Abstract
The transcription factors GATA-4 and GATA-6, members of the GATA family, play an important role in ovarian cell proliferation, differentiation and apoptosis. In this study, the full-length coding sequences of goose GATA-4 and GATA-6 were cloned and characterized. GATA-4 and GATA-6 consist of 1236 and 1104 nucleotides encoding proteins with 411 and 367 amino acids, respectively. The deduced amino acid sequences of both proteins include two adjacent zinc finger domains with the distinctive form (CVNC-X17-CNAC)-X29-(CANC-X17-CNAC) and share 84.76% identity within this domain. In silico prediction together with matching of the high affinity RRXS(T)Y motif revealed that the GATA-4 protein might be phosphorylated predominantly at S233, but no phosphorylation site was found in the GATA-6 protein. Real-time quantitative PCR analysis showed that GATA-4 and GATA-6 mRNAs were co-expressed in goose follicles, moderately expressed in granulosa cells and weakly expressed in theca cells. The expression level of GATA-4 mRNA in healthy follicles was significantly higher than in atretic follicles or postovulatory follicles (P<0.01), and the expression level of GATA-6 mRNA in healthy follicles was significantly lower than in atretic follicles or postovulatory follicles (P<0.01). The expression level of GATA-4 mRNA in granulosa cells was downregulated during follicle development; the peak of expression occurred in the 8-10 mm follicles, and the lowest expression was in the F1 follicles. GATA-6 was upregulated and reached its peak expression in the F1 follicles. These results indicate that the molecular structural differences in goose GATA-4 and GATA-6 may be related to their different roles during follicle development.
Keywords: Cloning, Expression, GATA-4, GATA-6, Tianfu goose
The GATA transcription factor family is composed of six members (GATA-1, -2, -3, -4, -5 and -6). All GATA proteins contain a conserved DNA-binding domain consisting of two multifunctional tandem zinc finger domains. The carboxy-terminal zinc finger is required for interacting with the nucleotide sequence element WGATAR or related DNA cis-elements, and the amino-terminal zinc finger contributes to the specificity and stability of the DNA-binding [1,2,3]. Based on the similarities in their zinc finger domain sequences and spatiotemporal expression patterns, the GATA proteins have been divided into two subgroups: GATA-1, -2 and -3 and GATA-4, -5 and -6. GATA-1, -2 and -3 are predominantly expressed in hematopoietic cells, and each member has been shown to be essential for hematopoietic lineage development [4, 5]. GATA-4, -5 and -6 are expressed in tissues that originate from the mesoderm and endoderm, such as the heart, liver, lungs, gut and gonads, and each member mediates tissue-specific gene expression, sex differentiation and cell proliferation [6,7,8,9].
Among these factors, only GATA-4 and GATA-6 mRNAs and/or proteins have been detected in the healthy ovaries of adult mice [10,11,12], pigs [13] and humans [14, 15]. In the mouse ovary, GATA-4 mRNA is strongly expressed in granulosa cells, moderately expressed in theca cells and weakly expressed in corpora lutea. GATA-6 mRNA can be detected in granulosa cells, is strongly expressed in corpora lutea and is not expressed in theca cells. In the human ovary, GATA-4 and GATA-6 mRNAs and GATA-4 protein localize in granulosa cells and theca cells of both preantral and antral follicles. In the pig ovary, GATA-4 and GATA-6 proteins are abundantly localized in granulosa cells. The above studies have focused on the localization of GATA-4 and GATA-6 in the ovary. Functional studies have indicated that phosphorylation increases GATA-4’s DNA-binding activity and its transactivation of target genes [16,17,18]. However, little is known about the possible roles of GATA-6 and whether it can activate putative target genes in the ovary.
Indigenous Chinese Tianfu geese have low egg production rates and produce one egg every two days during the egg-laying period between September and May of the next year. In addition, many growing follicles undergo atresia as they advance toward ovulation. Thus, many factors are involved in goose follicle maturation, and a clear understanding of the localization and possible functions of GATA-4 and GATA-6 during goose follicle development remain elusive. In this study, we cloned the full-length coding sequence of goose GATA-4 and GATA-6, and the phosphorylation sites in goose GATA-4 and GATA-6 proteins were predicted and mapped. In addition, we examined the mRNA expression levels of GATA-4 and GATA-6 in three types of follicles (healthy follicles, atretic follicles and postovulatory follicles) and in the granulosa and theca cells at different stages during follicle development. The results should help to further elucidate the functional roles of the GATA family in goose follicle development.
Materials and Methods
Goose granulosa and theca cell isolation
A maternal line of laying Tianfu geese (Anser cygnoides) was raised under natural temperature and light conditions at the experimental station of waterfowl breeding at Sichuan Agricultural University. Three geese with regular laying sequences were randomly selected as experimental samples and sacrificed by cervical bleeding post anesthesia at 2 h after oviposition. A pool of ovarian follicles was immediately collected from the abdominal cavities of the geese and placed in sterile normal saline. Then, the follicles were divided into groups according to their sizes (<2, 2–4, 4–6, 6–8 and 8–10 mm) and stages of differentiation (follicle hierarchy: F5, F4, F3, F2 and F1, according to previously reported nomenclature [19]). The outer connective tissue was removed from the follicles, and the follicles were bisected to allow the yolk and adhered granulosa cells to flow out. The granulosa and theca cells were isolated as described previously [20]. The same numbers and sizes (2–4 mm) of healthy and atretic follicles and all postovulatory follicles were selected. Then, they were washed three times with PBS, quickly frozen in liquid nitrogen and stored at –80 C for RNA extraction. All procedures in the current study were approved by the Beijing Animal Welfare Committee.
RNA isolation and cDNA synthesis
Total RNA was extracted from the granulosa cells, theca cells and three types of follicles using TRlzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. The first strand cDNA was synthesized from 1 μg of total RNA using a cDNA synthesis kit (Takara, Otsu, Japan) according to the manufacturer’s instructions. The newly synthesized cDNA was immediately stored at –80 C until further analysis of gene expression.
Cloning the full-length coding sequences of GATA-4 and GATA-6
Based on the GATA-4 and GATA-6 sequences of Gallus gallus (GATA-4, XM_420041.1; GATA-6, NM_205420.1), Meleagris gallopavo (GATA-4, XM_003204620.1; GATA-6, XM_003204995.1) and Taeniopygia guttata (GATA-4, XM_002186806.1; GATA-6, XM_002194963.1) in the GenBank database, we designed and selected three and four pairs of gene-specific primers, respectively, using the DNAMAN 7.0, Primer Premier 5.0 and Oligo 7.0 software. PCR amplification was performed using the following conditions: denaturation at 94 C for 5 min; 36 cycles of 94 C for 30 sec, annealing at 56 C (P1), 58 C (P2), 49.5 C (P3), 60 C (P4), 61 C (P5), 61.5 C (P6) or, 60.5 C (P7) and extension at 72 C for 50 sec (P1) or 40 sec (P2, P3, P4, P5, P6 and P7); final extension for 10 min at 72 C; and storage at 4 C. The primer sequences and PCR product lengths are listed in Table 1. The amplified products were verified by 1.5% agarose gel electrophoresis and purified with a gel extraction kit (Omega, Norcross, GA, USA). The purified products were ligated into the pMD-19T vector (Biomed, Beijing, P.R. China) and sequenced by Sangon Biotech (Shanghai) .
Table 1. Primers for PCR analysis.
| Gene | Primer | Primer sequence (5’ to 3’) | Product length (bp) | Accession number |
| GATA-4 | P1-F P1-R |
gtcacctcgcttctccttctc gcttacgcttccttgtctgaat |
617 | KC454275 |
| P2-F P2-R |
atgtaccagagcttagccatgg tccctcccgttagcagaa |
635 | ||
| P3-F P3-R |
tagcaatgagaaaagagg ttatgccgttatgatgtc |
401 | ||
| GATA-6 | P4-F P4-R |
ggctctacaccaaaatgaacg cagcccatcttgacctgaata |
464 | KC493562 |
| P5-F P5-R |
cgaggaaacgaaaacctaagaa tcacgccagagccagggcac |
323 | ||
| P6-F P6-R |
atgtaccagaccttggccatct gggtaggagccgttgagcg |
380 | ||
| P7-F P7-R |
cgctcaacggctcctaccc cgttgcagacgggctaca |
411 | ||
F, forward primer; R, reverse primer.
Bioinformatics analysis
We used the NCBI online ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html) and DNASTAR to predict the open reading frames of GATA-4 and GATA-6. The functional domains of GATA-4 and GATA-6 proteins were predicted by SMART (http://smart.embl-heidelberg.de/). The physicochemical parameters of the deduced proteins were determined by ProtParam (http://web.expasy.org/protparam/). The phosphorylation sites in the proteins were predicted by Scansite 3 (http://scansite3.mit.edu/#home). The protein structures and functional classification were predicted by the ProtFun 2.2 Server (http://www.cbs.dtu.dk/services/ProtFun/). Clustal X 2.0 and BOXSHADE 3.21 (http://www.ch.embnet.org/software/BOX_form.html) were used for multiple sequence alignments. MEGA 5.0 was used to construct the phylogenetic tree with the neighbor-joining (NJ) method.
Gene expression measurements
Real-time RT-PCR was conducted to detect the expression levels of goose GATA-4 and GATA-6 mRNAs in granulosa cells, theca cells and the three types of follicles using a SYBR PrimeScript RT-PCR kit (Takara, Otsu, Japan) in the CFX96™ Real-Time System (Bio-Rad, Hercules, CA, USA). Each real-time PCR reaction (in 25 μl) contained 12.5 μl of SYBR Green II, 10.5 μl of ddH2O, 0.5 μl of primers (Table 2) and 2 μl of normalized template cDNA. The cycling conditions consisted of 95 C for 3 min; 40 cycles of 95 C for 10 sec, 60 C (as the optimal annealing temperature) for 30 sec, 68 C for 3 min, 95 C for 1 min and 55 C for 1 min; and then final extension for 5 min. The specificity of the PCR products was confirmed by a melting curve analysis. To normalize the expression levels of GATA-4 and GATA-6 mRNAs in similar cDNA samples, β-actin and ribosomal 18S rRNAs were selected as the reference genes. Relative mRNA expression for GATA-4 and GATA-6 was determined using the 2(−ΔΔCt) method [21]. All reactions were performed in triplicate, and the data represent the means from three independent experiments.
Table 2. Primers for real-time PCR analysis.
| Gene | Primer sequence (5’ to 3’) | Product length (bp) |
|
GATA-4-F GATA-4-R |
acaaccaccgaggaaatgcg ggagactggctgatggctgac |
188 |
|
GATA-6-F GATA-6-R |
ccaggaagcgaaaacctaagaac cctgaagctgacggttgtgtg |
154 |
|
β-actin-F β-actin-R |
caacgagcggttcaggtgt tggagttgaaggtggtctcg |
92 |
|
18S-F 18S-R |
ttggtggagcgatttgtc atctcgggtggctgaacg |
129 |
F, forward primer; R, reverse primer; β-actin and ribosomal 18S rRNAs were selected as the reference genes.
Statistical analysis
All data were expressed as the mean ± SD. We conducted an ANOVA test to analyze the data, and the means were assessed for significance by Duncan’s multiple range tests using the SAS 9.0 software. A value of P<0.05 was considered a significant difference, and a value of P<0.01 was considered an extremely significantly difference.
Results
Cloning and sequence analysis of goose GATA-4 and GATA-6
By sequencing and assembling the data, we obtained the full-length coding sequences of goose GATA-4 and GATA-6. These sequence data have been submitted to the GenBank database under accession numbers KC454275 and KC493562, respectively.
The full-length coding sequence of goose GATA-4 consists of 1236 nucleotides that encode a 411 amino acid protein with an estimated molecular mass of 46.3 kDa and a theoretical isoelectric point of 9.37. A multiple alignment of the deduced amino acid sequence of GATA-4 is shown in Fig. 1A. The deduced goose GATA-4 amino acid sequence shares 93.51, 93.9 and 93.53% identity with Gallus gallus, Meleagris gallopavo and Taeniopygia guttata, respectively.
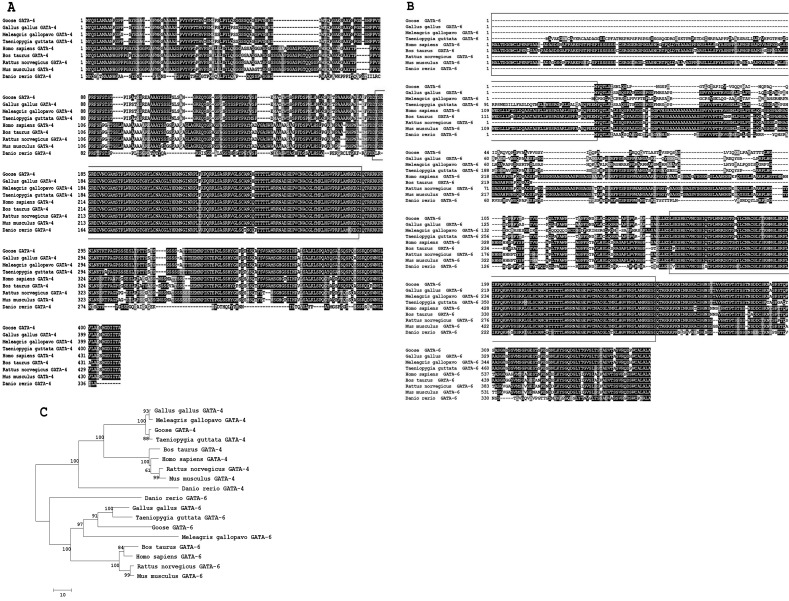
Fig. 1.
Amino acid sequence alignments of GATA-4 and GATA-6 sequences from the goose and other species. The GATA-4 and GATA-6 amino acid sequences used for analysis were extracted from the GenBank database for the following species: Gallus gallus (GATA-4, XP_420041.1; GATA-6, NP_990751.1); Meleagris gallopavo (GATA-4, XP_003204668.1; GATA-6, XP_003205043.1); Taeniopygia guttata (GATA-4, XP_002186842.1; GATA-6, XP_002194999.1); Bos taurus (GATA-4, NP_001179806.1; GATA-6, XP_001253597.2); Homo sapiens (GATA-4, NP_002043.2; GATA-6, NP_005248.2); Rattus norvegicus (GATA-4, NP_653331.1; GATA-6, NP_062058.1); Mus musculus (GATA-4, NP_032118.2; GATA-6, NP_034388.2); and Danio rerio (GATA-4, NP_571311.1; GATA-6, NP_571632.1). The black blocks represent identical residues between the sequences shown, while the red boxes represent zinc finger domains. A: A multiple amino acid sequence alignment of GATA-4. B: A multiple amino acid sequence alignment of GATA-6. The black boxes represent the 5’-upstream sequences in Taeniopygia guttata, Homo sapiens, Bos taurus and Mus musculus, which have an extra amino-terminal extension from the initiator methionine, thus belonging to the long type of GATA-6, and all other species belong to the short type of GATA-6. C: A phylogenetic tree of the amino acid sequences of GATA-4 and GATA-6 was constructed using the neighbor-joining method (1000 bootstrap replicates).
The full-length coding sequence of goose GATA-6 consists of 1104 nucleotides that encode a 367 amino acid protein with an estimated molecular mass of 38.8 kDa and a theoretical isoelectric point of 9.22. A multiple alignment of the deduced amino acid sequence of GATA-6 is shown in Fig. 1B. The deduced goose GATA-6 amino acid sequence shares 74.74, 61.44 and 54.63% identity with Gallus gallus, Meleagris gallopavo and Taeniopygia guttata, respectively. Goose GATA-6 has low identity with other birds, and the goose GATA-6 protein is the short type. In addition, a phylogenetic tree (Fig. 1C) showed that goose GATA-4 and GATA-6 are located in the same group with other avian species.
Primary structure analysis of goose GATA-4 and GATA-6 proteins
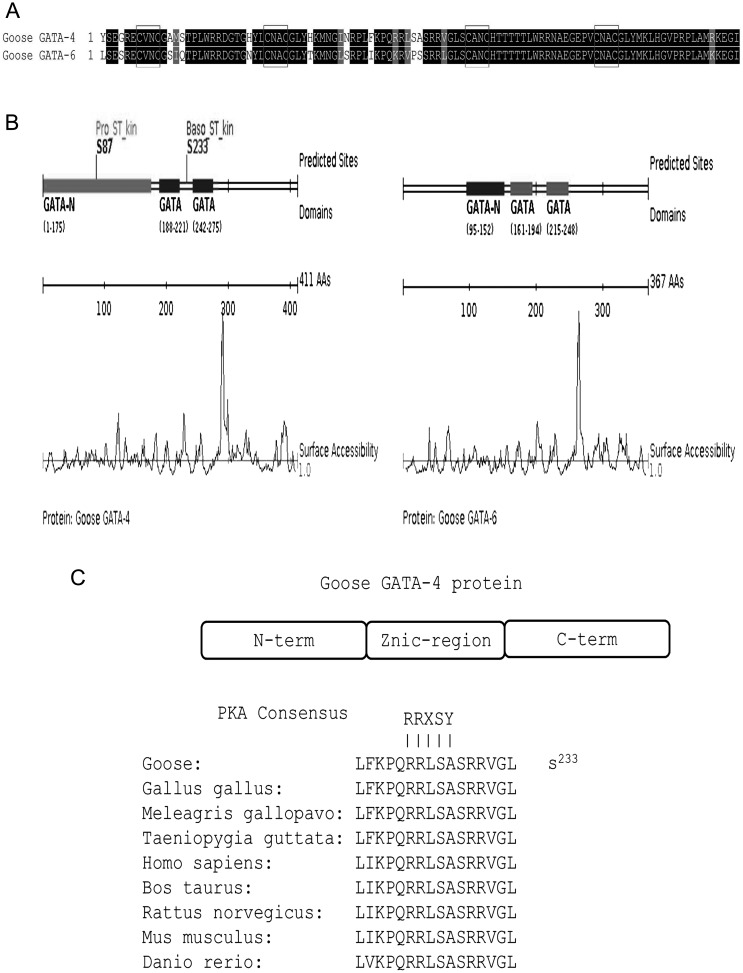
Goose GATA-4 and GATA-6 proteins contain a highly conserved DNA binding domain that consists of two zinc finger domains with the distinctive form (CVNC-X17-CNAC)-X29-(CANC-X17-CNAC) (Fig. 2A), and the proteins share 84.76% identity between the zinc finger domains. We found two phosphorylation sites at amino acid positions 87 (S87) and 233 (S233) in goose GATA-4, but no sites were found in goose GATA-6 by Scansite 3 (Fig. 2B). To map the relevant phospho-acceptor site, we examined the GATA-4 primary amino acid sequence for potential PKA phosphorylation sites, and the high affinity RRXS(T)Y motif was found at amino acid position 233 (S233) (Fig. 2C). This region is conserved among Gallus gallus, Meleagris gallopavo, Taeniopygia guttata, Homo sapiens, Bos taurus, Rattus norvegicus, Mus musculus and Danio rerio GATA-4 proteins. In contrast, no high-affinity RRXS(T)Y motif was found in the goose GATA-6 primary amino acid sequence.
Fig. 2.
A: Amino acid sequence alignment of the zinc finger domains between goose GATA-4 and GATA-6. The black blocks represent identical residues between the zinc finger domains shown, while the red boxes represent the distinct form (CVNC-X17-CNAC)-X29-(CANC-X17-CNAC). B: The phosphorylation sites in goose GATA-4 and GATA-6 proteins were predicted by Scansite 3. The GATA-4 protein contains two phosphorylation sites located at amino acid positions 87 (S87) and 233 (S233). No sites were found in goose GATA-6. C: Mapping of the phospho-residue reveals that the GATA-4 protein contains a species-conserved PKA consensus phosphorylation site located within the zinc finger region.
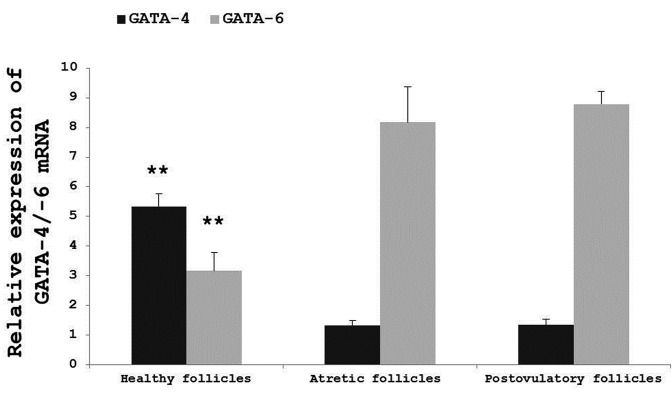
Expression patterns of GATA-4 and GATA-6 mRNAs in three types of follicles
A SYBR Green real-time RT-PCR analysis was performed to determine the expression patterns of GATA-4 and GATA-6 in three types of follicles (Fig. 3). The assay revealed that GATA-4 and GATA-6 mRNAs are co-expressed in goose follicles. It also revealed that the relative mRNA expression level of GATA-4 in healthy follicles was significantly higher than in atretic follicles or postovulatory follicles (P<0.01) and that the relative mRNA expression level of GATA-6 in healthy follicles was significantly lower than in atretic follicles or postovulatory follicles (P<0.01). Finally, no significant variation in the relative mRNA expression levels of GATA-4 and GATA-6 was identified between atretic follicles and postovulatory follicles.
Fig. 3.
Relative expression levels of GATA-4 and GATA-6 mRNAs in three types of follicles. The expression values were normalized to β-actin and 18S and were compared using the 2^(−ΔΔCt) method. The data are represented as the mean ± SD (n=3). **, # Values are significantly different compared with healthy follicles in GATA-4 and GATA-6 relative expression, respectively (P<0.01).
Expression levels of GATA-4 and GATA-6 mRNAs in granulosa and theca cells during goose follicle development
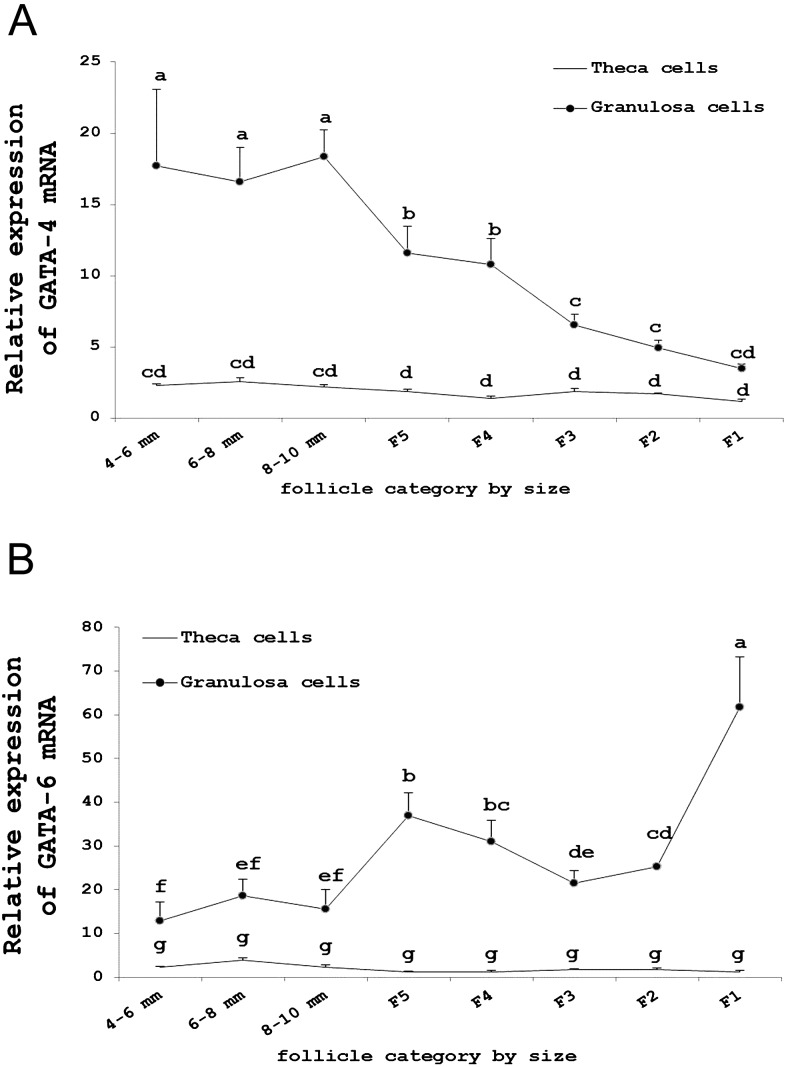
To detect the expression patterns of GATA-4 and GATA-6 in granulosa and theca cells during goose follicle development, we performed real-time PCR analysis (Fig. 4). In our study, GATA-4 and GATA-6 mRNAs were primarily expressed in granulosa cells and weakly expressed in theca cells.
Fig. 4.
A: Relative expression level of GATA-4 mRNA in granulosa and theca cells in different sized follicles. B: Relative expression level of GATA-6 mRNA in granulosa and theca cells in different sized follicles. The expression values were normalized to β-actin and 18S and were compared using the 2^(−ΔΔCt) method. The data are presented as the mean ± SD (n=3). The lowercase letters indicate significant differences (P<0.05).
The expression level of GATA-4 mRNA in granulosa cells was downregulated during follicle development; the expression reached its peak in 8–10 mm follicles and was lowest in the F1 follicles (Fig. 4A). However, GATA-6 was upregulated and reached its peak expression in the F1 follicles (Fig. 4B).
Discussion
Little is known about the molecular roles of GATA-4 and GATA-6 in geese. This study is the first to report the structures, expression patterns and possible functions of GATA-4 and GATA-6 during goose follicle development. We obtained the full-length coding sequences of goose GATA-4 and GATA-6, which consist of 1236 and 1104 nucleotides and encode 411 and 367 amino acid proteins, respectively. The predicted amino acid sequences of goose GATA-4 and GATA-6 contain two adjacent zinc finger domains and associated basic domains that are characteristics of this transcription factor family [3].
It has been well established that GATA-6 has an extra amino-terminal extension from the initiator methionine and can use two Met-codons in frame as translational initiation codons, forming a short open reading frame (uORF) in the entire upstream sequence; therefore, this led to the long-type and short-type GATA-6 proteins [22,23,24]. Through multiple amino acid sequence alignments of GATA-6, we found that the GATA-6 protein in the goose, Gallus gallus and Meleagris gallopavo is the short type, while the GATA-6 protein in Taeniopygia guttata has an extra amino-terminal extension from the initiator methionine and forms the long type. To date, the functional significance of the two forms of GATA-6 remains to be investigated.
However, the phosphorylation of GATA-4 has been the best studied posttranslational modification to date; in vitro experiments have revealed that the phosphorylation level of GATA-4 rises rapidly when induced by cAMP and that is mediated by PKA the pathway [16]. Phosphorylation increases the DNA-binding of GATA-4 and enhances its transcriptional properties on multiple gene promoters. Recent research has indicated that the GATA-4-dependent genes in the ovary include steroidogenic acute regulatory protein (StAR) [25], P-450 aromatase (CYP19A1) [26], P-450 side-chain cleavage enzyme (CYP11A1) [27], gonadotropin-releasing hormone receptor (GnRHR) [28], inhibin-α [29] and bcl-2 [30]. Through online prediction and mapping, goose GATA-4 might be phosphorylated predominantly on S233, which is located within a consensus motif (RRXS(T)Y) [31] for PKA phosphorylation. This phosphorylated site is highly conserved among several species. However, we did not find a phosphorylated site in goose GATA-6, and no reports have indicated GATA-6 target genes in the ovary. Although GATA factors universally recognize a GATA sequence element, subtle differences in the GATA core DNA motif might promote differential DNA-binding activity among co-expressed GATA family members within a specific tissue [32].
In this study, we demonstrated that GATA-4 and GATA-6 mRNAs are co-expressed in goose follicles. Compared with mammals, poultry do not have corpora lutea, which form in postovulatory follicles after ovulation. Our results indicate that the relative mRNA expression level of GATA-4 in healthy follicles is significantly higher than in atretic follicles or postovulatory follicles (P<0.01). The opposite expression pattern occurred for GATA-6 mRNA, and the level was lower than in atretic follicles or postovulatory follicles (P<0.01). The reverse expression patterns may indicate that GATA-4 and GATA-6 have different functions in goose follicles. Previous studies have reported that the GATA-4 expression level was lower in the corpus luteum compared with in follicles, whereas GATA-6 was abundantly expressed in both structures in rats and mice [10, 11], which agrees with the findings of studies conducted in humans and pigs [13, 14]. Moreover, GATA-4 has been associated with the inhibition of apoptosis in cardioblasts and cardiac muscle cells [33, 34]; therefore, goose GATA-4 may play a role in inhibiting ovarian cell apoptosis.
In addition, we found that GATA-4 and GATA-6 mRNAs are primarily expressed in granulosa cells and weakly expressed in theca cells. The expression level of GATA-4 mRNA decreased in granulosa cells during follicle development; the expression reached its highest point in 8–10 mm follicles and its lowest level in the F1 follicles. In both mouse and human granulosa cells, the expression level of GATA-4 is correlated with cell proliferation activity and low levels of apoptosis [11], and GATA-4 can regulate bcl-2 and inhibin-α, which are capable of blocking the induction of granulosa cell apoptosis [30, 35, 36]. In the goose ovary, selection of a single follicle into the preovulatory follicle occurs from a small cohort of the largest prehierarchical follicles measuring approximately 8 to 10 mm in diameter [37]. The expression level of GATA-4 in granulosa cells was highest at this stage. In the goose F1 follicle, the rapid downregulation of GATA-4 precedes physiological apoptosis in the ovulating follicle. Overexpression of GATA-4 protects human granulosa cell tumors from TRAIL-induced apoptosis in vitro, whereas the disruption of GATA4 function induces apoptosis and potentiates the apoptotic effect of TRAIL administration [38], suggesting that GATA-4 could act as a prognostic marker of granulosa cells [39].
The changes in the expression level of GATA-6 mRNA were the opposite of those of GATA-4 in granulosa cells. The expression level of GATA-6 mRNA increased during follicle development and reached its highest point in F1 follicles. GATA-6 has been shown to induce cell cycle quiescence and differentiation in vascular smooth muscle cells [40, 41]; thus, the highest expression level of GATA-6 in the F1 follicle may help to mediate the terminal differentiation of granulosa cells into postovulatory follicles. Previous studies indicated that the GATA-4 expression level in rodent granulosa cells is usually downregulated in follicles undergoing atresia, while GATA-6 expression is retained at this stage [10]. After ovulation, GATA-4 mRNA and protein dramatically decreases in the murine ovary, while the expression of GATA-6 mRNA is strongly expressed [11, 42]. However, whether the decreased expression of GATA-4 and increased expression of GATA-6 could be used as a marker for granulosa cell apoptosis is unclear.
In conclusion, the present study provided evidence for the existence of two members of the GATA family in goose follicles and indicated that both GATA-4 and GATA-6 are needed to ensure proper follicle growth. The molecular structural differences of goose GATA-4 and GATA-6 may be related to their different roles during follicle development. GATA-6 may be important for normal ovarian functions, and we identified that the goose GATA-6 protein is the short-type form. These results provide valuable insight to help understand the functions of GATA-6 in goose follicles and are helpful for identifying putative target genes and determining the binding sites in vitro.
Acknowledgments
The study was supported by the National Waterfowl Industrial Technology System (No. CARS-43-6) and the Program for Breeding Research of Sichuan Province (2011NZ0099-8), poultry genetic resources exploring and innovation (No. 13TD0034).
References
- 1.Viger RS, Guittot SM, Anttonen M, Wilson DB, Heikinheimo M. Role of the GATA family of transcription factors in endocrine development, function, and disease. Mol Endocrinol 2008; 22: 781–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Patient RK, McGhee JD. The GATA family (vertebrates and invertebrates). Curr Opin Genet Dev 2002; 12: 416–422 [DOI] [PubMed] [Google Scholar]
- 3.Lowry JA, Atchley WR. Molecular evolution of the GATA family of transcription factors: conservation within the DNA-binding domain. J Mol Evol 2000; 50: 103–115 [DOI] [PubMed] [Google Scholar]
- 4.Pandolfi PP, Roth ME, Karis A, Leonard MW, Dzierzak E, Grosveld FG, Engel JD, Lindenbaum MH. Targeted disruption of the GATA3 gene causes severe abnormalities in the nervous system and in fetal liver haematopoiesis. Nat Genet 1995; 11: 40–44 [DOI] [PubMed] [Google Scholar]
- 5.Orkin SH. GATA-binding transcription factors in hematopoietic cells. Blood 1992; 80: 575–581 [PubMed] [Google Scholar]
- 6.Laverriere AC, MacNeill C, Mueller C, Poelmann RE, Burch JB, Evans T. GATA-4/5/6, a subfamily of three transcription factors transcribed in developing heart and gut. J Biol Chem 1994; 269: 23177–23184 [PubMed] [Google Scholar]
- 7.Molkentin JD. The zinc finger-containing transcription factors GATA-4, -5, and -6. Ubiquitously expressed regulators of tissue-specific gene expression. J Biol Chem 2000; 275: 38949–38952 [DOI] [PubMed] [Google Scholar]
- 8.LaVoie HA. The role of GATA in mammalian reproduction. Exp Biol Med (Maywood) 2003; 228: 1282–1290 [DOI] [PubMed] [Google Scholar]
- 9.Bennett J, Wu YG, Gossen J, Zhou P, Stocco C. Loss of GATA-6 and GATA-4 in granulosa cells blocks folliculogenesis, ovulation, and follicle stimulating hormone receptor expression leading to female infertility. Endocrinology 2012; 153: 2474–2485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lavoie HA, McCoy GL, Blake CA. Expression of the GATA-4 and GATA-6 transcription factors in the fetal rat gonad and in the ovary during postnatal development and pregnancy. Mol Cell Endocrinol 2004; 227: 31–40 [DOI] [PubMed] [Google Scholar]
- 11.Heikinheimo M, Ermolaeva M, Bielinska M, Rahman NA, Narita N, Huhtaniemi IT, Tapanainen JS, Wilson DB. Expression and hormonal regulation of transcription factors GATA-4 and GATA-6 in the mouse ovary. Endocrinology 1997; 138: 3505–3514 [DOI] [PubMed] [Google Scholar]
- 12.Kyrönlahti A, Vetter M, Euler R, Bielinska M, Jay PY, Anttonen M, Heikinheimo M, Wilson DB. GATA4 deficiency impairs ovarian function in adult mice. Biol Reprod 2011; 84: 1033–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gillio-Meina C, Hui YY, LaVoie HA. GATA-4 and GATA-6 transcription factors: expression, immunohistochemical localization, and possible function in the porcine ovary. Biol Reprod 2003; 68: 412–422 [DOI] [PubMed] [Google Scholar]
- 14.Laitinen MP, Anttonen M, Ketola I, Wilson DB, Ritvos O, Butzow R, Heikinheimo M. Transcription factors GATA-4 and GATA-6 and a GATA family cofactor, FOG-2, are expressed in human ovary and sex cord-derived ovarian tumors. J Clin Endocrinol Metab 2000; 85: 3476–3483 [DOI] [PubMed] [Google Scholar]
- 15.Vaskivuo TE, Anttonen M, Herva R, Billig H, Dorland M, te Velde ER, Stenbäck F, Heikinheimo M, Tapanainen JS. Survival of human ovarian follicles from fetal to adult life: apoptosis, apoptosis-related proteins, and transcription factor GATA-4. J Clin Endocrinol Metab 2001; 86: 3421–3429 [DOI] [PubMed] [Google Scholar]
- 16.Tremblay JJ, Viger RS. Novel roles for GATA transcription factors in the regulation of steroidogenesis. J Steroid Biochem Mol Biol 2003; 85: 291–298 [DOI] [PubMed] [Google Scholar]
- 17.Tremblay JJ, Viger RS. GATA factors differentially activate multiple gonadal promoters through conserved GATA regulatory elements. Endocrinology 2001; 142: 977–986 [DOI] [PubMed] [Google Scholar]
- 18.Tremblay JJ, Viger RS. Transcription factor GATA-4 is activated by phosphorylation of serine 261 via the cAMP/protein kinase a signaling pathway in gonadal cells. J Biol Chem 2003; 278: 22128–22135 [DOI] [PubMed] [Google Scholar]
- 19.Gilbert AB. The Ovary in Physiology and Biochemistry of the Domestic Fowl. Academic Press, 1971; vol: 3.
- 20.Gilbert AB, Evans AJ, Perry MM, Davidson MH. A method for separating the granulosa cells, the basal lamina and the theca of the preovulatory ovarian follicle of the domestic fowl (Gallus domesticus). J Reprod Fertil 1977; 50: 179–181 [DOI] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2− ΔΔCT Method. Methods 2001; 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 22.Takeda M, Obayashi K, Kobayashi A, Maeda M. A unique role of an amino terminal 16-residue region of long-type GATA-6. J Biochem 2004; 135: 639–650 [DOI] [PubMed] [Google Scholar]
- 23.Brewer A, Gove C, Davies A, McNulty C, Barrow D, Koutsourakis M, Farzaneh F, Pizzey J, Bomford A, Patient R. The human and mouse GATA-6 genes utilize two promoters and two initiation codons. J Biol Chem 1999; 274: 38004–38016 [DOI] [PubMed] [Google Scholar]
- 24.Maeda M, Ohashi K, Ohashi-Kobayashi A. Further extension of mammalian GATA-6. Dev Growth Differ 2005; 47: 591–600 [DOI] [PubMed] [Google Scholar]
- 25.Silverman E, Yivgi-Ohana N, Sher N, Bell M, Eimerl S, Orly J. Transcriptional activation of the steroidogenic acute regulatory protein (StAR) gene: GATA-4 and CCAAT/enhancer-binding protein β confer synergistic responsiveness in hormone-treated rat granulosa and HEK293 cell models. Mol Cell Endocrinol 2006; 252: 92–101 [DOI] [PubMed] [Google Scholar]
- 26.Monga R, Ghai S, Datta TK, Singh D. Involvement of transcription factor GATA-4 in regulation of CYP19 gene during folliculogenesis and luteinization in buffalo ovary. J Steroid Biochem Mol Biol 2012; 130: 45–56 [DOI] [PubMed] [Google Scholar]
- 27.Sher N, Yivgi-Ohana N, Orly J. Transcriptional regulation of the cholesterol side chain cleavage cytochrome P450 gene (CYP11A1) revisited: binding of GATA, cyclic adenosine 3′,5′-monophosphate response element-binding protein and activating protein (AP)-1 proteins to a distal novel cluster of cis-regulatory elements potentiates AP-2 and steroidogenic factor-1-dependent gene expression in the rodent placenta and ovary. Mol Endocrinol 2007; 21: 948–962 [DOI] [PubMed] [Google Scholar]
- 28.Cheng CK, Yeung CM, Chow BK, Leung PC. Characterization of a new upstream GnRH receptor promoter in human ovarian granulosa-luteal cells. Mol Endocrinol 2002; 16: 1552–1564 [DOI] [PubMed] [Google Scholar]
- 29.Anttonen M, Parviainen H, Kyrönlahti A, Bielinska M, Wilson DB, Ritvos O, Heikinheimo M. GATA-4 is a granulosa cell factor employed in inhibin-α activation by the TGF-β pathway. J Mol Endocrinol 2006; 36: 557–568 [DOI] [PubMed] [Google Scholar]
- 30.Kyrönlahti A, Rämö M, Tamminen M, Unkila-Kallio L, Butzow R, Leminen A, Nemer M, Rahman N, Huhtaniemi I, Heikinheimo M, Anttonen M. GATA-4 regulates Bcl-2 expression in ovarian granulosa cell tumors. Endocrinology 2008; 149: 5635–5642 [DOI] [PubMed] [Google Scholar]
- 31.Kemp BE, Pearson RB. Protein kinase recognition sequence motifs. Trends Biochem Sci 1990; 15: 342–346 [DOI] [PubMed] [Google Scholar]
- 32.Sakai Y, Nakagawa R, Sato R, Maeda M. Selection of DNA binding sites for human transcriptional regulator GATA-6. Biochem Biophys Res Commun 1998; 250: 682–688 [DOI] [PubMed] [Google Scholar]
- 33.Grépin C, Nemer G, Nemer M. Enhanced cardiogenesis in embryonic stem cells overexpressing the GATA-4 transcription factor. Development 1997; 124: 2387–2395 [DOI] [PubMed] [Google Scholar]
- 34.Kitta K, Day RM, Kim Y, Torregroza I, Evans T, Suzuki YJ. Hepatocyte growth factor induces GATA-4 phosphorylation and cell survival in cardiac muscle cells. J Biol Chem 2003; 278: 4705–4712 [DOI] [PubMed] [Google Scholar]
- 35.Pru JK, Tilly JL. Programmed cell death in the ovary: insights and future prospects using genetic technologies. Mol Endocrinol 2001; 15: 845–853 [DOI] [PubMed] [Google Scholar]
- 36.Findlay JK, Drummond AE, Dyson M, Baillie AJ, Robertson DM, Ethier JF. Production and actions of inhibin and activin during folliculogenesis in the rat. Mol Cell Endocrinol 2001; 180: 139–144. [DOI] [PubMed] [Google Scholar]
- 37.Johnson PA. Follicle selection in the avian ovary. Reprod Domest Anim 2012; 47(Suppl 4): 283–287 [DOI] [PubMed] [Google Scholar]
- 38.Kyrönlahti A, Kauppinen M, Lind E, Unkila-Kallio L, Butzow R, Klefström J, Wilson DB, Anttonen M, Heikinheimo M. GATA4 protects granulosa cell tumors from TRAIL-induced apoptosis. Endocr Relat Cancer 2010; 17: 709–717 [DOI] [PubMed] [Google Scholar]
- 39.Anttonen M, Unkila-Kallio L, Leminen A, Butzow R, Heikinheimo M. High GATA-4 expression associates with aggressive behavior, whereas low anti-Müllerian hormone expression associates with growth potential of ovarian granulosa cell tumors. J Clin Endocrinol Metab 2005; 90: 6529–6535 [DOI] [PubMed] [Google Scholar]
- 40.Suzuki E, Evans T, Lowry J, Truong L, Bell DW, Testa JR, Walsh K. The human GATA-6 gene: structure, chromosomal location, and regulation of expression by tissue-specific and mitogen-responsive signals. Genomics 1996; 38: 283–290 [DOI] [PubMed] [Google Scholar]
- 41.Morrisey EE. GATA-6: the proliferation stops here: cell proliferation in glomerular mesangial and vascular smooth muscle cells. Circ Res 2000; 87: 638–640 [DOI] [PubMed] [Google Scholar]
- 42.Anttonen M, Ketola I, Parviainen H, Pusa AK, Heikinheimo M. FOG-2 and GATA-4 are coexpressed in the mouse ovary and can modulate mullerian-inhibiting substance expression. Biol Reprod 2003; 68: 1333–1340 [DOI] [PubMed] [Google Scholar]