Abstract
The presence of SXT/R391-related integrating conjugative elements (ICEs) in V. cholerae O1 and non-O1/non-O139 isolated from clinical and environmental samples in Haiti in 2010 was studied. The main finding of this work was the identification of the novel ICEVchHai2 among closely related V. cholerae non-O1/non-O139 clinical strains. The mosaic structure of this element confirms the role of ICEs as efficient recombination systems whereby new genetic material can be acquired and exchanged, according V. cholerae strains new accessory functions.
Keywords: Vibrio cholera, Non-O1/non-O139, Haiti, Integrative conjugative elements
Vibrio cholerae is an autochthonous inhabitant of riverine and estuarine environments. It is a facultative pathogen of humans, being the causative agent of cholera, a disease endemic in many developing countries (Colwell, 1996). Since October 2010, when the first case of cholera was diagnosed in Haiti, cholera remains a major health threat, with more than 600,000 cases to date (Ministère de la Santé Publique et de la Population, 2013).
Integrative conjugative elements (ICEs) of the SXT/R391 family are recognized for their role in bacterial genome plasticity and as vectors of antibiotic resistance and alternative metabolic pathways (Seth-Smith and Croucher, 2009). They share a conserved genetic scaffold of 52 genes encoding their own conjugation, integration, excision, and regulatory machinery (Wozniak et al., 2009). The backbone is scattered with accessory sequences located in conserved regions (hotspots) and exhibiting significant variability in genetic content, including resistance to antibiotics and heavy metals, toxin/antitoxin systems, restriction/modification systems, motility and biofilm formation (Bordeleau et al., 2010; Wozniak et al., 2009).
SXT/R391 ICEs are ubiquitous in clinical V. cholerae O1 and O139, as well as in other vibrios (Wozniak et al., 2009), and their acquisition has been recognized as a distinctive trait in the ongoing evolution of pandemic V. cholerae strains (Mutreja et al., 2011). Our knowledge of the distribution of SXT/R391 ICEs in environmental isolates is still very limited. To date, SXTR391 ICEs were detected in few Gammaproteobacteria and Vibrio spp. from aquaculture and marine environments (Badhai et al., 2013; Osorio et al., 2008; Rodriguez-Blanco et al., 2012) and in V. cholerae non-O1/non-O139 environmental isolates from Mexico and Mozambique (Taviani et al., 2008; Wozniak et al., 2009).
In this work, we report ICE analysis of V. cholerae O1 and non-O1/non-O139 isolated from clinical and environmental samples collected in the early phase of the cholera outbreak in Haiti in 2010 (Hasan et al., 2012). We analyzed 57 sequenced strains that had been isolated in 11 arrondissements of Haiti: 30 clinical V. cholerae O1 and 27 non-O1/non-O139 strains, the latter of clinical and environmental origin. ICE sequence and genetic organization were compared with other SXT/R391 ICEs in GenBank using tools previously described (Taviani et al., 2012).
Based on occurrence and genetic structure of their SXT/R391 ICEs, the Haitian strains can be divided into three clusters. All V. cholerae O1 clinical strains contain an ICE that is >99% similar to ICEVchInd5 (a.k.a. ICEVchHai1), as previously reported for V. cholerae O1 strains from the same epidemic (Ceccarelli et al., 2011c; Sjolund-Karlsson et al., 2011). This is not surprising considering that ICEVchInd5 and siblings are prevalent in epidemic V. cholerae O1 altered El Tor variants worldwide (Ceccarelli et al., 2011b; Ceccarelli et al., 2011a).
The non-O1/non-O139 strains, in comparison, showed two profiles. Twelve strains, both environmental and clinical, were devoid of SXT/R391 ICEs. Those clinical strains were isolated from the arrondissements of Croix-des-Bouquets, Delmas and Port-au-Prince in the Ouest department, and formed a closely related cluster (Hasan et al., 2012), whereas environmental isolates were from gray water, latrines and hospital waste in Cange, in the Central Plateau of Haiti.
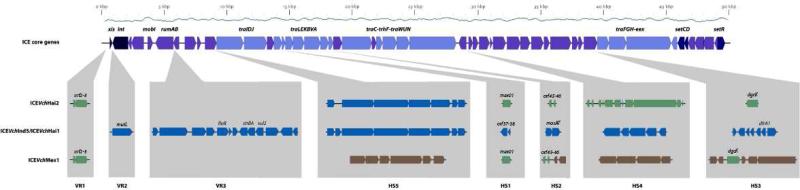
The main finding of this work is identification of the novel ICEVchHai2 in 15 non-O1/non-O139 V. cholerae clinical strains isolated both in the Artibonite and Ouest departments. The 15 ICEs share almost 100% similarity at the nucleotide level and are separated only by one single nucleotide polymorphism (SNP) among the whole sequence. As noted for all SXT-related ICEs, the genetic structure of ICEVchHai2 (GenBank Accession No. AJRO01000008, nt. 45958-128752, 82.795 bp) is a genetic mosaic shaped by inter-ICE recombination (Garriss et al., 2009; Wozniak et al., 2009). We identified gene sequences from other ICEs as well as unique genetic clusters (Figure 1). Like all environmental ICEs described to date in V. cholerae (Taviani et al., 2008; Wozniak et al., 2009), ICEVchHai2 lacks the antibiotic resistance cluster typically inserted in variable region 3 (VR3). Hotspot 1 and variable region 1 (VR1) have the same molecular rearrangement as ICEVchMex1. Among other genes, hotspot 3 of ICEVchMex1 encodes dgcK and dgcL involved in the c-di-GMP signaling pathways (Bordeleau et al., 2010). ICEVchHai2 contains only dgcK with an inverted orientation compared to ICEVchMex1, and the rest of hotspot 3 is deleted. Also, hotspot 2 appears to be the result of a deletion event. It contains only two genes (orf45, orf46) found in both R391 and ICEVchMex1, but the rest of the hotspot is missing. Hotspot 4, located between genes traN and s063, is characterized by a unique 9.4 kb region that includes nine ORFs. Bioinformatics analysis of its genetic content revealed three hypothetical proteins and two transposase genes. The remaining five genes are associated with known functions involved in DNA-mediated transposition, transformation or recombination, as well as deoxyribonuclease activity and nucleic acid binding (Table 1). Finally, hotspot 5 revealed 96% homology with the same region in ICEVchInd5, a 14.8-kb hypothetical operon of unknown function (Ceccarelli et al., 2011b).
Fig. 1. Structural comparison between ICEVchInd5 (ICEVchHai1), ICEVchHai2 and ICEVchMex1.
The upper map represents the set of core genes common to all SXT/R391 ICEs. Highlighted are genes involved in site-specific excision and integration (xis, int, mobI), error prone DNA repair (rumAB), entry exclusion (eex), ICE regulation (setCDR) and conjugative transfer (tra genes). The bottom map represents the specific regions of ICEVchInd5 (ICEVchHai1), ICEVchHai2, and ICEVchMex1 inserted the variable regions and hotspots. For a detailed description of hotspot 4 in ICEVchHai2 see Table 1.
Table 1. ORF content of hotspot 4 in ICEVchHai2.
Annotated genes refer to ICEVchHai2 in V. cholerae HC-1A2 (GenBank Accession No. AJRO01000008.1). Blast2Go was used to perform functional annotation with Gene Ontology (GO) and InterProScan.
| Gene | Putative role | Gene Length (nt) | GO terms |
|---|---|---|---|
| VCHC1A2_1617 | Transposase | 327 | Transposition, DNA-mediated |
| VCHC1A2_1618 | Polyribonucleotide nucleotidyltransferase | 141 | Transposition, DNA-mediated |
| VCHC1A2_1619 | trp Repressor family protein | 951 | Nucleic acid binding |
| VCHC1A2_1620 | Transposase dde domain protein | 759 | Transposition, DNA-mediated |
| VCHC1A2_1621 | Hypothetical protein VCHC1A2_1621 | 126 | - |
| VCHC1A2_1622 | smf family protein | 930 | DNA-mediated transformation |
| VCHC1A2_1623 | ATP-dependent DNA helicase | 4677 | DNA recombination |
| VCHC1A2_1624 | Hypothetical protein VCHC59A1_0788 | 489 | - |
| VCHC1A2_1625 | Hypothetical protein VCHC1A2_1625 | 117 | - |
| VCHC1A2_1626 | Endonuclease-1 | 678 | Deoxyribonuclease I activity |
The results of our analysis show that Haitian V. cholerae strains contain two SXT/R391 ICEs displaying different genetic organization: ICEVchInd5 and ICEVchHai2. ICEVchHai2 is described here for the first time, appears to circulate only among closely related V. cholerae non-O1/non-O139 clinical strains (Hasan et al., 2012) and shares several hotspots with ICEVchMex1, isolated in non-O1/non-O139 V. cholerae from sewage in Mexico (Wozniak et al., 2009). Yet a general phenomenon of recombination appears to have occurred in ICEVchHai2 compared with ICEVchMex1. The Haitian ICE shows deletions in homologous hotspot regions and bears new genes involved in recombination and nucleic acid binding and processing. These findings indicate that the ICEVchHai2 hotspots have been deleted or rearranged without compromising the integrity of core genes required for ICE mobility and ability to acquire DNA. Indeed, ICEVchHai2 may have a function yet to be determined, but confirms the role of SXT/R391 ICEs as efficient recombination systems whereby new genetic material can be acquired and exchanged, according new ICE variants with different accessory functions.
Acknowledgments
The National Institute of Health Grant No. 2RO1A1039129-11A2, the Department of Health and Human Services under contract number HHSN2722009000 and the National Science Foundation under Grant No. 1034836 funded this research. M.S. is supported by a fellowship from Cenci Bolognetti – Institut Pasteur Foundation, Italy. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Badhai J, Kumari P, Krishnan P, Ramamurthy T, Das SK. Presence of SXT integrating conjugative element in marine bacteria isolated from the mucus of the coral Fungia echinata from Andaman Sea. FEMS Microbiol. Lett. 2013;338:118–123. doi: 10.1111/1574-6968.12033. [DOI] [PubMed] [Google Scholar]
- Bordeleau E, Brouillette E, Robichaud N, Burrus V. Beyond antibiotic resistance: integrating conjugative elements of the SXT/R391 family that encode novel diguanylate cyclases participate to c-di-GMP signalling in Vibrio cholerae. Environ. Microbiol. 2010;12:510–523. doi: 10.1111/j.1462-2920.2009.02094.x. [DOI] [PubMed] [Google Scholar]
- Ceccarelli D, Spagnoletti M, Bacciu D, Cappuccinelli P, Colombo MM. New V. cholerae atypical El Tor variant emerged during the 2006 epidemic outbreak in Angola. BMC Microbiol. 2011a;11:130. doi: 10.1186/1471-2180-11-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccarelli D, Spagnoletti M, Bacciu D, Danin-Poleg Y, Mendiratta D, Kashi Y, Cappuccinelli P, Burrus V, et al. ICEVchInd5 is prevalent in epidemic Vibrio cholerae O1 El Tor strains isolated in India. Int. J. Med. Microbiol. 2011b;301:318–324. doi: 10.1016/j.ijmm.2010.11.005. [DOI] [PubMed] [Google Scholar]
- Ceccarelli D, Spagnoletti M, Cappuccinelli P, Burrus V, Colombo MM. Origin of Vibrio cholerae in Haiti. Lancet Infect. Dis. 2011c;11:260. doi: 10.1016/S1473-3099(11)70078-0. [DOI] [PubMed] [Google Scholar]
- Colwell RR. Global climate and infectious disease: the cholera paradigm. Science. 1996;274:2025–2031. doi: 10.1126/science.274.5295.2025. [DOI] [PubMed] [Google Scholar]
- Garriss G, Waldor MK, Burrus V. Mobile antibiotic resistance encoding elements promote their own diversity. PLoS Genet. 2009;5:e1000775. doi: 10.1371/journal.pgen.1000775. doi:1000710.1001371/journal.pgen.1000775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan NA, Choi SY, Eppinger M, Clark PW, Chen A, Alam M, Haley BJ, Taviani E, et al. Genomic diversity of 2010 Haitian cholera outbreak strains. Proc. Natl. Acad. Sci. U. S. A. 2012;109:E2010–E2017. doi: 10.1073/pnas.1207359109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ministère de la Santé Publique et de la Population, R.d.H. Rapports journaliers du MSPP sur l'évolution du choléra en Haiti. Rapport du 30 juin 2013, in. 2013:1–5. http://www.mspp.gouv.ht.
- Mutreja A, Kim DW, Thomson NR, Connor TR, Lee JH, Kariuki S, Croucher NJ, Choi SY, et al. Evidence for several waves of global transmission in the seventh cholera pandemic. Nature. 2011;477:462–465. doi: 10.1038/nature10392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorio CR, Marrero J, Wozniak RAF, Lemos ML, Burrus V, Waldor MK. Genomic and functional analysis of ICEPdaSpa1, a fish-pathogen-derived SXT-related integrating conjugative element that can mobilize a virulence plasmid. J. Bacteriol. 2008;190:3353–3361. doi: 10.1128/JB.00109-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Blanco A, Lemos ML, Osorio CR. Integrating Conjugative Elements (ICEs) as vectors of antibiotic, mercury and quaternary ammonium compounds resistance in marine aquaculture environments. Antimicrob. Agents Chemother. 2012;56:2619–2626. doi: 10.1128/AAC.05997-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth-Smith H, Croucher NJ. Genome watch: breaking the ICE. Nat. Rev. Microbiol. 2009;7:328–329. doi: 10.1038/nrmicro2137. [DOI] [PubMed] [Google Scholar]
- Sjolund-Karlsson M, Reimer A, Folster JP, Walker M, Dahourou GA, Batra DG, Martin I, Joyce K, et al. Drug-resistance mechanisms in Vibrio cholerae O1 outbreak strain, Haiti, 2010. Emerg. Infect. Dis. 2011;17:2151–2154. doi: 10.3201/eid1711.110720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taviani E, Ceccarelli D, Lazaro N, Bani S, Cappuccinelli P, Colwell RR, Colombo MM. Environmental Vibrio spp., isolated in Mozambique, contain a polymorphic group of integrative conjugative elements and class 1 integrons. FEMS Microbiol. Ecol. 2008;64:45–54. doi: 10.1111/j.1574-6941.2008.00455.x. [DOI] [PubMed] [Google Scholar]
- Taviani E, Spagnoletti M, Ceccarelli D, Haley BJ, Hasan NA, Chen A, Colombo MM, Huq A, et al. Genomic analysis of ICEVchBan8: an atypical genetic element in Vibrio cholerae. FEBS Lett. 2012;586:1617–1621. doi: 10.1016/j.febslet.2012.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak RAF, Fouts DE, Spagnoletti M, Colombo MM, Ceccarelli D, Garriss G, Déry C, Burrus V, et al. Comparative ICE genomics: insights into the evolution of the SXT/R391 family of ICEs. PLoS Genet. 2009;5:e1000786. doi: 10.1371/journal.pgen.1000786. doi: 1000710.1001371/journal.pgen.1000786. [DOI] [PMC free article] [PubMed] [Google Scholar]