Abstract
We measured the dynamics of an essential epigenetic modifier, HP1β, in human cells at different stages of differentiation using Fluorescence Recovery After Photobleaching (FRAP). We found that HP1β mobility is similar in human embryonic stem cells (hES) and iPS cells where it is more mobile compared to fibroblasts; HP1β is less mobile in senescent fibroblasts than in young (dividing) fibroblasts. Introduction of “reprogramming factors”, Oct4, Sox2, Klf4, cMyc and Lin28, into senescent fibroblasts and measuring the changes in HP1β mobility as reprogramming proceeds shows that the mobility of HP1β in senescent cells increases and by day 9 is the same as that found in young fibroblasts. Thus the dynamics of a key epigenetic modifier can be rejuvenated without de-differentiation through an embryonic stage. Future work will test whether other aspects of cellular physiology that age can be so rejuvenated without de-differentiation.
Somatic cell nuclear transfer (SCNT)1 has shown that a newborn clone can be derived from an old, differentiated, cell2,3. Likewise, senescent fibroblasts can be de-differentiated into a stable pluripotent ES-cell-like state (iPS cells)4 using ‘reprogramming factors' and then re-differentiated back to fibroblasts that have lost the senescent phenotype and have acquired the characteristics of young fibroblasts5. Based on these findings we have proposed that SCNT and iPS cell technology could be used to rejuvenate senescent cells without going through an embryonic stage because “developmental reprogramming” to the embryonic (ES and iPS cell) state may be molecularly separated from “age reprogramming”. A key aspect of age reprogramming concerns the epigenome, which includes the DNA modifications, histone modifications and chromatin structures that regulate the expression, integrity and organisation of the genome6,7; during age reprogramming age-related changes in the epigenome of an old cell are reprogrammed to a youthful one found in a young cell by a process we have termed epigenetic rejuvenation8,9.
Chromatin is the natural environment of nearly all eukaryotic genes and is known to undergo changes during ageing10,11. Cytological examination has revealed that chromatin exists in two distinct states of compaction called euchromatin and heterochromatin12. Heterochromatin represents the dense compartment and its formation is thought to be regulated by a number of factors including transcription factor binding13, histone modifications14,15, DNA methylation16 alteration of nucleosome positioning17, transcription of repetitive DNAs18,19 and the binding of non-histone chromosomal proteins such as Heterochromatin Protein 1 (HP1)20. HP1 proteins are evolutionarily conserved proteins whose presence, along with methylated lysine 9 of histone H3 (Me(3)H3K9), is a conserved characteristic of constitutive heterochromatin in organisms as diverse as fission yeast and man21. In mouse and man there are three HP1 isotypes, termed HP1α, HP1β and HP1γ22,23. The three mammalian isotypes exhibit a high degree of sequence and 3-D structural organisation similarity, but both antibody localisation studies and mutational analysis in mice have shown that they have non-redundant functions, with HP1β being essential24. Notably, HP1 proteins are found at telomeric heterochromatin as part of the shelterin complex that maintains the structural integrity of the telomere “cap”25. Erosion of the telomeres is known to take place during ageing and the short telomeres of old cells can be lengthened by passage through an embryonic iPS cell stage26. HP1 proteins are components of senescence-associated heterochromatin foci (SAHF) that are thought to sequester proliferation-promoting genes as cells exit the cell cycle and enter a state of cellular senescence27,28. Cellular senescence can be induced by several methods, including oncogene induced senescence29, combined interferon gamma and TNF treatment of cells30 and replicative exhaustion31. Of these, induction of senescence by replicative exhaustion has been used as a well-known in vitro model for studying ageing at the cellular and molecular level31. HP1 proteins can also regulate gene activity locally, both positively and negatively32, and are known to bind to the Oct3/433 and Nanog34 genes, which are key components of the pluripotency pathway in mammalian ES cells35,36. That HP1 proteins are part of the shelterin complex and SAHF and are likely involved in regulation of pluripotency genes has led to the suggestion that study of these proteins might provide insight into the molecular mechanisms underpinning epigenetic rejuvenation8,9.
HP1 proteins were one of the first proteins to be used in non-invasive FRAP studies to probe chromatin protein interactions in the living cell37. Numerous FRAP studies, in conjunction with kinetic modelling, have shown that at steady state equilibrium the nuclear HP1 pool can be separated into three kinetic fractions: a highly mobile “fast” fraction that freely diffuses through the nucleoplasm, a less mobile “slow” fraction that transiently binds to the HP1 ligand, Me(3)H3K9, and a smaller immobilised HP1 fraction whose ligand(s) is not known38,39,40,41,42,43,44. Studies in the mouse have shown that the mobility of HP1 is dependent upon the differentiation state of the cell with HP1 being “hyper” dynamic in undifferentiated ES cells compared to the same cells after 24 hrs of LIF withdrawal or differentiation into neuronal progenitors45. HP1 mobility in hES, human iPS cells and in senescent cells has not been determined.
Here we show, for the first time, that HP1β mobility is similar in hES and human iPS cells and is more mobile compared to primary human fibroblasts. Furthermore HP1β in senescent cells is significantly less mobile than in young (dividing) fibroblasts. To test the central idea of epigenetic rejuvenation we have measured, using FRAP, the dynamics of HP1β during the reprogramming of senescent fibroblasts. To do this we developed a construct based on the “piggyBac” construct46 that expresses five “reprogramming factors” along with a marker gene, dsRed, whose expression enables reprogramming to be followed at the single cell level. Introduction of the construct into senescent fibroblasts and following the changes in HP1β mobility as reprogramming proceeds shows that the mobility of HP1β in senescent cells increases and by day 9 is the same as that found in young fibroblasts. This occurs without any overt changes in the differentiated phenotype of the fibroblast. Thus the dynamics of a key epigenetic modifier can be rejuvenated without de-differentiation indicating that epigenetic rejuvenation, with respect to HP1β mobility, has been achieved using iPS cell technology.
Results
HP1β has the same mobility in hES and iPS cells and is less mobile in LF1 primary human fibroblasts
Using the five-factor “piggyBac” vector46 we reprogrammed LF1 primary human fibroblasts to produce 10 iPS cell lines of which one, line #2.4, was characterised further. Line #2.4 had a normal chromosomal complement (Fig. S1a), was alkaline phosphatase positive (Fig. S1b), showed reduced CpG methylation of the Oct4 and Nanog genes compared to the parental LF1 fibroblasts (Fig. S1c), expressed six different pluripotency markers in common with H9 cells (Fig. S1d) and could differentiate into all three germ layers (Fig. S1e). Endogenous HP1β and Me(3)K9H3 co-localised in hES and iPS line #2.4 nuclei (Fig. 1a; top two rows) and the protein levels of HP1α, HP1β and HP1γ were similar in hES and the iPS line #2.4 cell extracts (Fig. 1b). We introduced HP1β-GFP into H9 and iPS line #2.4 by transient lipofection and measured the FRAP of HP1β-GFP foci. The recovery data were similar and showed no significant difference (Table 1; Fig. 2a; p = 0.85).
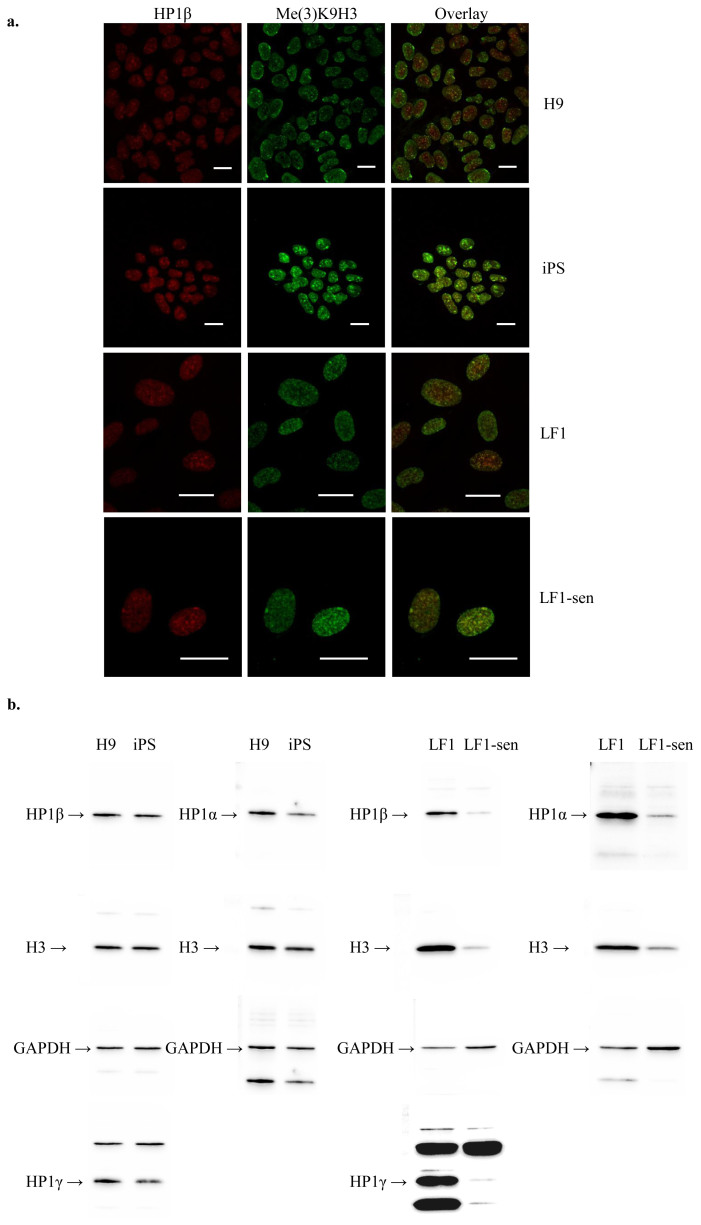
Figure 1. Nuclear localisation and detection of HP1β in H9 hES, iPS, LF1 and senescent LF1 cells.
(a) The left-hand column depicts the distribution of HP1β (red) and the middle column shows the distribution of tri-methylated lysine 9 of histone H3 in (Me(3)K9H3) (green). The merged overlay of the images in the left-hand and middle columns is given in the right hand column. For all cells types (going top to bottom) H9, iPS, LF1 and LF1 senescent cells the enriched foci of HP1β and Me(3)K9H3 overlap and are seen as yellow foci in the right-hand column. The scale bar in each panel is 20 μm. (b) The levels of HP1α, HP1β and HP1γ were similar in H9 hES and the iPS line #2.4 cell extracts (protein loading amount 20 μg). The levels of HP1α and HP1β were similar in LF1 cell extracts with HP1γ showing slightly greater levels. The protein levels of HP1 isotypes appeared higher in young LF1 cells compared to senescent LF1 cells (protein loading amount 30 μg). However, once corrected for the endogenous H3 levels, which reflected the amount of nuclear proteins isolated in the cell extracts, the protein levels of the three HP1 isotypes in both young and senescent cell extracts were similar. Each blot was probed multiple times; for uncropped blots and order of probing please see Supplementary Fig. S2.
Table 1. FRAP analysis of HP1β mobility in H9 hES, iPS, young LF1 and senescent LF1 cells. The parameters for H9 hES and iPS cells are not significantly different to each other (p = 0.85). The recovery parameters for LF1 cells are significantly different (p < 0.0001) from parameters for iPS and H9 hES cells. Senescent LF1 cells have longer halftimes for the “fast” and “slow” fractions compared to LF1 cells (p < 0.0001).
| H9 | iPS | LF1 | LF1 senescent | |
|---|---|---|---|---|
| T1/2 fast (s) | 0.27 ± 0.1 | 0.3 ± 0.09 | 0.33 ± 0.1 | 0.45 ± 0.1 |
| T1/2 slow (s) | 3.38 ± 0.6 | 3.6 ± 0.48 | 3.8 ± 0.3 | 5.2 ± 0.6 |
| Free (%) | 32 ± 5.1 | 33 ± 3.6 | 26 ± 2.7 | 26 ± 2.3 |
| Bound (%) | 50 ± 5.1 | 49 ± 3.6 | 57 ± 2.7 | 57 ± 2.3 |
| fim (%) | 18 ± 5.8 | 18 ± 3.7 | 17 ± 2.9 | 17 ± 3 |
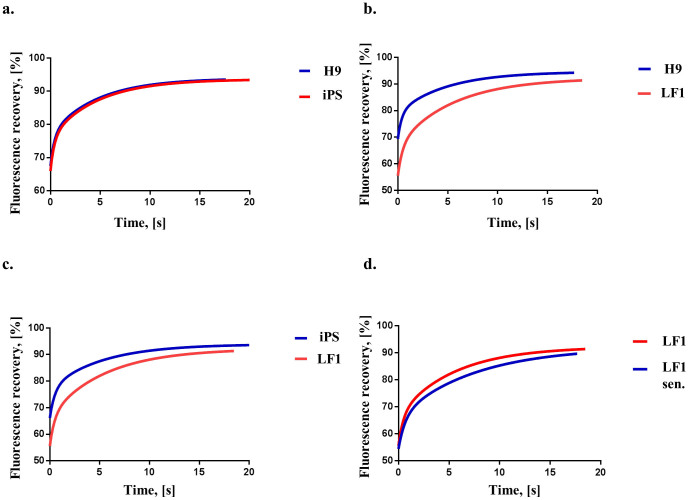
Figure 2. Pairwise comparison of HP1β FRAP curves from H9 hES, iPS, LF1 and senescent LF1 cells.
(a) H9 and iPS recovery curves; the recovery parameters for H9 hES and iPS cells are not significantly different to each other (p = 0.85). (b) H9 and LF1 recovery curves; the recovery parameters for LF1 cells are significantly different (p < 0.0001) from H9 cells (c) iPS and LF1 recovery curves; the recovery parameters for LF1 cells are significantly different (p < 0.0001) from iPS cells. (d) Young LF1 and senescent LF1 recovery curves; the recovery parameters for young LF1 cells are significantly different (p < 0.0001) from senescent LF1 cells. All recovery curves are calculated from at least 3 experiments. For details of the recovery curve parameters please see Table 1; the individual HP1β FRAP measurements are presented in Supplementary Fig. S6.
We next measured HP1β mobility in the parental LF1 primary human fibroblast cell line that was used for reprogramming into iPS cells. Measurement of the HP1β mobility in LF1 nuclei after transient lipofection showed that the recovery data was significantly different to that for hES and iPS line #2.4 (Table 1; Figs. 2b and 2c; both p < 0.0001).
HP1β is less mobile in senescent compared to young LF1 fibroblasts
Young LF1 fibroblasts were positive for the proliferation marker Ki-67 and negative for SA-β-galactosidase activity (Fig. S3a). LF1 fibroblasts were passaged until they reached senescence and had exhausted their proliferative capacity. Senescent LF1 cells were positive for SA-β-galactosidase activity and negative for Ki-67 (Fig. S3b). In both young and senescent LF1 cells endogenous HP1β co-localised with Me(3)K9H3 (Fig. 1a, bottom two rows). When the protein levels of HP1α, HP1β and HP1γ in cellular extracts from senescent LF1 cells were compared to young (dividing) LF1 cells it showed that the levels were lower (Figures 1b and S2b). The histone H3 control levels also decrease in senescent cells indicating that the nuclear:cytoplasmic ratio decreases in senescent cells, which may be related to the known increase in overall size of cells as they enter cellular senescence47. As shown in Table 1 measurement of the HP1β mobility in senescent LF1 nuclei after transient lipofection showed that the recovery data was significantly different to that for the young LF1 cells (Table 1; Fig. 2d; p < 0.0001).
Rejuvenation of HP1β mobility in senescent LF1 cells by introduction of reprogramming factors
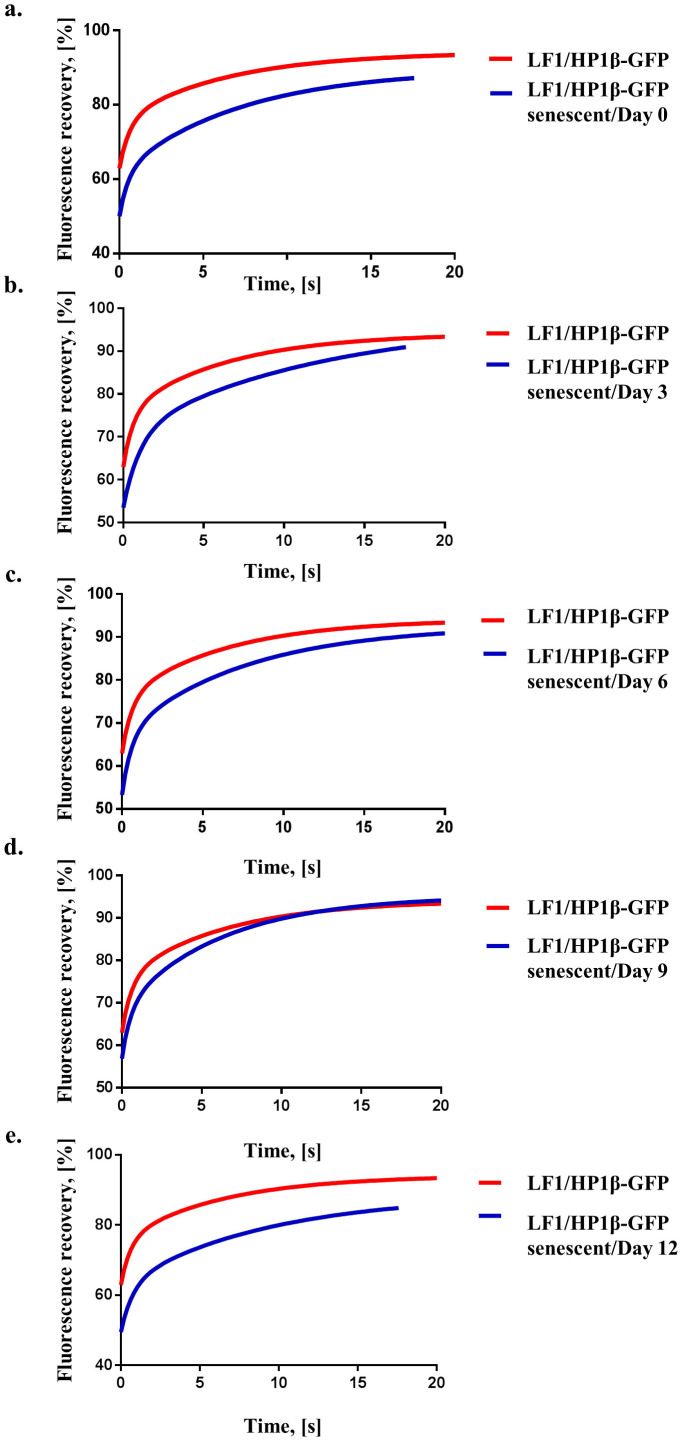
In order to be able to measure the mobility of HP1β during the reprogramming process we generated LF1 fibroblast cell lines that stably expressed HP1β-GFP (LF1/HPβ−GFP fibroblasts; Fig. S4). We chose a line that expressed “low” levels of HP1β−GFP for our experiments (see Fig. S4); it has previously been shown that HP1−GFP dynamics is insensitive to expression levels40,45. Use of LF1/HP1β−GFP fibroblasts avoided the need to transiently transfect HP1β−GFP into cells at various times during the reprogramming process. LF1/HP1β−GFP fibroblasts were passaged until they reached senescence (Fig. S3c). To follow the reprogramming of LF1/HP1β−GFP senescent fibroblasts after introduction of the 5-factor “piggyBac” vector we engineered the construct so that it expressed the dsRed marker gene along with the 5 reprogramming factors (see material and methods, Fig. S5). The newly-constructed vector, called “dsRed-OSKML”, enabled the identification of single cells that had taken up the vector. HP1β mobility was measured in senescent LF1/HP1β−GFP fibroblasts on day 0 (just before dsRed-OSKML was introduced by lipofection) and every three days (days 3, 6, 9 and 12) after lipofection of dsRed-OSKML. As shown in Figs. 3a–c the recovery curves for days 0, 3 and 6 were different to young (dividing) control LF1/HP1β−GFP fibroblasts (p < 0.0001). In particular, the mean T1/2 halftime values for the “slow” fraction were over 5 seconds for days 0 through to 6 compared to the young control LF1/HP1β−GFP fibroblasts which have a mean “slow” halftime of 4.4 seconds (Table 2). We also observed that the values for % immobile fraction varied considerably over the same time scale, with day 3 showing a mean value of 6% compared to 16% and 20% for the controls, young and senescent LF1/HP1β−GFP fibroblasts respectively. On day 9 the HP1β recovery data showed that HP1β was more mobile and was similar to that of control young LF1/HP1β−GFP fibroblasts (Fig. 3d, p = 0.12). There was also a reduction in SA β-galactosidase staining of the cells at day 9 with 49% of cells being positive for β-galactosidase compared to 90% for day 0 (Fig. S3d). Measurement of HP1β mobility on day 12 showed that it was less mobile and now showed similar kinetics to that for days 0 through to 6 and the recovery curve was again different to the control LF1/HP1β−GFP fibroblasts (Fig. 3e; p < 0.0001).
Figure 3. Comparison of HP1β FRAP curves from young LF1/HP1β−GFP cells and senescent LF1/HPβ−GFP cells undergoing reprogramming.
(a) The young and senescent LF1/HP1β−GFP fibroblast recovery curves measured on day 0 of reprogramming; the recovery parameters for young LF1/HP1β−GFP cells are significantly different (p < 0.0001) from senescent LF1/HP1β−GFP cells. (b) The young and senescent LF1/HP1β−GFP fibroblast recovery curves after 3 days of reprogramming; the recovery parameters for young LF1/HP1β−GFP cells are significantly different (p < 0.0001) from senescent LF1/HP1β−GFP cells after 3 days of reprogramming. (c) The young and senescent LF1/HP1β−GFP fibroblast recovery curves after 6 days of reprogramming; the recovery parameters for young LF1/HP1β−GFP cells are significantly different (p < 0.0001) from senescent LF1/HP1β−GFP cells after 6 days of reprogramming. (d) The young and senescent LF1/HP1β−GFP fibroblast recovery curves after 9 days of reprogramming; the recovery parameters for young LF1/HP1β−GFP cells are not significantly different (p = 0.12) from senescent LF1/HP1β−GFP cells after 9 days of reprogramming. (e) The young and senescent LF1/HP1β−GFP fibroblast recovery curves after 12 days of reprogramming; the recovery parameters for young LF1/HP1β−GFP cells are significantly different (p < 0.0001) from senescent LF1/HP1β−GFP cells after 12 days of reprogramming. All recovery curves are calculated from at least 3 experiments. For details of the recovery curve parameters please see Table 2; the individual HP1β FRAP measurements are presented in Supplementary Fig. S7.
Table 2. FRAP analysis of HP1β mobility during reprogramming of senescent LF1/HP1β−GFP cells. The left hand column represents the FRAP parameters for young LF1/HP1β−GFP cells. The column headed “Day 0” contains the FRAP parameters for senescent LF1/HP1β−GFP cells just before lipofection with dsRed-OSKML. Days 3, 6, 9 and 12 are the columns containing the parameters for senescent LF1/HP1β−GFP cells after 3, 6, 9 and 12 days of reprogramming (i.e., 3, 6, 9 and 12 days after lipofection with dsRed-OSKML). Using these FRAP parameters it can be seen in Figs. 3a, b, c and e that the recovery of HP1β in young LF1/HP1β−GFP fibroblasts is significantly faster (p < 0.0001) compared to the recovery of HP1β in Day 0 senescent LF1/HP1β−GFP fibroblasts and after 3, 6, and 12 days of reprogramming. There is no significant difference (p = 0.12) between the recovery parameters of young LF1/HP1β−GFP fibroblasts and senescent LF1/HP1β−GFP fibroblasts on day 9 of reprogramming (recovery curves see Fig. 3d).
| LF1/HP1β | Day 0 | Day 3 | Day 6 | Day 9 | Day 12 | |
|---|---|---|---|---|---|---|
| T1/2 fast (s) | 0.4 ± 0.1 | 0.37 ± 0.1 | 0.7 ± 0.15 | 0.4 ± 0.1 | 0.38 ± 0.14 | 0.5 ± 0.13 |
| T1/2 slow (s) | 4.4 ± 0.6 | 5.1 ± 0.6 | 8.3 ± 2.8 | 5.2 ± 0.76 | 4.4 ± 0.66 | 6.4 ± 1.24 |
| Free (%) | 34 ± 3.4 | 24 ± 2.56 | 37 ± 2.4 | 29 ± 2.7 | 28 ± 4.2 | 27 ± 2.5 |
| Bound (%) | 50 ± 3.4 | 55 ± 2.56 | 57 ± 2.4 | 55 ± 2.7 | 61 ± 4.2 | 51 ± 2.5 |
| fim (%) | 16 ± 2.3 | 20 ± 3.4 | 6 ± 8.4 | 16 ± 3.9 | 11 ± 4.8 | 22 ± 4.4 |
Discussion
The hallmarks of ageing11 are reversible. Old cells can be rejuvenated when they are reprogrammed by SCNT2,3 or via iPS cell induction5,26,48. Both methods require the passage of the old cell through an embryonic (ES or iPS cell) stage. By contrast we have proposed that “developmental reprogramming” to an embryonic stage can be separated from “age reprogramming” where the aged phenotype is reverted back to a youthful one; an old cell is rejuvenated while maintaining its specialised, differentiated, functions8,9. In this work we have tested whether these two apparently seamless aspects of reprogramming - developmental and age reprogramming - can be disentangled using as our experimental approach the mobility of the essential non-histone protein, HP1β20.
HP1β has the same kinetic parameters in hES and human iPS cells and is significantly more mobile in these embryonic cell types than in the LF1 fibroblasts from which the iPS cells were derived. This extends the work in the mouse, which has shown that HP1 is “hyper” dynamic in murine ES cells compared to differentiated progeny45. Taken together with our work on human cells these data indicate that the higher mobility of HP1 proteins in ES cells might serve as a parameter that characterises the epigenetic chromatin “ground state” of ES cells. While experimentally more involved than using antibodies for stem cell markers, measurement of HP1β mobility after transient transfection of HP1β-GFP into presumptive ES/iPS cells might provide a measure of the degree of reprogramming achieved and, further, whether the high mobility of HP1β characteristic of ES cells has been attained.
HP1β mobility in dividing, Ki-67 positive, LF1 cells is significantly greater than in non-dividing, SA β-galactosidase positive, senescent LF1 cells that have exited the cell cycle. Comparison of T1/2 for the “slow” fraction across the different cell types studied shows that senescent cells have a mean halftime that is over five seconds whereas all other halftimes are less than four seconds (Table 1). The decreased mobility of HP1β represents, we suggest, the senescent epigenetic chromatin “ground state” of old, non-dividing cells and provides a marker against which “epigenetic rejuvenation” upon reprogramming of senescent cells by iPS cell induction can be measured. Introduction of five reprogramming factors into senescent LF1 cells showed that the mobility of HP1β increased in senescent cells and reached the levels found in young LF1 cells by day 9; there was also a decrease in SA β-galactosidase positive cells on day 9 (Figure S3d). The increase in HP1β mobility was transient. HP1β mobility slowed and fell back to senescent levels by day 12 of reprogramming. Throughout the 12 days of reprogramming the senescent fibroblasts were maintained in fibroblast medium and they retained their fibroblast morphology; it takes 40 days for iPS cells to be generated from senescent cells and only after transfer to hES medium5.
The transient nature of the rejuvenation of HP1β mobility at day 9 is at around the time the epigenome is thought to become “unstable” in specialised cells undergoing reprogramming49,50. Instability of the rejuvenated epigenome (including histone/DNA modifications) may be an obstacle to the generation of stably rejuvenated cells. Stabilisation may require the development of culture conditions or chemicals that can maintain the rejuvenated state, by analogy with the regimes that have been developed for the “capture” of the pluripotent state51. It is of course possible that the intimate association of developmental reprogramming to age reprogramming does not allow the stabilisation of a rejuvenated state without passage through an embryonic (ES/iPS) stage. It is likely that cell biological processes (organelles), epigenome modifications and molecular pathways in the senescent cells are rejuvenated at different times to others during the reprogramming process and some are more refractory to age reprogramming and require passage through an embryonic (ES/iPS) stage. The mechanisms and degree of rejuvenation of senescent cells may also be dependent upon how the senescent state was induced; the “senescence” induced by interferon gamma and TNF treatement30 may be molecularly different to that induced, for example, by replicative exhaustion. It also remains to be shown that the cell biological processes, epigenome modifications and molecular pathways in a rejuvenated cell are the same as in young cell that has not been through the ageing process. If they are not the same, the ability to “age reprogram” particular aspects of cellular physiology is likely to be facilitated by understanding the novel pathways and mechanisms that operate in rejuvenated cells.
Much work needs to be done to validate age reprogramming as another approach to patient-specific regenerative therapies. But there are clear advantages to pursuing such an alternative to SCNT52 and iPS cell53 based therapies9. The present work indicates that developmental and age reprogramming may be separated, at least in terms of HP1β mobility, suggesting that such a pursuit could be fruitful.
Methods
Cells and reagents
LF1 human embryonic fibroblasts were a gift from John Sedivy (Brown University Providence, Rhode Island, USA) and were described previously54. Cells were maintained in Ham's F10 medium (PAN-Biotech, Germany) supplemented with 15% FBS (Life Technologies, Darmstadt, Germany).
H9 hES cells were obtained from Wisconsin International Stem Cell Bank (WiCell Research Institute Inc., Madison, WI). H9 hES and iPS cells were cultured on Matrigel (BD Biosciences, Germany) in mTeSR1 media (STEMCELL Technologies, Grenoble, France).
HP1β IgG rat monoclonal Ab (MAC353), Me(3)K9H3 polyclonal Ab were used as described55,56. Ki-67 monoclonal Ab (MIB-1) was from DAKO (Hamburg, Germany). Tra 1–60, Tra1–81, Nanog, Sox2, Pax6 Abs were from Stemgent (Miltenyi Biotec GmbH, Germany). HP1α (rabbit IgG), HP1γ (rabbit IgG), Histone H3 (mouse IgG), Oct4 (C30A3) Abs and Senescence β-Galactosidase Staining Kit were from Cell Signaling Technology (New England Biolabs, Germany). Sox 17, Brachyury Abs, BMP4, Activin A were from R&D (Wiesbaden-Nordenstadt, Germany). c-Myc and Lin28 polyclonal Abs were from Abcam (Cambridge, UK). Oct4 (C-10) and Klf4 Ab were from Santa Cruz Biotechnology (Heidelberg, Germany). GAPDH Ab (rabbit IgG) was from Sigma (Munich, Germany). Alkaline Phosphatase Staining Kit was from Cell Biolabs Inc. (Biocat, Heidelberg, Germany). SSEA4 Ab, ROCK kinase inhibitor Y27632 and Sodium Butyrate were from MerckMillipore (Schwalbach, Germany). bFGF and secondary fluorescent antibodies labeled with Alexa dyes were purchased from Life Technologies (Darmstadt, Germany).
DNA constructs
pPB-CAG.OSKML and pCMV-hyPBase constructs for iPS reprogramming46 were obtained from Wellcome Trust Sanger Institute (Cambridge, UK). dsRed-OSKML was engineered from pPB-CAG.OSKML by the introduction of an IRES - dsRed sequence derived from pMIdsRedExpress2. Specifically, IRES - dsRed was excised from pMIdsRedExpress2 with a double digest with XhoI and SalI enzymes. The resulting IRES – dsRed was sub-cloned into the unique XhoI site in pPB-CAG.OSKML, giving dsRed-OSKML.
EF1/HP1β-GFP construct was made by subcloning EF1 promoter into pEGFP-HP1β-C1 vector (described in38) at NheI/NdeI restriction sites after removal of the CMV promoter.
iPS cell reprogramming
LF1 human embryonic fibroblasts at low passage (p3) were seeded at density 0.65 × 105/well in Matrigel coated 6 well plates. The next day cells were transfected with OSKML and hyPBase vectors (2 μg each) using Lipofectamine LTX and Plus reagent (Life Technologies, Darmstadt, Germany). On Day 2 cells were put into mTeSR1 media which was replaced every day. In addition from Day 2 to Day 11 cells mTeSR1 media was supplemented with 0.25 mM Sodium Butyrate. iPS colonies were picked after Day 18 and transferred onto Matrigel coated 24 well plate for expansion.
For FRAP reprogramming studies LF1/HP1β-GFP fibroblasts were plated on 35 mm ibidi microscopy dishes (ibidi GmbH, Martinsried, Germany) at density 1 × 105/dish. The next day cells were transfected with with dsRed-OSKML and hyPBase vectors (2 μg each) using Lipofectamine LTX and Plus reagent. Transfection efficiency was 44 ± 11% (Figure S5b). Cells were maintained in LF1 media (Ham's F10 with 15% FBS) throughout the whole experiment. Before imaging cell media was replaced for Phenol Red free Ham's F10, 1% FBS and 20 mM HEPES.
Alkaline phosphatase staining
The assay was performed using Alkaline Phosphatase Staining Kit from Cell Biolabs Inc. (Biocat, Heidelberg, Germany) according to the manufacturer's recommendations.
SA β-galactosidase staining
The assay was performed using Senescence-associated β-Galactosidase Staining Kit from Cell Signaling Technology (New England Biolabs, Germany) according to the manufacturer's recommendations. On day 0 and day 9 we determined the % SA β-Galactosidase positive ± s.d. on a slide by counting the number SA β-Galactosidase positive in 5 fields each of the dimension of 2062.5 μm × 1562.5 μm (area 3.22 mm2). 100 cells were counted in each field.
Karyotyping
Metaphase chromosome spreads and karyotyping analysis was performed at the Intitute of Human Genetics University Clinic Freiburg.
Bisulfite sequencing analysis
Isolation of DNA from H9 hES, iPS and LF1 cells was performed using DNeasy kit (Qiagen, Germany). Samples were analysed for DNA methylation by pyrosequencing analysis at EpigenDx Inc. (Hopkinton, MA, USA). Methylation levels were analysed at 7 CpGs for Oct4 (−2234, −2140, +5, +31, +41, +59 from Oct4 transcriptional start site (TSS)) and at 2 CpGs for Nanog (−346 and −337 from Nanog TSS) genes.
Differentiation into germ layers
For ectoderm differentiation iPS cells were dissociated using Accutase and seeded onto Matrigel coated ibidi dishes (ibidi GmbH, Martinsried, Germany) in STEMdiff Neural Induction Medium (STEMCELL Technologies, Grenoble, France) supplemented with 10 μM ROCK inhibitor. The next day medium was replaced with STEMdiff Neural Induction Medium without ROCK inhibitor and subsequently was changed daily for 10 days. On Day 11 cells were stained by immunofluorescence for Pax6 ectoderm marker.
Differentiation into mesoderm layer was performed as following. Cells were passaged as clumps using Dispase onto Matrigel coated ibidi dish (ibidi GmbH, Martinsried, Germany) in mTeSR1 media. The next day cells were fed with Apel Medium (STEMCELL Technologies, Grenoble, France) supplemented with 0.5 ng/ml BMP4. On Day 2 Apel Medium was supplemented with 10 ng/ml BMP4, 10 ng/ml Activin A and 10 ng/ml bFGF. On Day 3 medium was replaced with Apel Medium containing 10 ng/BMP4, 10 ng/ml Activin A and 5 ng/ml bFGF. Immunofluorescence staining for mesoderm marker Brachyury was performed on day 4.
iPS cells were differentiated into endoderm germ layer using STEMdiff Definitive Endoderm kit (STEMCELL Technologies, Grenoble, France) according to the manufacturer's instructions. On day 5 of differentiation cells were fixed and stained for the Sox17 endoderm marker.
Transfection
Transient transfection of LF1 cells was done using Lipofectamine LTX and Plus reagent (Life Technologies, Darmstadt, Germany). Cells were plated on 35 mm ibidi dishes (ibidi GmbH, Martinsried, Germany) at a density of 1.5 × 105/dish. 24 h later, cells were transiently transfected with 1 μg of HP1β-GFP plasmid according to the manufacturer's recommendations. 48 h later cells were imaged at the confocal microscope in Phenol Red free Ham's F10, 1% FBS and 20 mM HEPES.
Transfections of H9 hES and iPS cells were done using Stemfect Enhanced hES transfection kit (Miltenyi Biotec GmbH, Germany). Cells were passaged as clumps using dispase onto Matrigel coated ibidi dishes (ibidi GmbH, Martinsried, Germany) in mTeSR1 media. 24 h later, cells were transiently transfected with 13 μg of HP1β-GFP plasmid according to the manufacturer's recommendations. 48 h later cells were imaged at the confocal microscope in Phenol Red free DMEM (Life Technologies).
LF1 cells expressing HP1β
Transfection of LF1 cells was done using Amaxa Nucleofection (Lonza, Germany). 1 × 106 cells were transfected with 1 μg of HP1β-GFP plasmid per 100 μl Ingenio electroporation solution (Mirius Bio LLC, WI, USA) in 0.2 cm cuvettes with U023 program settings according to the manufacturer's recommendations. After 72 h cells were cultured in the presence of G418 [400 μg/ml] (Life Technologies) until control un-transfected cells were completely eliminated. Subsequently HP1β-GFP cells were flow sorted for different fractions with low, medium and high levels of HP1β-GFP expression. Cells were maintained in culture in the presence of G418 [80 μg/ml].
Immunoblotting
Cell lysates were prepared in RIPA lysis buffer supplemented with protease inhibitors. Protein concentrations were determined by BCA assay (Pierce, Thermo Scientific). Cell lysates were separated by SDS-PAGE and transferred onto nitrocellulose membrane (Millipore). Proteins were stained with primary and secondary Abs and detected by ECL (Pierce, Thermo Scientific). Chemiluminescence was detected by Fusion Fx7 Imaging System (Peqlab, Germany).
Immunofluorescence staining
Cells were seeded on ibidi 35 mm tissue culture dishes (ibidi GmbH, Martinsried, Germany) at 1.5 × 105/well. Cells were fixed with 4% paraformaldehyde for 20 min at RT and permeabilized with 0.05% Triton X-100. After 1 h blocking staining with specific Abs was performed for 1 h RT or overnight at 4°C. After washing cells were incubated with fluorescent secondary Abs for 1 h RT. Nuclei were stained with Hoechst during the washing steps. Cells were mounted in Pro Long Gold mounting media (Life Technologies).
FRAP analysis and confocal microscopy
Confocal imaging was performed with Zeiss LSM 5 Live DUO inverted confocal microscope using LCI- Plan Neofluar 63×/1.3 DIC immersion corr. objective or LD LCI Plan Apochromat 25×/0.8 immersion corr. objective. FRAP experiments were performed using LCI- Plan Neofluar 63×/1.3 DIC immersion corr. objective and 488 laser line with a LF505 emission filter for GFP. A 1 μm2 circular ROI within nucleus in heterochromatic region was set up for bleaching, 4 μm2 ROI for bleaching correction, and additionally 4 μm2 control ROI outside of cell area was acquired for background subtraction. The laser intensity was set up at 75%. Ten images were taken before the bleach pulse (15 laser iterations with bleaching intensity output 100%) and 60 images were taken after the bleaching with image acquisition each 0.3 s with 5% laser transmission. All FRAP experiments were performed in temperature controlled CO2 supplied chamber. All data were background subtracted and normalized using Excel software and analyzed by non-linear regression analysis with GraphPad Prism Software (San Diego, USA). Recovery parameters were estimated from the fitted recovery curves.
Statistics
When indicated experimental data (minimum of three biological replicates) was analyzed for statistical significance employing the Extra sum-of-squares F-test with GraphPad Prism Software (San Diego, USA). Significance is described in figure legends. All western blots and confocal images are representatives of biological replicates.
Author Contributions
M.M., P.B.S. conceived the experiments. M.M. designed and performed the experiments, and analysed the data. M.M., P.B.S. discussed the data and wrote the paper.
Supplementary Material
Supplementary Information
Acknowledgments
This study was supported by the Excellence Initiative of the German Federal and State Governments (EXC 294). We would like to thank Dr.M. Leipoldt for karyotyping analysis.
References
- Gurdon J. B. The cloning of a frog. Development 140, 2446–8 (2013). [DOI] [PubMed] [Google Scholar]
- Wilmut I., Schnieke A. E., McWhir J., Kind A. J. & Campbell K. H. Viable offspring derived from fetal and adult mammalian cells. Nature 385, 810–3 (1997). [DOI] [PubMed] [Google Scholar]
- Mizutani E., Ono T., Li C., Maki-Suetsugu R. & Wakayama T. Propagation of senescent mice using nuclear transfer embryonic stem cell lines. Genesis 46, 478–83 (2008). [DOI] [PubMed] [Google Scholar]
- Takahashi K. & Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–76 (2006). [DOI] [PubMed] [Google Scholar]
- Lapasset L. et al. Rejuvenating senescent and centenarian human cells by reprogramming through the pluripotent state. Genes & Development 25, 2248–2253 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera C. M. & Ren B. Mapping human epigenomes. Cell 155, 39–55 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickmore W. A. & van Steensel B. Genome architecture: domain organization of interphase chromosomes. Cell 152, 1270–84 (2013). [DOI] [PubMed] [Google Scholar]
- Singh P. B. & Zacouto F. Nuclear reprogramming and epigenetic rejuvenation. J Biosci 35, 315–9 (2010). [DOI] [PubMed] [Google Scholar]
- Manukyan M. & Singh P. B. Epigenetic rejuvenation. Genes Cells 17, 337–43 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah P. P. et al. Lamin B1 depletion in senescent cells triggers large-scale changes in gene expression and the chromatin landscape. Genes Dev 27, 1787–99 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Otin C., Blasco M. A., Partridge L., Serrano M. & Kroemer G. The hallmarks of aging. Cell 153, 1194–217 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S. W. Heterochromatin. Science 151, 417–25 (1966). [DOI] [PubMed] [Google Scholar]
- Bulut-Karslioglu A. et al. A transcription factor-based mechanism for mouse heterochromatin formation. Nat Struct Mol Biol 19, 1023–30 (2012). [DOI] [PubMed] [Google Scholar]
- Kourmouli N. et al. Heterochromatin and tri-methylated lysine 20 of histone H4 in animals. J Cell Sci 117, 2491–501 (2004). [DOI] [PubMed] [Google Scholar]
- Schotta G. et al. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev 18, 1251–62 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T., Tsujimoto N. & Li E. The PWWP domain of Dnmt3a and Dnmt3b is required for directing DNA methylation to the major satellite repeats at pericentric heterochromatin. Mol Cell Biol 24, 9048–58 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallrath L. L. & Elgin S. C. Position effect variegation in Drosophila is associated with an altered chromatin structure. Genes Dev 9, 1263–77 (1995). [DOI] [PubMed] [Google Scholar]
- Casanova M. et al. Heterochromatin Reorganization during Early Mouse Development Requires a Single-Stranded Noncoding Transcript. Cell Rep 4, 1156–67 (2013). [DOI] [PubMed] [Google Scholar]
- Grewal S. I. & Elgin S. C. Transcription and RNA interference in the formation of heterochromatin. Nature 447, 399–406 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billur M., Bartunik H. D. & Singh P. B. The essential function of HP1 beta: a case of the tail wagging the dog? Trends Biochem Sci 35, 115–23 (2010). [DOI] [PubMed] [Google Scholar]
- Grewal S. I. & Elgin S. C. Heterochromatin: new possibilities for the inheritance of structure. Curr Opin Genet Dev 12, 178–87 (2002). [DOI] [PubMed] [Google Scholar]
- Maison C. & Almouzni G. HP1 and the dynamics of heterochromatin maintenance. Nat Rev Mol Cell Biol 5, 296–304 (2004). [DOI] [PubMed] [Google Scholar]
- Jones D. O., Cowell I. G. & Singh P. B. Mammalian chromodomain proteins: their role in genome organisation and expression. Bioessays 22, 124–37 (2000). [DOI] [PubMed] [Google Scholar]
- Singh P. B. HP1 proteins--what is the essential interaction? Genetika 46, 1424–9 (2010). [PubMed] [Google Scholar]
- de Lange T. How shelterin solves the telomere end-protection problem. Cold Spring Harb Symp Quant Biol 75, 167–77 (2010). [DOI] [PubMed] [Google Scholar]
- Marion R. M. & Blasco M. A. Telomere rejuvenation during nuclear reprogramming. Curr Opin Genet Dev 20, 190–6 (2010). [DOI] [PubMed] [Google Scholar]
- Narita M. et al. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell 113, 703–16 (2003). [DOI] [PubMed] [Google Scholar]
- Sadaie M. et al. Redistribution of the Lamin B1 genomic binding profile affects rearrangement of heterochromatic domains and SAHF formation during senescence. Genes Dev 27, 1800–8 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgoulis V. G. & Halazonetis T. D. Oncogene-induced senescence: the bright and dark side of the response. Curr Opin Cell Biol 22, 816–27 (2010). [DOI] [PubMed] [Google Scholar]
- Braumuller H. et al. T-helper-1-cell cytokines drive cancer into senescence. Nature 494, 361–5 (2013). [DOI] [PubMed] [Google Scholar]
- Sikora E., Arendt T., Bennett M. & Narita M. Impact of cellular senescence signature on ageing research. Ageing Res Rev 10, 146–52 (2011). [DOI] [PubMed] [Google Scholar]
- Kwon S. H. & Workman J. L. The changing faces of HP1: From heterochromatin formation and gene silencing to euchromatic gene expression: HP1 acts as a positive regulator of transcription. Bioessays 33, 280–9 (2011). [DOI] [PubMed] [Google Scholar]
- Feldman N. et al. G9a-mediated irreversible epigenetic inactivation of Oct-3/4 during early embryogenesis. Nat Cell Biol 8, 188–94 (2006). [DOI] [PubMed] [Google Scholar]
- Sridharan R. et al. Proteomic and genomic approaches reveal critical functions of H3K9 methylation and heterochromatin protein-1gamma in reprogramming to pluripotency. Nat Cell Biol 15, 872–82 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers I. et al. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell 113, 643–55 (2003). [DOI] [PubMed] [Google Scholar]
- Mitsui K. et al. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 113, 631–42 (2003). [DOI] [PubMed] [Google Scholar]
- Dundr M. & Misteli T. Measuring dynamics of nuclear proteins by photobleaching. Curr Protoc Cell Biol 18, 13.5.1–13.5.18 (2003). [DOI] [PubMed] [Google Scholar]
- Cheutin T. et al. Maintenance of stable heterochromatin domains by dynamic HP1 binding. Science 299, 721–5 (2003). [DOI] [PubMed] [Google Scholar]
- Festenstein R. et al. Modulation of heterochromatin protein 1 dynamics in primary Mammalian cells. Science 299, 719–21 (2003). [DOI] [PubMed] [Google Scholar]
- Cheutin T., Gorski S. A., May K. M., Singh P. B. & Misteli T. In vivo dynamics of Swi6 in yeast: evidence for a stochastic model of heterochromatin. Mol Cell Biol 24, 3157–67 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmiedeberg L., Weisshart K., Diekmann S., Meyer Zu Hoerste G. & Hemmerich P. High- and low-mobility populations of HP1 in heterochromatin of mammalian cells. Mol Biol Cell 15, 2819–33 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dialynas G. K. et al. Methylation-independent binding to histone H3 and cell cycle-dependent incorporation of HP1beta into heterochromatin. J Biol Chem 281, 14350–60 (2006). [DOI] [PubMed] [Google Scholar]
- Dialynas G. K. et al. Plasticity of HP1 proteins in mammalian cells. J Cell Sci 120, 3415–24 (2007). [DOI] [PubMed] [Google Scholar]
- Muller K. P. et al. Multiscale analysis of dynamics and interactions of heterochromatin protein 1 by fluorescence fluctuation microscopy. Biophys J 97, 2876–85 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meshorer E. et al. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev Cell 10, 105–16 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusa K., Rad R., Takeda J. & Bradley A. Generation of transgene-free induced pluripotent mouse stem cells by the piggyBac transposon. Nat Methods 6, 363–9 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman P. D., Meek R. L. & Daniel C. W. Aging of human fibroblasts in vitro. Correlations between DNA synthetic ability and cell size. Exp Cell Res 93, 184–90 (1975). [DOI] [PubMed] [Google Scholar]
- Suhr S. T. et al. Mitochondrial rejuvenation after induced pluripotency. PLoS One 5, e14095 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A. & Nagy K. The mysteries of induced pluripotency: where will they lead? Nat Methods 7, 22–4 (2010). [DOI] [PubMed] [Google Scholar]
- Efe J. A. et al. Conversion of mouse fibroblasts into cardiomyocytes using a direct reprogramming strategy. Nat Cell Biol 13, 215–222 (2011). [DOI] [PubMed] [Google Scholar]
- Buehr M. et al. Capture of authentic embryonic stem cells from rat blastocysts. Cell 135, 1287–98 (2008). [DOI] [PubMed] [Google Scholar]
- Tachibana M. et al. Human embryonic stem cells derived by somatic cell nuclear transfer. Cell 153, 1228–38 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka S. Strategies and new developments in the generation of patient-specific pluripotent stem cells. Cell Stem Cell 1, 39–49 (2007). [DOI] [PubMed] [Google Scholar]
- Brown J. P., Wei W. & Sedivy J. M. Bypass of senescence after disruption of p21CIP1/WAF1 gene in normal diploid human fibroblasts. Science 277, 831–4 (1997). [DOI] [PubMed] [Google Scholar]
- Wreggett K. A. et al. A mammalian homologue of Drosophila heterochromatin protein 1 (HP1) is a component of constitutive heterochromatin. Cytogenet Cell Genet 66, 99–103 (1994). [DOI] [PubMed] [Google Scholar]
- Cowell I. G. et al. Heterochromatin, HP1 and methylation at lysine 9 of histone H3 in animals. Chromosoma 111, 22–36 (2002). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information