights into the spliceosome by single molecule fluorescence microscopy. Curr. Opin. Chem. Biol. 2011;15:864–870. doi: 10.1016/j.cbpa.2011.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lohman TM, Bjornson KP. Mechanisms of helicase-catalyzed DNA unwinding. Ann. Rev. Biochem. 1996;65:169–214. doi: 10.1146/annurev.bi.65.070196.001125. [DOI] [PubMed] [Google Scholar]
- 122.Murray NE. Type I restriction systems: Sophisticated molecular machines (a legacy of Bertani and Weigle) Microbiology and Molecular Biology Reviews. 2000;64:412–434. doi: 10.1128/mmbr.64.2.412-434.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Uptain SM, Kane CM, Chamberlin MJ. Basic mechanisms of transcript elongation and its regulation. Ann. Rev. Biochem. 1997;66:117–172. doi: 10.1146/annurev.biochem.66.1.117. [DOI] [PubMed] [Google Scholar]
- 124.Kunkel TA, Bebenek R. DNA replication fidelity. Ann. Rev. Biochem. 2000;69:497–529. doi: 10.1146/annurev.biochem.69.1.497. [DOI] [PubMed] [Google Scholar]
- 125.Semlow DR, Staley JP. Staying on message: ensuring fidelity in pre-mRNA splicing. Trends Biochem. Sci. 2012;37:263–273. doi: 10.1016/j.tibs.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Fersht AR, Kaethner MM. Enzyme hyper-specificity - Rejection of threonine by valyl-transfer-RNA synthetase by mis-acylation and hydrolytic editing. Biochemistry. 1976;15:3342–3346. doi: 10.1021/bi00660a026. [DOI] [PubMed] [Google Scholar]
- 127.Rodnina MV, Wintermeyer W. Ribosome fidelity: tRNA discrimination, proofreading and induced fit, Trends Biochem. Trends Biochem. Sci. 2001;26:124–130. doi: 10.1016/s0968-0004(00)01737-0. [DOI] [PubMed] [Google Scholar]
- 128.Siegel V, Walter P. The affinity of signal recognition particle for presecretory proteins is dependent on nascent chain-length. EMBO J. 1988;7:1769–1775. doi: 10.1002/j.1460-2075.1988.tb03007.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lakkaraju AKK, Mary C, Scherrer A, Johnson AE, Strub K. SRP keeps polypeptides translocation-competent by slowing translation to match limiting ER-targeting sites. Cell. 2008;133:440–451. doi: 10.1016/j.cell.2008.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ogg SC, Walter P. SRP samples nascent chains for the presence of signal sequences by interacting with ribosomes at a discrete step during translation elongation. Cell. 1995;81:1075–1084. doi: 10.1016/s0092-8674(05)80012-1. [DOI] [PubMed] [Google Scholar]
- 131.Zhang D, Shan S-O. Translation elongation regulates substrate selection by the signal recognition particle. J. Biol. Chem. 2012;287:7652–7660. doi: 10.1074/jbc.M111.325001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kramer G, Boehringer D, Ban N, Bukau B. The ribosome as a platform for co-translational processing, folding and targeting of newly synthesized proteins, Nat. Nat. Struct. Mol. Biol. 2009;16:589–597. doi: 10.1038/nsmb.1614. [DOI] [PubMed] [Google Scholar]
- 133.Raue U, Oellerer S, Rospert S. Association of protein biogenesis factors at the yeast ribosomal tunnel exit is affected by the translational status and nascent polypeptide sequence. J. Biol. Chem. 2007;282:7809–7816. doi: 10.1074/jbc.M611436200. [DOI] [PubMed] [Google Scholar]
- 134.Rospert S, Dubaquie Y, Gautschi M. Nascent-polypeptide-associated complex. Cell. Mol. Life Sci. 2002;59:1632–1639. doi: 10.1007/PL00012490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.del Alamo M, Hogan DJ, Pechmann S, Albanese V, Brown PO, Frydman J. Defining the Specificity of Cotranslationally Acting Chaperones by Systematic Analysis of mRNAs Associated with Ribosome-Nascent Chain Complexes. PLOS Biol. 2011;9 doi: 10.1371/journal.pbio.1001100. e1001100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zhang Y, Berndt U, Goelz H, Tais A, Oellerer S, Woelfle T, Fitzke E, Rospert S. NAC functions as a modulator of SRP during the early steps of protein targeting to the endoplasmic reticulum. Mol. Biol. Cell. 2012;23:3027–3040. doi: 10.1091/mbc.E12-02-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]