Abstract
Successful intracellular delivery of nucleic acid therapeutics relies on multi-aspect optimization, one of which is formulation. While there has been ample innovation on chemical design of polymeric gene carriers, the same cannot be said for physical processing of polymer-DNA nanocomplexes (polyplexes). Conventional synthesis of polyplexes by bulk mixing depends on the operators’ experience. The poorly controlled bulk-mixing process may also lead to batch-to-batch variation and consequent irreproducibility. Here, we synthesize polyplexes by using a three-dimensional hydrodynamic focusing (3D-HF) technique in a single-layered, planar microfluidic device. Without any additional chemical treatment or post processing, the polyplexes prepared by the 3D-HF method show smaller size, slower aggregation rate, and higher transfection efficiency, while exhibiting reduced cytotoxicity compared to the ones synthesized by conventional bulk mixing. In addition, by introducing external acoustic perturbation, mixing can be further enhanced, leading to even smaller nanocomplexes. The 3D-HF method provides a simple and reproducible process for synthesizing high-quality polyplexes, addressing a critical barrier in the eventual translation of nucleic acid therapeutics.
Keywords: Polyplex, Gene Delivery, 3D Hydrodynamic Focusing, Transfection
Gene therapy has shown significant promise in the treatment of many acquired and inherited diseases, but development of nonviral gene carriers for efficient delivery remains a major challenge.1 Many synthetic polymer-based carriers, responsible for condensing nucleic acids into nanocomplexes (polyplexes), have demonstrated high efficiency.2-5 Currently, polyplexes are prepared by adding a polymer solution to a DNA solution, followed by vigorous pipetting or vortex mixing of the resulting solution. The polyplexes form spontaneously due to electrostatic interaction between the cationic polymer and the negatively charged nucleic acid. Such bulk mixing produces polyplexes that are often metastable, showing poor uniformity, batch-to-batch variability, as well as subsequent aggregation, all of which render poor biological reproducibility.6,7 Here, we synthesize polyplexes through a microfluidic three-dimensional hydrodynamic focusing (3D-HF) method, where uniform mixing is enhanced by the reduced diffusion length via a microfluidic channel, resulting in the production of polyplexes with high uniformity and improved biological performance.
In conventional bulk mixing, the self-assembly of nanocomplexes occurs instantaneously following the introduction of polymer and DNA solutions. The quality of the polyplexes is therefore determined by the mixing uniformity. Although rapid vortexing or repeated pipetting may improve the mixture uniformity, the operator’s experience, or even the sequence in which reagents is added, can greatly alter the physical properties of the resulting polyplexes.8-10 Recently, microfluidic devices11-13 amenable to automated operation have attracted increasing interest due to their ability to minimize human factors and synthesize uniform products.14-27 The reaction conditions (e.g., reagent ratio, flow rate, ionic concentration) can be finely tuned, leading to highly controllable parameterization throughout the complexation process.28,29 Ho et al. showed that the self-assembly of polyplexes through microfluidics-assisted confinement (MAC) in picoliter droplets produces more homogenous and compact polyplexes.5,9 Although promising, the MAC approach requires the use of oil and surfactant to generate and stabilize a water-in-oil emulsion.5,9,30 Further, microfluidic devices typically operate under the low Reynolds number, or laminar flow regime, which suppresses turbulent mixing, leaving molecular diffusion as the dominant mixing mechanism. Particularly important for reactions of fast kinetics, such as charge neutralization of polyelectrolytes,31 rapid and uniform mixing is paramount. Towards this end, hydrodynamic focusing has been used to enhance mixing and provide homogenous parameters within the reaction region.32-46 As the central solution is focused by the sheath of outer fluids, the diffusion length decreases and diffusion occurs all around the central stream, resulting in faster mixing and improved polyplex homogeneity.33,34 Two-dimensional (2D) hydrodynamic focusing, which focuses the central solution in the horizontal plane only, has been used to prepare polyplexes35 and lipolexes.36 However, transverse diffusive broadening observed in existing two-phase laminar flows37 compromises the quality of the resulting polyplexes. A better design to ensure effective mixing in microscale continuous flow settings is needed.38 Compared with 2D hydrodynamic focusing, 3D hydrodynamic focusing (3D-HF) can further reduce the reaction volume and enhance the vertical diffusion by squeezing the center stream in the vertical direction. In recent years, different designs with intrinsic 3D structures for 3D-HF have been proposed, which require complicated fabrication and offer relatively low reproducibility.39-42 Rhee et al. have demonstrated a single-layer 3D-HF device for polymer nanoparticles synthesis. In their design, three sequential inlets with precisely controlled size and alignment were used for vertical focusing, and a conventional cross junction was used for horizontal focusing.43 Rhee’s 3D-HF device showed advantages in synthesis over 2D-HF devices, but the stringent control required for the inlet drilling and the low flow rate diminish the appeal of this design. Previously, we have developed a “microfluidic drifting” technique to achieve 3D-HF in a single-layer, planar microfluidic structure for on-chip flow cytometry application,44-46 through standard soft lithography without multi-layer assembly requirements, rendering it ideal for low-cost and large-scale production. However, our first generation of “microfluidic drifting” based 3D-HF device was not suitable for nanoparticle synthesis due to a relatively low ratio between central and total flow rate (~1:17), which may result in diluted concentration of the synthesized polyplexes.
In this report, we redesigned the “microfluidic drifting” based 3D-HF device for polyplexes synthesis application. The new device has 180° a curved channel section, and the ratio between central and total flow rate is 1:3. This 3D-HF approach can synthesize polyplexes (Fig. 1) with improved properties compared to bulk mixing. In addition, we introduce external acoustic perturbation47-55 to further enhance the mixing in the focused stream.
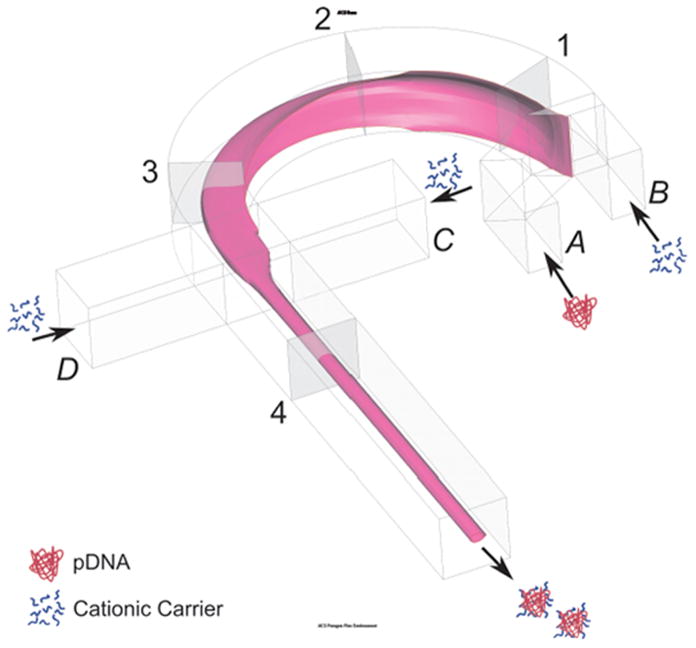
Figure 1.
The Schematic of the microfluidic device for polyplex synthesis by 3D-HF. The DNA solution is injected through inlet A, while the polymer solution is injected from the inlet B, C, and D.
In this study we demonstrate that the 3D-HF-synthesized polyplexes show smaller size, narrower size distribution and higher colloidal stability compared to the bulk-prepared counterpart. In particular, 3D-HF accompanied by acoustic perturbation produce the smallest polyplexes. Aided by acoustic perturbation, the 3D-HF-synthesized polyplexes also show higher transfection efficiency and lower cytotoxicity. Therefore, acoustic-assisted 3D-HF represents a new approach of producing high-quality polyplexes in a reproducible and scalable manner.
RESULTS AND DISCUSSION
Reducing Polyplex Size by 3D-HF and Acoustic Perturbation
Several flow rates were tested for optimization based on the size of the synthesized polyplexes, while keeping the flow rate ratio between inlets A:B:C:D as 3:4:1:1, as shown in Fig. 1. The polyplex size (Z average diameter, Zave) and polydispersity index (PdI) were measured (Fig. 2). The computational fluid dynamics (CFD) simulation of the experimental parameters showed the confinement of the DNA solution in both horizontal and vertical directions at various flow rates (see Supporting Information, Fig. S1). As the flow rate increased, the size of the highly concentrated DNA region decreased, while the region of lower DNA concentration increased, indicating the enhancement of mixing. According to previous research, faster mixing can generate polyplex with smaller sizes.5,9 The experimental result showed the expected trend that the size of the polyplexes decreased with flow rate (Fig. 2a). However, as the total flow rate increased from 270 μL/min to 360 μL/min, the decrease in particle size was less than 30 nm, and further increase of flow rate might cause leakage of the microfluidic channel. Therefore, the total flow rate of 360 μL/min was used in subsequent studies. At this flow rate, the 3D-HF-prepared polyplexes showed smaller size compared to those prepared by bulk mixing (Zave,3D = 263.0 nm versus Zave,bulk = 419.1 nm), while the size distribution was comparable in both conditions (PDI3D = 0.131 versus PDIbulk = 0.142 , n = 3, p = 0.789).
Figure 2.
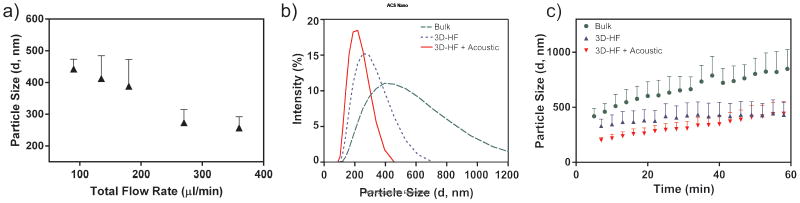
Comparison of polyplexes prepared by bulk mixing and 3D-HF. (a) Size of polyplexes as a function of flow rate. (b) Intensity-based size distribution obtained under the reaction condition of 2 μL Turbofect reagent per μg of pDNA (Zave,3D,acoustic = 200.0 nm, Zave,3D = 263.0 nm, Zave,bulk = 419.1 nm; PDI3D,acoustic = 0.067, PDI3D = 0.131, PDIbulk = 0.142); (c) aggregation kinetics.
Besides 3D-HF, acoustic perturbation can also enhance mixing. The acoustic oscillation of the microfluidic channel can cause liquid motion or vortices, which are known as acoustic streaming.56 Even at micrometer scale, turbulence can still be induced actively by acoustic oscillation, resulting in fast mixing. Therefore, we hypothesized that acoustic perturbation in conjunction with 3D-HF method could generate even smaller polyplexes. Fig. 2b showed the size distribution of polyplexes prepared by bulk mixing, 3D-HF, and acoustic perturbation assisted 3D-HF. As expected, introduction of acoustic perturbation further decreased the particle size and polydispersity index (Zave,3D,acoustic = 200.0 nm, PDI3D,acoustic = 0.067).
Aggregation Kinetics Studies
Aside from nanocomplex heterogeneity in size, significant aggregation is often observed in bulk preparations, presumably due to a corona of excess polycation and an uneven surface coverage.57 As shown in Fig. 2c, comparison of the aggregation kinetics of polyplexes prepared by bulk mixing or 3D-HF suggests that the latter produces more stable particles without any treatment, such as pegylation or addition of anti-caking agent. The aggregation kinetics over 4 hours (the time required for transfection) shows the same trend (See Supporting Information, Fig. S3). The improved size uniformity and slower aggregation rate exhibited by polyplexes prepared with 3D-HF, especially with acoustic perturbation, is most likely due to enhanced mixing, which in turn leads to a more uniform surface property, thereby reducing the aggregation or flocculation that typically occurs in solution.
Estimation of Shear Rate and Temperature Increment
Even though the mixing performance can be enhanced by increasing the flow rate and external acoustic field, the potential degradation of DNA becomes a concern due to the increased shear stress and introduction of acoustic power. High shear rate (for example, 40 milliseconds of shear rate at 3.5×105/s) 58 or long time of relatively low shear stress (~100 seconds of shear rate at 2.1×104/s)59 may break the phosphodiester backbone and physically fragment plasmids DNA (pDNA) into small pieces.58-60 Ultrasound may also fragment DNA via mechanical or thermal degradation due to cavitation.61 Degradation of DNA by these mechanisms will negate the benefits of the proposed polyplex manufacturing process.
We first evaluated the shear stress under our experimental settings, based on the applied flow rate and time of operation. At the highest flow rate of 360 μL/min, the residence time of the DNA in the microfluidic channel would be shorter than 400 milliseconds. The velocity distribution was simulated (See Supporting Information, Fig. S2). The shear rate (γ̇) was calculated as γ̇ = ∂v/∂h, where v is the velocity of the fluid and h is the distance from the channel wall. The maximum shear rate was calculated as 8.1×104/s near the channel wall, and the shear rate decreased from the wall to the center of the channel. The relatively low shear rate58 and the short time interval59 suggest that the DNA degradation due to shear stress would be negligible in our experiments.
Secondly, the two main mechanisms of DNA degradation by acoustic field are cavitation and direct mechanical or thermal degradation. When acoustic pressure is in the order of one atmosphere or higher, gas bubbles appear and oscillate vigorously, resulting in mechanical stress that can be several orders of magnitude higher than that in fluid without bubble.62 However, acoustic cavitation is negligible in our experiment because no bubble was observed due to the extremely low acoustic pressure (< 100 Pa). Another concern is the heat generated by the piezoelectric transducer, which is made of Lead zirconium titanate (PZT), and the absorption of acoustic energy in the fluid. This has been an issue extensively investigated in other ultrasound applications; the relationship between heat generation in PZT materials and acoustic frequency is known.63 The low frequency (55 Hz) applied in our process will produce temperature increase less than 1°C. As an analogy, this low frequency is similar to the frequency of a normal bench-top shaker. It should not damage the pDNA, as confirmed through an agarose gel electrophoresis experiment (see Supporting Information, Fig. S4), which shows that 3D-HF with acoustic perturbation causes no significant DNA fragmentation or degradation. In summary, the external acoustic perturbation provides an additional “active” mixing in the proposed 3D-HF mechanism, while maintaining the integrity of the DNA.
Biological Performance of the Synthesized Polyplexes
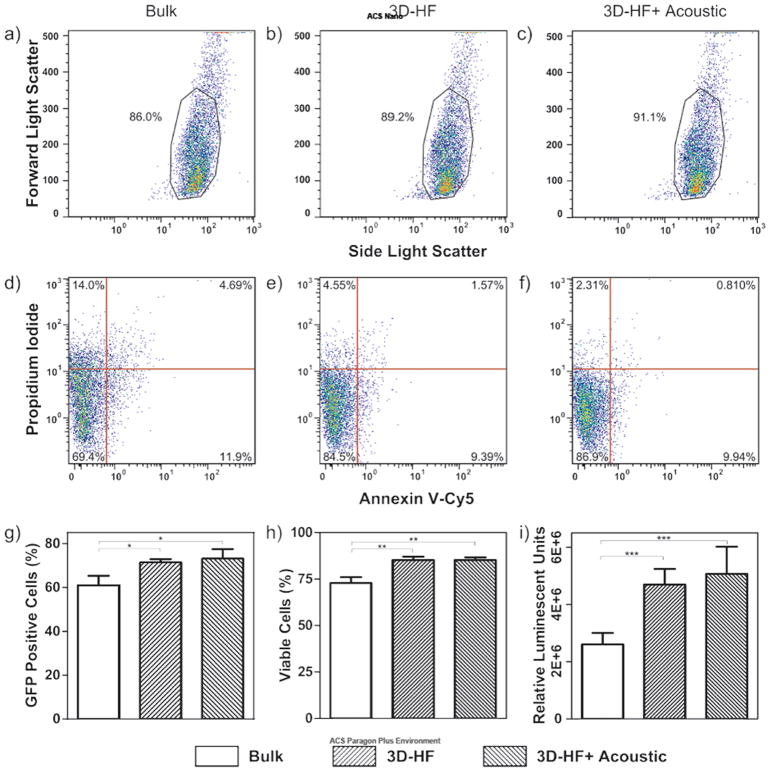
Fig. 3 and Fig. 4 compare the biological performance of the polyplexes prepared by bulk mixing and 3D-HF with and without acoustic perturbation. The reporter pDNA vectors pmax-GFP encoding green fluoresenct protein (GFP) was used for polyplexes synthesis. Qualitatively, the cells showed comparable transfection efficiency in all cases (Fig. 3). Quantitatively, 3D-HF polyplexes prepared with or without acoustic perturbation achieved ~75% transfection compared to the 60% by bulk-mixed polyplexes (Fig. 4g). The improvement in transfection efficiency again confirmed that there was no obvious DNA damage during polyplexes synthesis by 3D-HF with and without acoustic perturbation. Similar distribution in forward scatter/side scatter (FSC/SSC) plots (Fig. 4 a-c) suggests that the cells maintained comparable morphology after transfection with polyplexes prepared in all cases. Further investigation showed that polyplexes prepared by 3D-HF induced less cell death (PI+) and apoptosis (PI-, Annexin V+)64 than polyplexes prepared by the bulk mixing method (Fig. 4 d-f), which is also shown in Fig. 4h.
Figure 3.
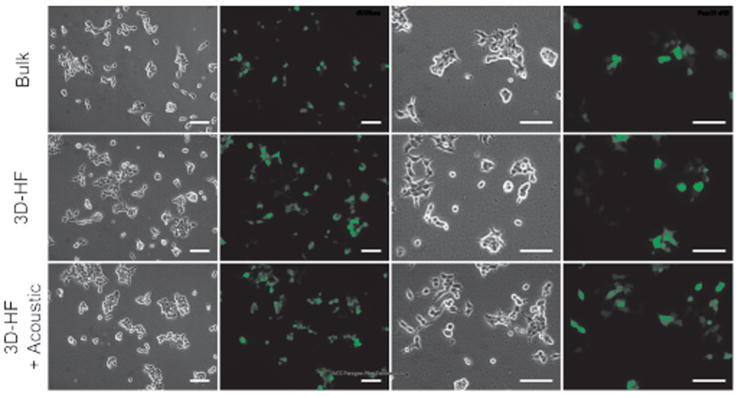
Microscopic observation of GFP transfection. At 24 h post-transfection, the human embryonic kidney (HEK293T) cells transfected by turbofect polyplexes were examined by fluorescence microscopy. Visually, the cells showed comparable transfection efficiency in all cases. Scale bar = 100 μm.
Figure 4.
Quantification of transfection efficiency and cytotoxicity. (a-c) The FSC/SSC plots suggest that the cells maintained comparable morphology after transfection with polyplexes prepared in all cases. (d-f) Bivariate plots showing the fluorescence of PI and Annexin V-Cy5 staining were used to quantitatively evaluate cytotoxicity. The polyplexes prepared by 3D-HF method with and without acoustic perturbation induced less cell death and apoptosis. (g) Quantification of GFP expression level (n = 3). *p < 0.026. (h) Quantification of cell viability. (n = 3). **p < 0.0042 (i) Quantification of Luciferase Assay (n = 3). ***p < 0.0004. (Unpaired t test, CI 95%, two-tailed p-value).
To complement the transfection parameter of percent cells transfected, we also measured the total gene expression level using the firefly luciferase (pLuc) reporter gene. The results are shown in Fig. 4i. Consistent with the GFP-transfection results, polyplexes prepared by 3D-HF showed a two-fold greater luminescence intensity as compared to the bulk mixing case. No statistical significance was found between the 3D-HF groups with or without acoustic perturbation.
The acoustic-assisted 3D-HF method showed the ability to synthesize polyplexes with better control of physical properties, such as size and colloidal stability, over the traditional bulk mixing method. Although we have not addressed the scale-up issue in this study, the throughput of the proposed process should be amenable to optimization of the device and flow parameters, or at the very least through integration of multiple 3D-HF devices in parallel for mass production.65,66
CONCLUSIONS
We have successfully demonstrated polyplex synthesis using a 3D-HF method, along with external acoustic perturbation, in a single-layered device. The polyplexes prepared by this 3D-HF method show smaller size, a slower aggregation rate, higher transfection efficiency, and lower cytotoxicity compared to the ones prepared by the bulk mixing method. Furthermore, acoustic perturbation further decreases the particle size. The 3D-HF method can produce high-quality polyplexes in an operator-independent, simple, and scalable manner. The improved reproducibility and efficacy derived from this 3D-HF synthesis may contribute to the future development of translational nucleic acid therapeutics.
MATERIALS AND METHODS
Materials
The reporter pDNA vectors pmax-GFP encoding green fluorescent protein (GFP) (Amaxa, Cologne, Germany) and VR1255C encoding firefly luciferase (Luc) (Vical, San Diego, CA) were used to quantify transfection efficiency. Turbofect (poly(2-hydroxypropyleneimine), pHP) transfection reagent was purchased from Thermo Scientific. Opti-MEM reduced-Serum Medium was purchased from Life Technology. All materials were used without any further treatment.
Device Preparation
The 3D-HF microfluidic channel was a single-layer polydimethylsiloxane (PDMS) microchannel fabricated using soft lithography techniques, as reported previously by our group.44,45 Briefly, the mold was patterned on a silicon wafer with photoresist (SU8-2050). The surface of the mold was modified to render hydrophobicity by coating it with 1H,1H,2H,2H-perfluorooctyl-trichlorosilane (Sigma Aldrich). The PDMS mixture was prepared by mixing the base and curing agent (Sylgard 184 Silicone Elastomer from Dow Corning) at the weight ratio of 10:1, then poured onto the mold, degassed in a vacuum chamber, and later cured at 65 °C for 30 min. Subsequently, the half-baked PDMS channel was removed from the mold. The inlets and outlets were drilled with a Harris Uni-Core puncher. The channel was treated with oxygen plasma, and bonded to a micro cover glass slide. Then, the whole microfluidic channel was cured at 65 °C overnight. To fabricate channels for the acoustics-assisted 3D-HF, a piezoelectric transducer (model no. SMBLTF120W60, STEINER & MARTINS, INC) was attached to the bottom of the microfluidic device using epoxy (Permatex 84101). Later, tubing was inserted into the inlets and outlets and sealed with epoxy. The microfluidic chip was connected to syringes via tubing, and the flow rate was controlled by the neMESYS syringe pump system (cetoni GmbH, Germany). Before synthesizing the polymer-DNA nanocomplexes, the channel was washed with 70% ethanol in water, then rinsed with water, and exposed to UV light for 1 h.
Preparation of Polyplexes by Microfluidics
The DNA stock solution and the turbofect transfection reagent were diluted in Opti-MEM Reduced-Serum Medium to 13.2 μg/mL and 13.2 μL/mL, respectively. As demonstrated in Fig. 1, the DNA solution was focused after injection through inlet A. The polymer solution was injected from inlets B, C, and D. Additionally, a very long channel length allowed for a longer diffusion time and ensured that the reaction could be completed within the microfluidic system. The polyplexes were collected at the outlet directly without further purification or separation. Several flow rates were tested for optimization based on the size of the synthesized polyplexes, while keeping the flow rate ratio between inlets A:B:C:D as 3:4:1:1. The optimized flow rate was then used to synthesize polyplexes with the introduction of acoustic perturbation (20 Vpp, 55 Hz).
The concentration distribution of DNA solution was simulated at different flow rates using a commercial CFD simulation software (CFD-ACE+, ESI-CFD), by assuming the diffusion coefficient of the 390 kD pGFP as 0.5 × 10-12 m2/s and the initial concentration as 1.905× 10-9 M.67 The simulation did not consider the reaction between the DNA and polymer.
Preparation of Polyplexes by Standard Pipetting Method
The DNA and turbofect concentrations were kept the same as the ones used in microfluidic experiments. As instructed by the manufacturer’s protocol, 1 mL of the turbofect solution was added to 0.5 mL DNA solution, followed by vigorous pipetting.
Size Characterization by Dynamic Light Scattering
The polyplex size (Z average diameter, Zave) and polydispersity index (PdI) were directly measured using the Zetasizer NanoS system (Malvern Instruments, Herrenberg, Germany). All measurements were carried out at 25 °C, using the refractive index (1.330) and viscosity (0.8872 cP) of water for data analysis. Each sample was measured at three minute intervals for a total of one hour. The reported standard deviation was calculated as σ2=PDI×(Zave)2 with the assumption of a Gaussian distribution.9
Cell Culture and Transfection
The human embryonic kidney (HEK293T) cell line was cultured at 37 °C under 5% CO2 in Eagle’s Minimum Essential Medium (EMEM) containing 10% heat-inactivated fetal bovine serum (HI-FBS) and 1% Penicillin/Streptomycin medium. HEK293T cells were seeded at 1 × 105 cells/well in 12-well plates and cultured overnight at 37 °C under 5% CO2 with 1 mL/well full growth media for 24 hours. Then the full growth media was replaced with 400 μL Opti-MEM containing polyplex of 1.5 μg DNA in each well. After 4 hours incubation, the transfection media was replaced with full growth media, and the cells were incubated for 24 hours and 36 hours for pGFP and pLuc expression analysis, respectively.
For the pGFP transfection study, the transfected cells were studied by fluorescence microscopy and flow cytometry. The apoptosis assay was carried out through flow cytometry after Annexin-V (AV) and Propidium Iodide (PI) staining.64 For the pLuc transfection study, the transfected cells were lysed in 400 μL of 1X Glo Lysis Buffer (E2661, Promega). Then the lysate was transferred to a 96-well plate and mixed with an equal amount of Steady-Glo Assay Reagent (E2510, Promega). The luminescence intensity was measured through the Fluoroskan Ascent FL after 20 minutes.
Supplementary Material
Acknowledgments
The authors thank Prof. V. Kalia, Prof. V. Kapur, Prof. Y. Mao, R. Cote, C.Y.K. Chan, D. Ahmed, F. Guo, J. Rufo and J. Beck for help with experiments and discussion. This research was supported by National Institutes of Health (Director’s New Innovator Award, DP2OD 72 9-01, HL109442, EB015300, and AI096305), the National Science Foundation, and the Penn State Center for Nanoscale Science (MRSEC) under grant DMR-0820404. Y.-P. Ho is grateful to the support of Danish Research Councils (11-116325/FTP) and Lundbeck Foundation (R95-A10275). C.L. Grigsby was supported by the Whitaker International Program and the US Fulbright Program. Components of this work were conducted at the Penn State node of the NSF-funded National Nanotechnology Infrastructure Network.
Footnotes
Supporting Information Available: (1) Concentration distribution of the DNA solution in cross-section area at location 1, 2, 3, and 4 in Fig. 1 by Simulation; (2) Velocity distribution in the microfluidic channel by simulation; (3) Aggregation kinetics over 4 hours after synthesis and (4) Agarose gel electrophoresis of DNA. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Thomas M, Klibanov AM. Non-Viral Gene Therapy: Polycation-Mediated DNA Delivery. Appl Microbiol Biot. 2003;62:27–34. doi: 10.1007/s00253-003-1321-8. [DOI] [PubMed] [Google Scholar]
- 2.Parhamifar L, Larsen AK, Hunter AC, Andresen TL, Moghimi SM. Polycation Cytotoxicity: a Delicate Matter for Nucleic Acid Therapy-Focus on Polyethylenimine. Soft Matter. 2010;6:4001–4009. [Google Scholar]
- 3.Grigsby CL, Leong KW. Balancing Protection and Release of DNA: Tools to Address a Bottleneck of Non-Viral Gene Delivery. J R Soc Interface. 2010;7:S67–S82. doi: 10.1098/rsif.2009.0260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang XA, Zheng YR, Chen HH, Leong KW, Wang TH, Mao HQ. Dual-Sensitive Micellar Nanoparticles Regulate DNA Unpacking and Enhance Gene-Delivery Efficiency. Adv Mater. 2010;22:2556–2560. doi: 10.1002/adma.200903933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grigsby CL, Ho Y-P, Lin C, Engbersen JFJ, Leong KW. Microfluidic Preparation of Polymer-Nucleic Acid Nanocomplexes Improves Nonviral Gene Transfer. Sci Rep. 2013;3:3155. doi: 10.1038/srep03155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sharma VK, Thomas M, Klibanov AM. Mechanistic Studies on Aggregation of Polyethylenimine-DNA Complexes and Its Prevention. Biotechnol Bioeng. 2005;90:614–620. doi: 10.1002/bit.20444. [DOI] [PubMed] [Google Scholar]
- 7.Xu YH, Hui SW, Frederik P, Szoka FC. Physicochemical Characterization and Purification of Cationic Lipoplexes. Biophys J. 1999;77:341–353. doi: 10.1016/S0006-3495(99)76894-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boussif O, Lezoualch F, Zanta MA, Mergny MD, Scherman D, Demeneix B, Behr JP. A Versatile Vector for Gene and Oligonucleotide Transfer into Cells in Culture and in vivo: Polyethylenimine. P Natl Acad Sci USA. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ho YP, Grigsby CL, Zhao F, Leong KW. Tuning Physical Properties of Nanocomplexes through Microfluidics-Assisted Confinement. Nano Lett. 2011;11:2178–2182. doi: 10.1021/nl200862n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boussif O, Zanta MA, Behr JP. Optimized Galenics Improve in vitro Gene Transfer with Cationic Molecules up to 1000-fold. Gene Ther. 1996;3:1074–1080. [PubMed] [Google Scholar]
- 11.Whitesides GM. The Origins and the Future of Microfluidics. Nature. 2006;442:368–373. doi: 10.1038/nature05058. [DOI] [PubMed] [Google Scholar]
- 12.Mao X, Huang TJ. Microfluidic Diagnostics for the Developing World. Lab Chip. 2012;12:1412–1416. doi: 10.1039/c2lc90022j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neuži P, Giselbrecht S, Länge K, Huang TJ, Manz A. Revisiting Lab-on-a-Chip Technology for Drug Discovery. Nat Rev Drug Discov. 2012;11:620–632. doi: 10.1038/nrd3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vladisavljevic GT, Henry JV, Duncanson WJ, Shum HC, Weitz DA. Fabrication of Biodegradable Poly(Lactic Acid) Particles in Flow-Focusing Glass Capillary Devices. Prog Coll Pol Sci S. 2012;139:111–114. [Google Scholar]
- 15.Abate AR, Romanowsky MB, Agresti JJ, Weitz DA. Valve-Based Flow Focusing for Drop Formation. Appl Phys Lett. 2009;94:023503. [Google Scholar]
- 16.Valencia PM, Farokhzad OC, Karnik R, Langer R. Microfluidic Technologies for Accelerating the Clinical Translation of Nanoparticles. Nat Nanotechnol. 2012;7:623–629. doi: 10.1038/nnano.2012.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fei ZZ, Hu X, Choi HW, Wang SN, Farson D, Lee LJ. Micronozzle Array Enhanced Sandwich Electroporation of Embryonic Stem Cells. Anal Chem. 2010;82:353–358. doi: 10.1021/ac902041h. [DOI] [PubMed] [Google Scholar]
- 18.Rajagopalan KK, Chen J, Lu B, Wang S. Manufacturing DNA Nanowires with Air Blowing Assembly on Micropatterned Surface. Nano LIFE. 2013;1:1350001–9. [Google Scholar]
- 19.Kürsten D, Cao J, Funfak A, Müller P, Köhler JM. Cultivation of Chlorella vulgaris in Microfluid Segments and Microtoxicological Determination of Their Sensitivity against CuCl2 in the Nanoliter Range. Eng Life Sci. 2011;11:580–587. [Google Scholar]
- 20.Jebrail MJ, Assem N, Mudrik JM, Dryden MDM, Lin K, Yudin A, Wheeler AR. Combinatorial Synthesis of Peptidomimetics Using Digital Microfluidics. J Flow Chem. 2012;2:103–107. [Google Scholar]
- 21.Jebrail MJ, Ng AHC, Rai V, Hili R, Yuding AK, Wheeler AR. Synchronized Synthesis of Peptide-Based Macrocycles by Digital Microfluidics. Angew Chemie Int Ed. 2010;49:8625–8629. doi: 10.1002/anie.201001604. [DOI] [PubMed] [Google Scholar]
- 22.Wang H, Liu K, Chen K-J, Lu Y, Wang S, Lin W-Y, Guo F, Kamei K-I, Chen Y-C, Ohashi M, et al. A Rapid Pathway toward a Superb Gene Delivery System: Programming Structural and Functional Diversity into a Supramolecular Nanoparticle Library. ACS Nano. 2010;4:6235–6243. doi: 10.1021/nn101908e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu K, Wang H, Chen K-J, Guo F, Lin W-Y, Chen Y-C, Phung DL, Tseng H-R, Shen CK-F. A Digital Microfluidic Droplet Generator Produces Self-Assembled Supramolecular Nanoparticles for Targeted Cell Imaging. Nanotechnology. 2010;21:445603. doi: 10.1088/0957-4484/21/44/445603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zeng Q, Guo F, Yao L, Zhu HW, Zheng L, Guo ZX, Liu W, Chen Y, Guo SS, Zhao XZ. Milliseconds Mixing in Microfluidic Channel Using Focused Surface Acoustic Wave. Sensor Actuat B-Chem. 2011;160:1552–1556. [Google Scholar]
- 25.Ji X-H, Cheng W, Guo F, Liu W, Guo S-S, He Z-K, Zhao X-Z. On-Demand Preparation of Quantum Dot-Encoded Microparticles Using a Droplet Microfluidic System. Lab Chip. 2011;11:2561–2568. doi: 10.1039/c1lc20150f. [DOI] [PubMed] [Google Scholar]
- 26.Ji X-H, Zhang N-G, Cheng W, Guo F, Liu W, Guo S-S, He Z-K, Zhao X-Z. Integrated Parallel Microfluidic Device for Simultaneous Preparation of Multiplex Optical-Encoded Microbeads with Distinct Quantum Dot Barcodes. J Mater Chem. 2011;21:13380–13387. [Google Scholar]
- 27.Yang S, Guo F, Kiraly B, Mao X, Lu M, Leong KW, Huang TJ. Microfluidic Synthesis of Multifunctional Janus Particles for Biomedical Applications. Lab Chip. 2012;12:2097–2102. doi: 10.1039/c2lc90046g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kasper JC, Schaffert D, Ogris M, Wagner E, Friess W. The Establishment of An Up-Scaled Micro-Mixer Method Allows the Standardized and Reproducible Preparation of Well-Defined Plasmid/LPEI Polyplexes. Eur J Pharm Biopharm. 2011;77:182–185. doi: 10.1016/j.ejpb.2010.11.012. [DOI] [PubMed] [Google Scholar]
- 29.Debus H, Beck-Broichsitter M, Kissel T. Optimized Preparation of pDNA/poly(ethylene imine) Polyplexes Using A Microfluidic System. Lab Chip. 2012;12:2498–2506. doi: 10.1039/c2lc40176b. [DOI] [PubMed] [Google Scholar]
- 30.Zec H, Rane TD, Wang TH. Microfluidic Platform for on-Demand Generation of Spatially Indexed Combinatorial Droplets. Lab Chip. 2012;12:3055–3062. doi: 10.1039/c2lc40399d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ho YP, Chen HH, Puleo CM, Yeh HC, Leong KW, Wang TH. Quantitative Kinetic Analysis of DNA Nanocomplex Self-Assembly with Quantum Dots FRET in A Microfluidic Device. Proc Ieee Micr Elect. 2008:30–33. [Google Scholar]
- 32.Chang Z, Serra CA, Bouquey M, Kraus I, Li S, Köhler JM. Multiscale Materials from Microcontinuous-Flow Syntheis: ZnO and Au Nanoparticle-Filled uniform and Homogeneous Polymer Microbeads. Nanotechnology. 2010;21:015605. doi: 10.1088/0957-4484/21/1/015605. [DOI] [PubMed] [Google Scholar]
- 33.Knight JB, Vishwanath A, Brody JP, Austin RH. Hydrodynamic Focusing on a Silicon Chip: Mixing Nanoliters in Microseconds. Phys Rev Lett. 1998;80:3863–3866. [Google Scholar]
- 34.Chang CC, Huang ZX, Yang RJ. Three-Dimensional Hydrodynamic Focusing in Two-layer Polydimethylsiloxane (PDMS) Microchannels. J Micromech Microeng. 2007;17:1479–1486. [Google Scholar]
- 35.Kim J, Hwang I, Britain D, Chung TD, Sun Y, Kim DH. Microfluidic Approaches for Gene Delivery and Gene Therapy. Lab Chip. 2011;11:3941–3948. doi: 10.1039/c1lc20766k. [DOI] [PubMed] [Google Scholar]
- 36.Koh CG, Zhang XL, Liu SJ, Golan S, Yu B, Yang XJ, Guan JJ, Jin Y, Talmon Y, Muthusamy N, et al. Delivery of Antisense Oligodeoxyribonucleotide Lipopolyplex Nanoparticles Assembled by Microfluidic Hydrodynamic Focusing. J Control Release. 2010;141:62–69. doi: 10.1016/j.jconrel.2009.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ismagilov RF, Stroock AD, Kenis PJA, Whitesides G, Stone HA. Experimental and Theoretical Scaling Laws for Transverse Diffusive Broadening in Two-Phase Laminar Flows in Microchannels. Appl Phys Lett. 2000;76:2376–2378. [Google Scholar]
- 38.Zhao CX, He LZ, Qiao SZ, Middelberg APJ. Nanoparticle Synthesis in Microreactors. Chem Eng Sci. 2011;66:1463–1479. [Google Scholar]
- 39.Sundararajan N, Pio MS, Lee LP, Berlin AA. Three-dimensional Hydrocynamic Focusing in Polydimethylsiloxane (PDMS) Microchannels. J Microelectromech Syst. 2004;13:559–567. [Google Scholar]
- 40.Wolff A, Perch-Nielsen IR, Larsen UD, Friis P, Goranovic G, Poulsen CR, Kutter JP, Tellenman P. Integrating Advanced Functionality in a Microfabricated High-throughput Fluorescent-activated Cell Sorter. Lab Chip. 2003;3:22–27. doi: 10.1039/b209333b. [DOI] [PubMed] [Google Scholar]
- 41.Huang S, Tan W, Tseng F, Takeuchi S. A Monolithically Three-dimensional Flow-focusing Device for Formation of Single/Double Emulsions in Closed/Open Microfluidic Systems. J Micromech Microeng. 2006;16:2336. [Google Scholar]
- 42.Scott R, Sethu P, Harnett CK. Three-dimensional Hydrodynamic Focusing in a Microfluidic Coulter Counter. Rev Sci Instrum. 2008;79:046104. doi: 10.1063/1.2900010. [DOI] [PubMed] [Google Scholar]
- 43.Rhee M, Valencia PM, Rodriguez MI, Langer R, Farokhzad OC, Karnik R. Synthesis of Size-Tunable Polymeric Nanoparticles Enabled by 3D Hydrodynamic Flow Focusing in Single-Layer Microchannels. Adv Mater. 2011:H79–H83. doi: 10.1002/adma.201004333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mao XL, Lin SCS, Dong C, Huang TJ. Single-Layer Planar on-Chip Flow Cytometer Using Microfluidic Drifting Based Three-Dimensional (3D) Hydrodynamic Focusing. Lab Chip. 2009;9:1583–1589. doi: 10.1039/b820138b. [DOI] [PubMed] [Google Scholar]
- 45.Mao XL, Waldeisen JR, Huang TJ. “Microfluidic Drifting” - Implementing Three-Dimensional Hydrodynamic Focusing with A Single-Layer Planar Microfluidic Device. Lab Chip. 2007;7:1260–1262. doi: 10.1039/b711155j. [DOI] [PubMed] [Google Scholar]
- 46.Mao S, Nawaz AA, Lin S-CS, Lapsley MI, Zhao Y, McCoy JP, El-Deiry WS, Huang TJ. An Integrated, Multiparametric Flow Cytometry Chip Using “Microfluidic Drifting” Based Three-Dimensional Hydrodynamic Focusing. Biomicrofluidics. 2012;6:024113. doi: 10.1063/1.3701566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oberti S, Neild A, Ng TW. Microfluidic Mixing under Low Frequency Vibration. Lab Chip. 2009;9:1435–1438. doi: 10.1039/b819739c. [DOI] [PubMed] [Google Scholar]
- 48.Ahmed D, Mao XL, Shi JJ, Juluri BK, Huang TJ. A Millisecond Micromixer via Single-Bubble-Based Acoustic Streaming. Lab Chip. 2009;9:2738–2741. doi: 10.1039/b903687c. [DOI] [PubMed] [Google Scholar]
- 49.Ahmed D, Mao XL, Juluri BK, Huang TJ. A Fast Microfluidic Mixer Based on Acoustically Driven Sidewall-Trapped Microbubbles. Microfluid Nanofluid. 2009;7:727–731. [Google Scholar]
- 50.Huang PH, Lapsley MI, Ahmed D, Chen YC, Wang L, Huang TJ. A Single-Layer, Planar, Optofluidic Switch Powered by Acoustically Driven, Oscillating Microbubbles. Appl Phys Lett. 2012;101:141101. doi: 10.1063/1.4742864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xie YL, Ahmed D, Lapsley MI, Lin SCS, Nawaz AA, Wang L, Huang TJ. Single-Shot Characterization of Enzymatic Reaction Constants Km and kcat by an Acoustic-Driven, Bubble-Based Fast Micromixer. Anal Chem. 2012;84:7495–7501. doi: 10.1021/ac301590y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ahmed D, Chan CY, Lin SCS, Muddana HS, Nama N, Benkovic SJ, Huang TJ. Tunable, Pulsatile Chemical Gradient Generation via Acoustically Driven Oscillating Bubbles. Lab Chip. 2013;13:328–331. doi: 10.1039/c2lc40923b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ding X, Li P, Lin S-CS, Stratton ZS, Nama N, Guo F, Slotcavage D, Mao X, Shi J, Costanzo F, et al. Surface Acoustic Wave Microfluidics. Lab Chip. 2013;13:3626–3649. doi: 10.1039/c3lc50361e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shi J, Mao X, Ahmed D, Colletti A, Huang TJ. Focusing Microparticles in a Microfluidic Channel with Standing Surface Acoustic Wave (SSAW) Lab Chip. 2008;8:221–223. doi: 10.1039/b716321e. [DOI] [PubMed] [Google Scholar]
- 55.Ding X, Lin S-CS, Lapsley MI, Li S, Guo X, Chan CY, Chiang IK, Wang L, McCoy JP, Huang TJ. Standing Surface Acoustic Wave (SSAW) Based Multichannel Cell Sorting. Lab Chip. 2012;12:4228–4231. doi: 10.1039/c2lc40751e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhu X, Kim ES. Microfluidic Motion Generation with Acoustic Waves. Sensor Actuat a-Phys. 1998;66:355–360. [Google Scholar]
- 57.Hunter RJ. Introduction to Modern Colloid Science. Oxford University Press; New York: 1994. [Google Scholar]
- 58.Levy MS, Collins IJ, Yim SS, Ward JM, Titchener-Hooker N, Shamlou PA, Dunnill P. Effect of Shear on Plasmid DNA in Solution. Bioprocess Eng. 1999;20:7–13. [Google Scholar]
- 59.Zhang H, Kong S, Booth A, Boushaba R, Levy MS, Hoare M. Prediction of Shear Damage of Plasmid DNA in Pump and Centrifuge Operations Using an Ultra Scale-Down Device. Biotechnol Progr. 2007;23:858–865. doi: 10.1021/bp070066z. [DOI] [PubMed] [Google Scholar]
- 60.Wu ML, Freitas SS, Monteiro GA, Prazeres DMF, Santos JAL. Stabilization of Naked and Condensed Plasmid DNA against Degradation Induced by Ultrasounds and High-Shear Vortices. Biotechnol Appl Bioc. 2009;53:237–246. doi: 10.1042/BA20080215. [DOI] [PubMed] [Google Scholar]
- 61.Elsner HI, Lindblad EB. Ultrasonic Degradation of DNA. DNA. 1989;8:697–701. doi: 10.1089/dna.1989.8.697. [DOI] [PubMed] [Google Scholar]
- 62.Carstensen EL. Acoustic Cavitation and the Safety of Diagnostic Ultrasound. Ultrasound Med Biol. 1987;13:597–606. doi: 10.1016/0301-5629(87)90057-3. [DOI] [PubMed] [Google Scholar]
- 63.Zheng JH, Takahashi S, Yoshikawa S, Uchino K, deVries JWC. Heat Generation in Multilayer Piezoelectric Actuators. J Am Ceram Soc. 1996;79:3193–3198. [Google Scholar]
- 64.Moghimi SM, Symonds P, Murray JC, Hunter AC, Debska G, Szewczyk A. A Two-Stage Poly(ethylenimine)-Mediated Cytotoxicity: Implications for Gene Transfer/Therapy. Mol Ther. 2005;11:990–995. doi: 10.1016/j.ymthe.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 65.Kang X, Luo C, Wei Q, Xiong C, Chen Q, Chen Y, Ouyang Q. Mass Production of Highly Monodisperse Polymeric Nanoparticles by Parallel Flow Focusing System. Microfluid Nanofluid. 2013:1–9. [Google Scholar]
- 66.Nisisako T, Torii T. Microfluidic Large-Scale Integration on A Chip for Mass Production of Monodisperse Droplets and Particles. Lab Chip. 2008;8:287–293. doi: 10.1039/b713141k. [DOI] [PubMed] [Google Scholar]
- 67.Prazeres DMF. Prediction of Diffusion Coefficients of Plasmids. Biotechnol Bioeng. 2008;99:1040–1044. doi: 10.1002/bit.21626. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.