Abstract
We evaluated the new C. DIFF QUIK CHEK COMPLETE (CD COMPLETE; TechLab, USA), which is a rapid membrane enzyme immunoassay that uses a combination of glutamate dehydrogenase (GDH) antigen and toxin A and B detection. A total of 608 consecutive loose stool specimens collected from the patients with suspected Clostridium difficile infection (CDI) from August to December 2012 were subjected to the CD COMPLETE and VIDAS Clostridium difficile A & B (VIDAS CDAB; bioMérieux, France). Their performances were compared with a toxigenic culture as a reference. Stool specimens that were culture-negative and CD COMPLETE- or VIDAS CDAB-positive were analyzed by using an enrichment procedure. In comparison to the toxigenic cultures, sensitivity, specificity, positive predictive values (PPV), and negative predictive values (NPV) were 63.6%, 98.0%, 76.1%, and 96.4%, respectively, for the CD COMPLETE-toxin and 75.5%, 97.4%, 72.5%, and 97.8%, respectively, for the VIDAS CDAB. In comparison to the enriched C. difficile cultures, the sensitivity, specificity, PPV, and NPV for the CD COMPLETE-GDH were 91.0%, 92.4%, 70.5%, and 98.1%, respectively. The CD COMPLETE is a reliable method for the diagnosis of CDI and provides greater sensitivity than toxin enzyme immunoassay alone. Furthermore, the CD COMPLETE-GDH has advantages over direct culture in detecting C. difficile.
Keywords: Glutamate dehydrogenase, Toxin A, Toxin B, Clostridium difficile, Enzyme immunoassays, Culture
Clostridium difficile is a major causative agent of pseudomembranous colitis, antibiotic-associated diarrhea, and colitis. Toxins A (enterotoxin; TcdA) and B (cytotoxin; TcdB) are well-known primary virulence factors of C. difficile. C. difficile infections (CDI) have recently increased in both number and severity [1]. Thus, rapid and accurate diagnosis of CDI is of great importance, and laboratories are challenged to provide cost-effective, rapid, and accurate results [2, 3, 4]. The diagnosis of CDI should be based on a combination of symptoms and a positive stool test result for C. difficile toxins or toxigenic C. difficile. Toxigenic culture is considered to be the reference method for the diagnosis of CDI, but is labor-intensive and time-consuming, which has limited its use in clinical laboratories [5]. Enzyme immunoassays (EIA) rapidly detect toxins A and B, but their sensitivity varies greatly according to the assay kits [2, 3, 6]. Glutamate dehydrogenase (GDH), a common antigen of the C. difficile cell wall, is a sensitive marker for the detection of C. difficile, approaching 100% sensitivity [2, 6, 7]. The GDH test detects non-toxigenic and toxigenic C. difficile as well as certain other clostridial species; therefore, it must be used in combination with a C. difficile toxin-detecting assay. It has been recommended that GDH be used as the first-line screening test, followed by the demonstration of toxigenic C. difficile in GDH-positive stool specimens [2, 4, 8, 9]. In this study, we evaluated the C. DIFF QUIK CHEK COMPLETE (CD COMPLETE; TechLab, Blacksburg, VA, USA) assay, which uses a combination of GDH antigen detection and toxin A and B detection, and compared it to the currently used VIDAS Clostridium difficile A & B (VIDAS CDAB; bioMérieux, Marcy l'Etoile, France) assay and toxigenic cultures for the diagnosis of CDI.
A total of 608 loose stool specimens were collected from suspected CDI patients in a tertiary care teaching hospital from August to December 2012. The CD COMPLETE and VIDAS CDAB assays were performed on the same day as specimen reception according to the manufacturer's instructions. In brief, for the CD COMPLETE assay, 25 µL or an equivalent volume of stool specimen was added to a tube containing the diluent and conjugate (TechLab), and the mixture was transferred to the device sample well. After incubation for 15 min at room temperature, wash buffer and then substrate (TechLab) were added to the reaction window. Results were read after 10 min. GDH antigen and/or toxins were reported as positive, if a visible band was seen on the device display window (GDH+/tox+; GDH+/tox-; GDH-/tox-). Discrepancies in results between GDH and toxin A/B (AB) were resolved by using the toxigenic culture as a reference.
In toxigenic culture, alcohol-shocked stool specimens were inoculated on C. difficile selective agar (CDSA; Becton, Dickinson and Company, Sparks, MD, USA) and incubated at 37℃ in an anaerobic chamber (Forma Scientific, Marietta, OH, USA) for 48 hr. The species were identified by using Gram staining and the ATB 32A system (bioMérieux). The identified C. difficile isolates were used to detect toxin genes by PCR as described previously [10, 11]. C. difficile VPI 10463 (A+B+), 3608/03 (A-B-), and 1470 (A-B+) were used as controls for the PCR assays. PCR ribotyping of the C. difficile isolates was performed according to a previously described method with minor modifications [12].
Stool specimens that were culture-negative and CD COMPLETE- or VIDAS CDAB-positive were analyzed by using an enrichment procedure in pre-reduced taurocholate and cycloserine-cefoxitin brain heart infusion broth (TCC broth), which was incubated for 48 hr at 37℃ in an anaerobic atmosphere as previously described [13]. Each TCC broth was plated onto TCC agar. The McNemar test and chi square test were used for statistical analyses. This study was approved by the Yonsei University Health System (YUHS) Institutional Review Board (IRB #4-2012-0231).
C. difficile was isolated in 100 of 608 stool specimens (16.4%) by direct and enriched cultures. In direct culture, 62 C. difficile isolates (10.2%) were recovered and 55 (88.7%) isolates were toxigenic. Discordant results were seen in 89 stool specimens (culture-negative and CD COMPLETE- or VIDAS CDAB-positive). Among them, 80 specimens were inoculated in selective enriched broth because of insufficient sample volume, with 38 (47.5%) of the specimens giving a positive C. difficile result, and 26 (68.4%) isolates were toxigenic.
In comparison to the direct toxigenic cultures, sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) for the CD COMPLETE-toxin assay were 63.6%, 98.0%, 76.1%, and 96.4%, respectively, and 75.5%, 97.4%, 72.5%, and 97.8%, respectively, for the VIDAS CDAB assay (Table 1). Sensitivities and NPV were significantly different (P=0.025 and P=0.028, respectively).
Table 1.
Performance of C. DIFF QUIK CHEK COMPLETE-toxin AB and VIDAS Clostridium difficile A & B compared to toxigenic culture
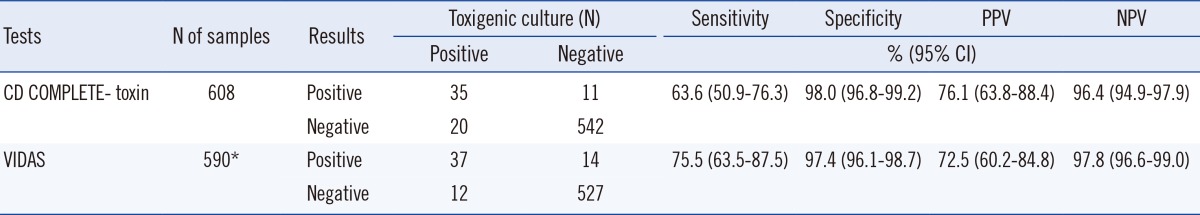
*Thirteen equivalent results and 5 samples not analyzed were excluded.
Abbreviations: CD COMPLETE-toxin, C. DIFF QUIK CHEK COMPLETE-toxinAB; VIDAS, VIDAS Clostridium difficile A & B; PPV, positive predictive value; NPV, negative predictive value; CI, confidence interval.
In comparison to the enriched C. difficile cultures, sensitivity, specificity, PPV, and NPV for the CD COMPLETE-GDH assay were 91.0%, 92.4%, 70.5%, and 98.1%, respectively (Table 2). The sensitivity, specificity, PPV, and NPV for the CD COMPLETE (GDH+toxin) assay were 93.0%, 92.2%, 70.5%, and 98.5%, respectively. CD COMPLETE-GDH assay showed significantly higher sensitivity, specificity, PPV, and NPV than the VIDAS CDAB assay (P<0.001).
Table 2.
Performance of C. DIFF QUIK CHEK COMPLETE-GDH compared with enriched toxigenic culture

*Nine samples insufficient for enrichment culture were excluded.
Abbreviations: GDH, C. DIFF QUIK CHEK COMPLETE-GDH; PPV, positive predictive value; NPV, negative predictive value; CI, confidence interval.
There were 81 (69 A+B+, 12 A-B+) isolates and 22 different ribotype patterns identified by PCR ribotyping. PCR ribotypes 018, 017, and 014/020 were frequent and accounted for 25.9%, 14.8%, and 13.6%, respectively, of the total. The detection rates of PCR ribotypes 018, 017, and 014/020 were 66.7%, 25.0%, and 36.4%, respectively, for the CD COMPLETE-toxin assay and 76.2%, 25.0%, and 36.4%, respectively, for the VIDAS CDAB assay (Table 3).
Table 3.
Characteristics of prevalent toxigenic Clostridium difficile ribotypes
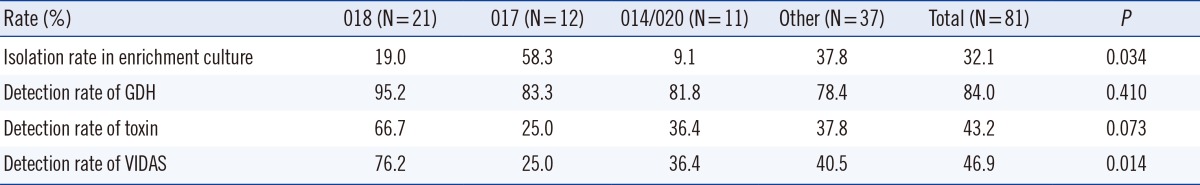
Abbreviations: GDH, C. DIFF QUIK CHEK COMPLETE-GDH; toxin, C. DIFF QUIK CHEK COMPLETE-toxin AB; VIDAS, VIDAS Clostridium difficile A & B.
Among the 80 stool specimens in this study that were grown in enrichment culture because of discordant results, 73 were CD COMPLETE-GDH-positive and culture-negative. Among them, 38 (47.5%) specimens were positive for C. difficile, and 26 isolates (26/38, 68.4%) were toxigenic, with 14 different identified ribotype patterns including ribotype 017 (N=7), 018 (N=4), 014/020 (N=2), and others (N=13). The majority (58.3%) of the specimens isolated in the enrichment procedure were ribotype 017.
Similarly, in a recent study evaluating real-time PCR using the toxigenic culture as a reference method, toxigenic culture-negative and real-time PCR-positive specimens turned out to be positive for toxigenic C. difficile by the enriched culture process [13, 14]. The use of enrichment broth was more sensitive than direct plating to agar, but was too time-consuming for routine diagnostic cultures [15]. Therefore, enrichment cultures or additional real-time PCR tests are recommended for GDH-positive, culture-negative samples.
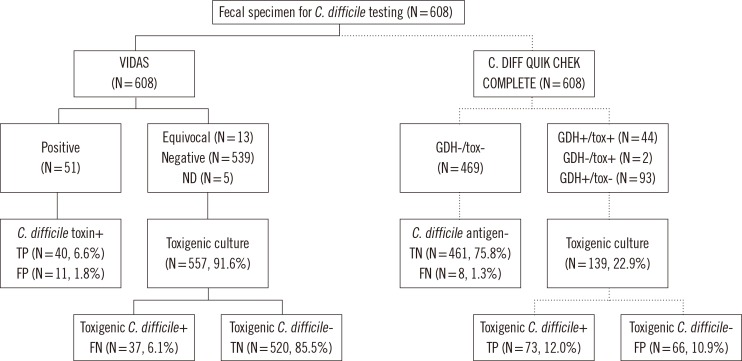
The GDH test is recommended as an initial screening test because of its very high sensitivity [2, 4, 7, 9, 16], reported to be 79.5-100%, and NPV, reported to be 94.6-100% [17]. The GDH test has high sensitivity and NPV, and thus would be a powerful test in a dual testing algorithm when combined with a test to detect toxin. It was reported that specimens that were negative for GDH were not tested further because of the reported high NPV of the assay [16, 17, 18]. Our laboratory has performed VIDAS CDAB assays and toxigenic cultures for the diagnosis of CDI. We simulated the two-step testing algorithm by applying the VIDAS CDAB and CD COMPLETE assays, followed by enriched toxigenic culture (Fig. 1). If CD COMPLETE assay had been used as an initial screening test with no further tests in 461 of 608 (75.8%) cases of GDH (-)/AB (-), 8 of 608 (1.3%) would have been false negatives. When the GDH test had not been routinely used, 26 of 608 (4.3%) would have been false negatives in this study. Therefore, the use of an initial screening with the CD COMPLETE assay instead of the VIDAS CDAB assay has diagnostic and economic advantages in our laboratory.
Fig. 1.
Flow chart for the two-step testing algorithm to detect toxigenic Clostridium difficile and their simulation results.
Abbreviations: GDH, C. DIFF QUIK CHEK COMPLETE-GDH; tox, C. DIFF QUIK CHEK COMPLETE-toxin; VIDAS, VIDAS Clostridium difficile A & B; ND, not done; TP, true positive; FP, false positive; TN, true negative; FN, false negative.
The toxin EIA test is not suitable as a stand-alone test for the diagnosis of CDI because of its low sensitivity and PPV, although it is the most common diagnostic laboratory method [2, 3, 6]. The performance of CD COMPLETE-toxin was comparable with VIDAS CDAB in this study.
Recently, Tenover et al. [19] found that the sensitivities of EIAs, GDH, and Xpert C. difficile assays may vary according to ribotype. However, Goldenberg et al. [20] reported that there were no differences in the detection of GDH and PCR according to specific ribotypes. In this study, the isolation rate in enrichment culture and the detection rates of VIDAS tended to vary according to ribotype (P<0.05). The detection rates of CD COMPLETE-GDH, -toxin, and VIDAS were higher in ribotype 018, which was the most prevalent ribotype, than in other ribotypes. Both our study and that by Tenover et al. [19] used stool samples, while Goldenberg et al. [20] used cultured isolates. Goldenberg et al. [20] also suggested that this conflicting result could be explained by natural variations in the amount of organisms in the stool samples tested.
In conclusion, in comparison to both the direct and enriched culture specimens, the CD COMPLETE assay is a reliable method for the diagnosis of CDI, and provides greater sensitivity than the toxin EIA alone. Furthermore, the CD COMPLETE-GDH assay has advantages over direct culture in the detection of C. difficile.
Footnotes
No potential conflicts of interest relevant to this article were reported.
References
- 1.Kelly CP, LaMont JT. Clostridium difficile-more difficult than ever. N Engl J Med. 2008;359:1932–1940. doi: 10.1056/NEJMra0707500. [DOI] [PubMed] [Google Scholar]
- 2.Ticehurst JR, Aird DZ, Dam LM, Borek AP, Hargrove JT, Carroll KC. Effective detection of toxigenic Clostridium difficile by a two-step algorithm including tests for antigen and cytotoxin. J Clin Microbiol. 2006;44:1145–1149. doi: 10.1128/JCM.44.3.1145-1149.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eastwood K, Else P, Charlett A, Wilcox M. Comparison of nine commercially available Clostridium difficile toxin detection assays, a real-time PCR assay for C. difficile tcdB, and a glutamate dehydrogenase detection assay to cytotoxin testing and cytotoxigenic culture methods. J Clin Microbiol. 2009;47:3211–3217. doi: 10.1128/JCM.01082-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Culbreath K, Ager E, Nemeyer RJ, Kerr A, Gilligan PH. Evolution of testing algorithms at a university hospital for detection of Clostridium infections. J Clin Microbiol. 2012;50:3073–3076. doi: 10.1128/JCM.00992-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bauer MP, Kuijper EJ, van Dissel JT European Society of Clinical Microbiology and Infectious Diseases. European Society of Clinical Microbiology and Infectious Diseases (ESCMID): treatment guidance document for Clostridium difficile infection (CDI) Clin Microbiol Infect. 2009;15:1067–1079. doi: 10.1111/j.1469-0691.2009.03099.x. [DOI] [PubMed] [Google Scholar]
- 6.Bruins MJ, Verbeek E, Wallinga JA, Bruijnesteijn van Coppenraet LE, Kuijper EJ, Bloembergen P. Evaluation of three enzyme immunoassays and a loop-mediated isothermal amplification test for the laboratory diagnosis of Clostridium difficile infection. Eur J Clin Microbiol Infect Dis. 2012;31:3035–3039. doi: 10.1007/s10096-012-1658-y. [DOI] [PubMed] [Google Scholar]
- 7.Swindells J, Brenwald N, Reading N, Oppenheim B. Evaluation of diagnostic tests for Clostridium difficile infection. J Clin Microbiol. 2010;48:606–608. doi: 10.1128/JCM.01579-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crobach MJ, Dekkers OM, Wilcox MH, Kuijper EJ. European Society of Clinical Microbiology and Infectious Diseases (ESCMID): data review and recommendations for diagnosing Clostridium difficile-infection (CDI) Clin Microbiol Infect. 2009;15:1053–1066. doi: 10.1111/j.1469-0691.2009.03098.x. [DOI] [PubMed] [Google Scholar]
- 9.Surawicz CM, Brandt LJ, Binion DG, Ananthakrishnan AN, Curry SR, Gilligan PH, et al. Guidelines for diagnosis, treatment, and prevention of Clostridium difficile infections. Am J Gastroenterol. 2013;108:478–498. doi: 10.1038/ajg.2013.4. [DOI] [PubMed] [Google Scholar]
- 10.Terhes G, Urbán E, Sóki J, Hamid KA, Nagy E. Community-acquired Clostridium difficile diarrhea caused by binary toxin, toxin A, and toxin B gene-positive isolates in Hungary. J Clin Microbiol. 2004;42:4316–4318. doi: 10.1128/JCM.42.9.4316-4318.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spigaglia P. Comparative analysis of Clostridium difficile clinical isolates belonging to different genetic lineages and time periods. J Med Microbiol. 2004;53:1129–1136. doi: 10.1099/jmm.0.45682-0. [DOI] [PubMed] [Google Scholar]
- 12.O'Neill GL, Ogunsola FT, Brazier JS, Duerden BI. Modification of a PCR ribotyping method for application as a routine typing scheme for Clostridium difficile. Anaerobe. 1996;2:205–209. [Google Scholar]
- 13.Barbut F, Monot M, Rousseau A, Cavelot S, Simon T, Burghoffer B, et al. Rapid diagnosis of Clostridium difficile infection by multiplex real-time PCR. Eur J Clin Microbiol Infect Dis. 2011;30:1279–1285. doi: 10.1007/s10096-011-1224-z. [DOI] [PubMed] [Google Scholar]
- 14.Kim H, Jeong SH, Kim M, Lee Y, Lee K. Detection of Clostridium difficile toxin A/B genes by multiplex real-time PCR for the diagnosis of C. difficile infection. J Med Microbiol. 2012;61:274–277. doi: 10.1099/jmm.0.035618-0. [DOI] [PubMed] [Google Scholar]
- 15.Arroyo LG, Rousseau J, Willey BM, Low DE, Staempfli H, McGeer A, et al. Use of a selective enrichment broth to recover Clostridium difficile from stool swabs stored under different conditions. J Clin Microbiol. 2005;43:5341–5343. doi: 10.1128/JCM.43.10.5341-5343.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reller ME, Lema CA, Perl TM, Cai M, Ross TL, Speck KA, et al. Yield of stool culture with isolate toxin testing versus a two-step algorithm including stool toxin testing for detection of toxigenic Clostridium difficile. J Clin Microbiol. 2007;45:3601–3605. doi: 10.1128/JCM.01305-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shetty N, Wren MW, Coen PG. The role of glutamate dehydrogenase for the detection of Clostridium difficile in faecal samples: a meta-analysis. J Hosp Infect. 2011;77:1–6. doi: 10.1016/j.jhin.2010.07.024. [DOI] [PubMed] [Google Scholar]
- 18.Karre T, Sloan L, Patel R, Mandrekar J, Rosenblatt J. Comparison of two commercial molecular assays to a laboratory-developed molecular assay for diagnosis of Clostridium difficile infection. J Clin Microbiol. 2011;49:725–727. doi: 10.1128/JCM.01028-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tenover FC, Novak-Weekley S, Woods CW, Peterson LR, Davis T, Schreckenberger P, et al. Impact of strain type on detection of toxigenic Clostridium difficile: comparison of molecular diagnostic and enzyme immunoassay approaches. J Clin Microbiol. 2010;48:3719–3724. doi: 10.1128/JCM.00427-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldenberg SD, Gumban M, Hall A, Patel A, French GL. Lack of effect of strain type on detection of toxigenic Clostridium difficile by glutamate dehydrogenase and polymerase chain reaction. Diagn Microbiol Infect Dis. 2011;70:417–419. doi: 10.1016/j.diagmicrobio.2011.03.012. [DOI] [PubMed] [Google Scholar]