Abstract
During human pregnancy, a subset of placental cytotrophoblasts (CTBs) differentiates into cells that aggressively invade the uterus and its vasculature, anchoring the progeny and rerouting maternal blood to the placenta. In preeclampsia (PE), CTB invasion is limited, reducing placental perfusion and/or creating intermittent flow. This syndrome, affecting 4%–8% of pregnancies, entails maternal vascular alterations (e.g., high blood pressure, proteinuria, and edema) and, in some patients, fetal growth restriction. The only cure is removal of the faulty placenta, i.e., delivery. Previously, we showed that defective CTB differentiation contributes to the placental component of PE, but the causes were unknown. Here, we cultured CTBs isolated from PE and control placentas for 48 hours, enabling differentiation and invasion. In various severe forms of PE, transcriptomics revealed common aberrations in CTB gene expression immediately after isolation, including upregulation of SEMA3B, which resolved in culture. The addition of SEMA3B to normal CTBs inhibited invasion and recreated aspects of the PE phenotype. Additionally, SEMA3B downregulated VEGF signaling through the PI3K/AKT and GSK3 pathways, effects that were observed in PE CTBs. We propose that, in severe PE, the in vivo environment dysregulates CTB gene expression; the autocrine actions of the upregulated molecules (including SEMA3B) impair CTB differentiation, invasion and signaling; and patient-specific factors determine the signs.
Introduction
In humans, villous cytotrophoblasts (vCTBs) of the placenta differentiate, establishing the maternal-fetal interface (ref. 1 and Supplemental Figure 1A; supplemental material available online with this article; doi: 10.1172/JCI66966DS1). In floating villi, vCTBs fuse into syncytiotrophoblasts (STBs) with functions that include transport and hormone production. In anchoring villi, vCTBs acquire tumor-like properties that enable invasion of the decidua and the adjacent third of the myometrium (interstitial invasion). They also breach uterine spiral arterioles, transiently replacing much of the maternal endothelial lining and intercalating within the muscular walls (endovascular invasion) (2). Consequently, high-resistance spiral arterioles are transformed into low-resistance, high-capacitance vessels that divert uterine blood flow to the floating villi. In contrast, CTBs breach and line only the termini of veins. At a molecular level, CTB invasion is accompanied by dramatic phenotypic changes in which these ectodermal derivatives mimic many aspects of the vascular cell surface, e.g., an adhesion molecule repertoire that includes VE-cadherin (3), Ephs/ephrins that confer an arterial identity (4), and Notch family members that play important roles in vessel functions (5). In parallel, the cells modulate the expression of a wide range of angiogenic/vasculogenic molecules, including VEGF family members (6).
Given this organ’s complexity, the explosive nature by which it develops, and its many critical functions, it is not surprising that some of the most clinically significant pregnancy complications are associated with placental anomalies. Preeclampsia (PE), a syndrome that adversely affects the mother (by altering vascular function) and the fetus (by intrauterine growth restriction [IUGR]), is a prime example. A 2-stage theory of PE pathogenesis has been proposed — abnormal placentation followed by maternal responses that eventually lead to the clinical presentation (7). In support of this concept, color Doppler ultrasound suggests that in PE deficient endovascular invasion precedes the signs (8). This finding, which is generally associated with the severe forms of this syndrome that occur during the late second trimester/early third trimester period, is consistent with the major placental pathologies, which include shallow uterine invasion (Supplemental Figure 1B and ref. 9). Anchoring villi, in particular invasive CTBs, are most consistently impacted. Interstitial invasion is often shallow, and endovascular invasion is incomplete with fewer spiral arterioles modified in toto (9–11). At a molecular level, many aspects of the unusual phenotypic switch to a vascular-type cell that normally accompanies CTB invasion fail in PE (5, 6, 12), with attendant increases in apoptosis (13).
What is known about the molecular underpinnings of the 2 stages of PE? A great deal of attention has been focused on the second stage — factors that play a role in the maternal signs. Among other molecules, sFlt (14), endoglin (15), and adrenomedullin (16) have been implicated. In contrast, the mechanisms that precipitate the first stage — defective CTB differentiation/invasion — are largely unknown. We reasoned that PE-associated changes in CTB gene expression would give us important clues about the causes of this syndrome. Thus, we isolated CTBs from the placentas of women who were diagnosed with different forms of severe PE (sPE) with or without IUGR, including superimposed hypertension and HELLP syndrome (HELLP is a syndrome characterized by hemolysis, elevated liver enzymes, and low platelet count). Global gene expression profiling immediately after isolation revealed a common set of upregulated mRNAs despite the different diagnoses. Surprisingly, they were downregulated to control levels over 48 hours. The differentially expressed genes included molecules previously identified as dysregulated in PE and many that were not known to be involved in this pregnancy complication. We hypothesized that their autocrine actions could contribute to the CTB phenotypic alterations that are the hallmark of PE. We proved this theory using the molecule SEMA3B and the VEGF signaling pathways it engages. Together, these results suggested that the CTB defects in sPE, which can be attributed to the in vivo environment, are reversible, evidence that therapeutic interventions may be possible. We also showed that a common signature of misexpressed genes that spans all the sPE subtypes we studied results in the diverse maternal signs that define these variants.
Results
Ex vivo normalization of sPE CTB gene expression.
We used global transcriptional profiling to explore mRNA changes that underlie CTB defects in various forms of sPE. Villous CTBs were isolated from sPE placentas and placentas of preterm labor patients with no signs of infection (nPTL), which served as gestation-matched controls. Our previous work showed that CTB invasion is essentially normal in the latter group (17), and we confirmed this finding for the samples in this study. However, we cannot eliminate the possibility of other placental pathologies relative to normal controls, which are not possible to collect at the relevant gestational ages. The newborns did not differ by birth weight or gestational age at delivery (Supplemental Table 1). The patients with sPE and nPTL had comparable BMIs and ages, but women with sPE had higher systolic/diastolic blood pressures and proteinuria. To better understand the CTB phenotype in the context of sPE variants, we included patients diagnosed with the most clinically significant forms of this condition that necessitated preterm delivery (18, 19): women with sPE with or without IUGR, superimposed hypertension, or HELLP syndrome with or without IUGR. We profiled the gene expression patterns of the case and control groups before plating (0 hour) and at 12, 24, and 48 hours after culture using the Affymetrix HG-U133Plus 2.0 GeneChip platform. With LIMMA, we identified numerous genes that were differentially expressed — a common CTB fingerprint — in nearly all the sPE samples at 1 or more time points (≥2 fold; P ≤ 0.05; Figure 1). Surprisingly, after 48 hours in culture most were expressed at control levels. Unsupervised hierarchical clustering showed that the gene expression patterns of sPE and nPTL samples, which segregated into their respective groups at 0 hours, merged at 48 hours (data not shown). The initially upregulated molecules included factors previously associated with PE (e.g., growth hormone 2, corticotropin-releasing hormone, inhibin A, KISS-1, ADAM-12) (20–23), a transcriptional regulator (HOPX), and an angiogenic factor (SEMA3B). In addition to growth hormone 2, other placenta-specific products, including PLAC1 and PLAC4, and 7 pregnancy-specific glycoprotein (PSG) family members, were also upregulated as was an enzyme involved in fat metabolism (oleoyl-ACP hydrolase). Many fewer genes were downregulated. A subset of the results was confirmed by qRT-PCR (Supplemental Figure 2). The fact that a common set of dysregulated genes was associated with a broad spectrum of the maternal signs suggested that a complex interplay between abnormal placentation and patient-specific factors ultimately determined the clinical features. The finding that most sPE-related aberrations in CTB gene expression normalized when the cells were cultured for 2 days supported the theory that an unfavorable in vivo environment contributed to placental defects in this syndrome. Next, we asked whether any of the dysregulated genes were autocrine regulators of the CTB phenotype that is the hallmark of PE.
Figure 1. sPE-associated aberrations in CTB gene expression returned to control values after 48 hours of culture.
RNA was analyzed immediately after the cells were isolated (0 hour) and after 12, 24, and 48 hours in culture. The relative gene expression levels for CTBs isolated from placentas of patients who delivered due to nPTL (n = 5) or sPE (n = 5) are shown as a heat map, ranging from high (red) to low (blue). The sPE CTBs were from the following cases (tiled from left to right): (a) HELLP syndrome and IUGR; (b) sPE; (c) sPE and IUGR; (d) superimposed sPE; and (e) HELLP syndrome. One sample of nPTL CTBs collected at 48 hours was omitted for technical reasons. The fold changes for each time point (sPE vs. nPTL) are shown on the right. ns, no significant difference (LIMMA); t, no significant difference in expression (sPE vs. nPTL) by 48 hours (maSigPro).
Upregulated trophoblast expression of SEMA3B in sPE.
In PE, trophoblast expression of angiogenic factors is dysregulated. This phenomenon plays a central role in restricting CTB invasion (6) and in the etiology of the maternal signs, including elevated blood pressure and proteinuria (14, 15). In this context, we addressed the functions of SEMA3B. SEMA3 family members play important roles in neuronal wiring (24), and SEMA3B is an angiogenesis inhibitor and tumor suppressor. As a first step, we profiled SEMA3B mRNA expression in a variety of human cells and organs. Placenta gave the strongest signal (Figure 2A). Northern blot analyses of mRNA from control (first, second, and third trimester) and experimental placentas from patients with sPE showed that the abundance of SEMA3B mRNA increased as a function of gestational age and was highest in sPE samples (Figure 2B). The 2 bands likely reflect alternative splicing. In situ hybridization of placental chorionic villi demonstrated that SEMA3B mRNA expression, which was limited to trophoblasts, was lower in normal second trimester and nPTL samples as compared with sPE chorionic villi (Figure 2C). Immunoblot analyses of CTB lysates showed that expression of SEMA3B was either undetectable or low during the second and third trimesters of normal pregnancy and in cells isolated from nPTL placentas (Figure 2D). In contrast, higher levels of SEMA3B were detected in sPE CTBs immediately after isolation. For these experiments, SEMA3A-Fc served as a negative control and recombinant SEMA3B protein served as a positive control for antibody specificity. Immunolocalization showed that trophoblasts of chorionic villi from control nPTL placentas had much lower anti-SEMA3B immunoreactivity as compared with samples of similar gestational ages from sPE placentas (Figure 2E). Within the basal plate, extravillous CTBs in the setting of sPE also exhibited stronger staining for SEMA3B as compared with the nPTL samples (Figure 2E).
Figure 2. SEMA3B expression was high in the placenta and upregulated in sPE.
(A) Binding of a 32P-SEMA3B probe to a multiple tissue expression array revealed high placental expression (coordinate B8). (B) Northern hybridization of polyA+ RNA extracted from chorionic villi and pooled from 3 placentas showed that SEMA3B expression increased over gestation and was highest in sPE (n = 3 replicates). (C) In situ hybridization (3 placentas per group) confirmed enhanced SEMA3B mRNA expression in the STB layer of the chorionic villi in sPE (25 weeks) as compared with normal pregnancy (23 weeks) and nPTL (34 weeks). (D) Immunoblotting of CTB lysates (15 μg per lane) showed that SEMA3B protein expression was low to undetectable in control cells from normal placentas (15–39 weeks). In all cases, expression was higher in sPE (26–33 weeks) as compared with nPTL (30, 33 weeks). A protein of the expected Mr was detected in COS-1 cells transfected with SEMA3B but not in those transfected with SEMA3A-Fc. Vertical lines denote noncontiguous lanes from the same gel. The relative intensity of the bands quantified by densitometry is also shown. The values for each sample type were averaged and expressed relative to the α-actin loading controls. The entire experiment was repeated twice. (E) Staining tissue sections with anti-SEMA3B showed a sPE-associated upregulation of immunoreactivity associated with the trophoblast components of chorionic villi and among extravillous CTBs within the basal plate (n = 5 per group). Trophoblasts were identified by staining adjacent tissue sections with anti–cytokeratin-8/18 (data not shown). Scale bars: 100 μm (C and E). NB, Northern blot; GA, gestational age; RP, recombinant protein.
Neuropilin expression and SEMA3B actions.
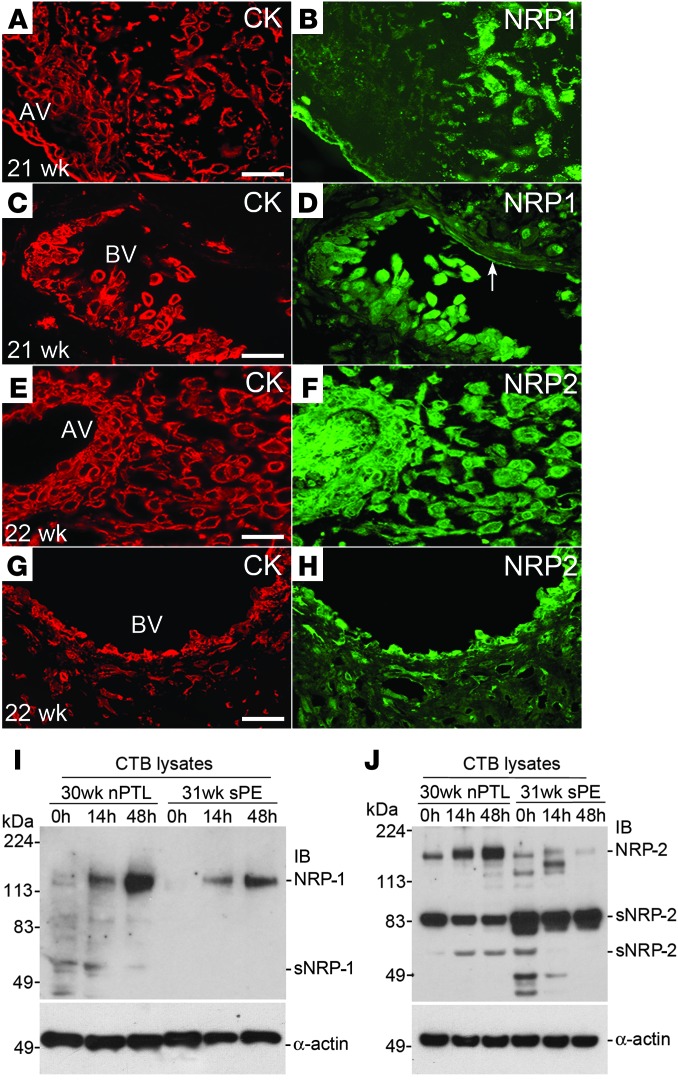
Next, we assessed the expression of the SEMA3B receptors, neuropilin-1 (NRP-1) and NRP-2 (24), in tissue sections of the maternal-fetal interface. Costaining with anti–cytokeratin-8/18 identified trophoblasts (Figure 3, A, C, E, and G). Immunolocalization analyses of normal second trimester samples showed that NRP-1, which was expressed by villous trophoblasts, was upregulated as the CTBs invaded the uterine wall (Figure 3B). Strong staining was also detected in association with endovascular CTBs and the endothelial lining of uterine vessels (Figure 3D, arrow). NRP-2 immunoreactivity was associated with villous trophoblasts and invasive CTBs as well as the villous stroma (Figure 3F). In the uterine wall, NRP-2 expression was strongly upregulated on endovascular CTBs (Figure 3H) and a subset of endothelial cells (data not shown).
Figure 3. NRP-1 and NRP-2 (protein) expression at the maternal-fetal interface in normal pregnancy and in sPE.
Tissue sections were double stained with anti–cytokeratin-8/18 (CK), which reacts with all trophoblast subpopulations, and anti–NRP-1 or NRP-2. (A and B) NRP-1 expression was detected in association with villous trophoblasts. Within the uterine wall, immunoreactivity associated with invasive CTBs was upregulated as the cells moved from the surface to the deeper regions. (C and D) Endovascular CTBs that lined a maternal blood vessel (BV) were also stained. (E–H) Anti–NRP-2 reacted with trophoblast and nontrophoblast cells in anchoring villi (AV) as well as interstitial and endovascular CTBs. Essentially the same staining patterns, but with weaker intensity, were observed in sPE (data not shown). CTBs were isolated from the placentas of control nPTL cases and from the placentas of women who experienced sPE. (I) Over 48 hours in culture, NRP-1 expression was upregulated in both instances but to a lesser degree in sPE. (J) Control nPTL CTBs also upregulated NRP-2. Expression of this receptor was reduced in sPE and the soluble form was more abundant. (A–J) The data shown are representative of the analysis of a minimum of 3 samples from different placentas. Scale bars: 100 μm. AV, anchoring villi.
Immunoblot analyses of CTB lysates from control nPTL placentas showed upregulation of NRP-1 expression over 48 hours of culture, which was blunted in sPE (Figure 3I). As for NRP-2, control nPTL CTBs also upregulated this receptor, and soluble forms were detected (Figure 3J). In sPE, NRP-2 was expressed at reduced levels, and the relative abundance of the major soluble form of this receptor increased as compared with the control nPTL CTBs. Together these data suggested that placenta-derived SEMA3B could have autocrine effects on CTBs and paracrine actions on uterine endothelial cells.
We tested this hypothesis in the context of VEGF actions using our in vitro model of CTB invasion. Previously, we showed that CTBs produce large amounts of VEGF and that its autocrine actions include promoting invasion and inhibiting apoptosis (6). Blocking VEGF signals (anti–VEGF-A) or the addition of recombinant SEMA3B protein reduced invasion by approximately 60% as compared with cells that were cultured with a control CD6-Fc protein (Figure 4A). In contrast, removal of both ligands (anti-VEGF plus NRP-1–Fc or NRP-2–Fc) restored invasion to control levels. The removal of VEGF-A or the addition of SEMA3B doubled the rate of apoptosis and the absence of both ligands returned levels to below control values (Figure 4B). Taken together, these results suggested that SEMA3B opposed the actions of VEGF to restrain CTB invasion by promoting apoptosis of these cells.
Figure 4. Exogenous SEMA3B mimicked the effects of sPE on CTBs and endothelial cells and inhibited angiogenesis.
(A) The addition of anti-VEGF or SEMA3B protein significantly inhibited CTB invasion as compared with the addition of a control protein, CD6-Fc. The removal of both ligands (anti–VEGF/NRP1-Fc and anti–VEGF/NRP-2–Fc) restored invasion to control levels. (B) The variables tested in A had the opposite effects on CTB apoptosis, suggesting that increased programmed cell death contributed to decreased invasion. (C) Exogenous VEGF stimulated the migration of UtMVECs, which was inhibited by SEMA3B. (D) The results in C were quantified relative to the addition of CD6-Fc. (E) In UtMVECs, VEGF promoted survival and SEMA3B increased apoptosis relative to control levels. (F) In the chick chorioallantoic membrane angiogenesis assay, VEGF promoted angiogenesis by approximately 3 fold and SEMA3B inhibited this process approximately 5 fold relative to the effects of CD6-Fc. Arrows mark the edge of the filter paper used to apply the protein. The area of the CAM beneath the filter paper is shown in the bottom row. Scale bar: 200 μm (top row); 100 μm (bottom row). n = 6 replicates (A–D); n = 3 replicates (E and F). Mean ± SEM; 2-tailed Student’s t test. *P < 0.05, **P < 0.01, ***P < 0.001.
As to vascular effects, a monolayer of uterine microvascular endothelial cells (UtMVECs) was disrupted with a scratch, and the effects of SEMA3B, in terms of migration, were tracked by video microscopy. The results of a typical experiment are shown in Figure 4C. As with many ECs, the addition of VEGF strongly promoted directed UtMVEC migration; exogenous SEMA3B decreased levels to approximately 50% of control values, with a loss of directionality. Figure 4D summarizes the results of 3 experiments. Under the same conditions, the opposite effects were observed with regard to apoptosis; VEGF was protective and SEMA3B was a strong inducer (Figure 4E). These findings suggested that SEMA3B is primarily antiangiogenic, as was previously proposed. To test this theory, we used the chick chorioallantoic membrane assay, in which filter paper discs delivered VEGF, SEMA3B, or CD6-Fc (Figure 4F, top row). Removal of the discs showed that SEMA3B markedly inhibited angiogenesis as compared with the positive control VEGF or CD6-Fc (Figure 4F, bottom row). Together, these results suggested that the autocrine effects of enhanced SEMA3B expression recapitulated aspects of the CTB phenotype in PE, with paracrine actions including impaired UtMVEC functions.
Exogenous SEMA3B alters CTB signaling, phenocopying sPE effects.
Next, we investigated the CTB signaling pathways that were involved. First, we asked whether SEMA3B opposed VEGF signaling by inhibiting the activation of PI3K, as measured by the production of phosphatidylinositol 3,4,5-triphosphate (PIP3). The addition of SEMA3B to first or second trimester CTBs reduced PIP3 concentrations to levels that were comparable to those after addition of the PI3K inhibitor, wortmannin (Figure 5A). DMSO, the vehicle, had no effect. Addition of VEGF increased PIP3 production 2.5 fold over control levels. These results suggested SEMA3B as a negative regulator of PI3K (25).
Figure 5. SEMA3B inhibited PI3K/AKT and GSK3β signaling in CTBs and the same effects were observed in sPE.
(A) SEMA3B and wortmannin (WM) inhibited PI3K activity, which was stimulated by VEGF. DMSO was used as a vehicle control. Mean ± SEM, 2-tailed Student’s t test. *P < 0.05, **P < 0.01. (B) The addition of SEMA3B to UtMVECs resulted in the dissociation of the p85 and the p110α subunits of PI3K, which was rescued by the addition of VEGF. (C) In COS-1 cells, SEMA3B inhibited AKT Ser473 phosphorylation (activation), which increased during CTB differentiation/invasion (0–12 hours). The addition of SEMA3B inhibited AKT phosphorylation, which was enhanced by exogenous VEGF. (D) In COS-1 cells, SEMA3B inhibited GSK3β Ser9 phosphorylation (inactivation), which increased during CTB differentiation/invasion (0–12 hours). Exogenous SEMA3B inhibited GSK3β phosphorylation, which was enhanced by VEGF. GSK3α Ser21 phosphorylation was variable. (E) In CTBs, sPE correlated with dissociation of the p85 and p110α (and γ) subunits of PI3K relative to control cells isolated from normal third trimester placentas. (F) In freshly isolated CTBs, sPE was associated with decreased phosphorylation of AKT Ser473 and GSK3β Ser9. α-Actin was used as a loading control. (G) In chorionic villi, sPE was associated with phosphorylation (inactivation) of β-catenin. (A–D) The same results were obtained in 3 separate experiments that used different preparations of cells. (F and G) The results shown are representative of analyses of a total of 6 CTB isolates from different placentas of women diagnosed with sPE.
Then we sought to explain the mechanisms involved. A previous study demonstrated that VEGF-mediated VEGFR-2 phosphorylation creates a docking site for the p85 subunit of PI3K (26). However, preliminary experiments showed that SEMA3B did not interfere with VEGFR-2 phosphorylation (data not shown). Thus, we studied the interactions between the regulatory subunits of PI3K. UtMVECs were cultured in medium containing SEMA3B and VEGF or in the absence of one or the other factor. Cell lysates were immunoprecipitated with an antibody that specifically recognized the p85 regulatory subunit of PI3K. The pull downs were immunoblotted with anti–VEGFR-2, anti–NRP-2, and anti-p110α PI3K (Figure 5B); NRP-1 was not expressed (data not shown). VEGFR-2 and NRP-2 levels remained constant under all the test conditions. The addition of SEMA3B resulted in the dissociation of p85 and p110α, which was rescued by the addition of VEGF. Taken together, these results suggested that SEMA3B inhibited PI3K activity (Figure 5A) by preventing the association of p85 and p110α, to our knowledge a novel mechanism.
Downstream of PI3K activation, AKT is phosphorylated at Thr308 and/or Ser473 (27, 28). Thus, we were interested in the effects of SEMA3B on this process (Figure 5C). In initial experiments, we failed to detect SEMA3B-associated changes in phosphorylation of Thr308. Thus, we focused on modification of Ser473. First, COS-1 cells were either transfected with an empty vector or SEMA3B. No Ser473 phosphorylation was observed in the latter case. Next, we evaluated AKT phosphorylation as a function of CTB differentiation in culture. Lysates of cells isolated from first and second trimester placentas were assayed immediately upon isolation (0 hour) and after 12 hours in culture, during which time a band with strong anti–p-Ser473 reactivity appeared. The time course was rapid. After initial CTB adhesion (1 hour), the addition of wortmannin for 30 minutes downregulated Ser473 phosphorylation as did addition of SEMA3B; addition of VEGF had the opposite effect. In each case, the results were compared with the total amount of AKT, which was determined by stripping the blots and reprobing with an antibody that recognized all forms of this molecule. Together, these data suggested that, in CTBs, SEMA3B strongly downregulated AKT signaling.
AKT inactivates GSK3α and GSK3β by phosphorylating Ser21 and Ser9 (29, 30), respectively. In COS-1 cells, expressing SEMA3B abolished phosphorylation of GSK3α and GSK3β (Figure 5D). In first and second trimester CTBs, GSK3β phosphorylation on Ser9 increased during 12 hours of culture, whereas GSK3α phosphorylation on Ser21 was variable (Figure 5D). LiCl, a GSK3 inhibitor, increased phosphorylation of Ser9 (data not shown), with wortmannin having the opposite effect (Figure 5D). Consistent with the AKT results, the addition of exogenous SEMA3B decreased phosphorylation of GSK3β, which was increased by the addition of VEGF. Immunoblot analysis with an antibody that recognized the GSK3 protein backbone showed that levels did not change under any of the experimental conditions. Since GSK3, often a negative regulator, intersects several critical signaling pathways (30, 31), it is likely that overexpression of SEMA3B has important consequences.
Based on our analysis of SEMA3B/VEGF effects on PI3K/AKT and GSK3 signaling, we predicted that this pathway would be dysregulated in sPE. CTBs from control placentas throughout gestation and from patients who were diagnosed with sPE were analyzed immediately after isolation. As with UtMVECs (Figure 5B), an IP/immunoblot strategy showed disassociation of the p85 and p110α subunits of PI3K in sPE (Figure 5E). In this case, the expression of P110γ was also detected and relative expression was reduced in sPE. The results of the AKT and GSK3 analyses were interpreted using the expression of α-actin as a control for protein loading (Figure 5F). The 2 bands observed in the 32-week sPE sample were attributed to proteolysis, which is sometimes observed in these samples. Two patterns were seen in sPE. In one (26 week), no differences were detected at the protein level, but phosphorylation was markedly decreased. The other was characterized by downregulation at both levels (25 and 32 week). In the case of AKT, p-Ser473 was either undetectable or nearly absent in the sPE samples (Figure 5F). Likewise, p-Ser9 of GSK3β was lower in abundance. Finally, we reasoned that an increase in GSK3 activity would have important effects on pathways that we know are critical to CTB invasion. In chorionic villi from sPE placentas, we observed a large increase in the phosphorylated form of β-catenin, which leads to ubiquitination and proteosomal degradation of this molecule (Figure 5G). Given that activation of β-catenin is associated with tumorigenesis (32), inhibiting this pathway could restrict CTB invasion perhaps by altering cell adhesion or Wnt signaling (33).
Based on these data, we propose a model that integrates SEMA3B and VEGF functions in normal pregnancy and in sPE (Figure 6). We found that SEMA3B competed with VEGF binding to neuropilins. High SEMA3B levels led to the dissociation of the p85 and p110α subunits of PI3K, a novel mechanism. The downstream consequences included inactivation of AKT and activation of GSK3, which led to apoptosis and degradation of β-catenin. Together, these data suggested an autocrine mechanism by which elevated SEMA3B levels contributed to the sPE-associated phenotype of invasive CTBs in terms of the signaling pathways that we analyzed.
Figure 6. Model of SEMA3B effects on CTBs in sPE and normal pregnancy.
Discussion
The causes of PE remain poorly understood and are under intense investigation. Here, we focused on the placental component of this pregnancy complication, thought to be the first link in the chain of events leading to the syndrome. As such, we reasoned that the findings might give us a better understanding of the instigating factors. We targeted the CTB subpopulation that invades the uterine wall for analysis, as defects in this process, particularly the remodeling of the arterial side of the uterine vasculature, are the hallmark of this condition. As anticipated from our study design, global transcriptional profiling revealed the gene expression patterns of CTBs from the placentas of affected patients. The molecules encoded by the dysregulated genes — the sPE signature — included factors that have been associated with this syndrome, others that have not been studied in relationship to the placental component of sPE, and novel participants. Additionally, a significant number were placenta-specific gene products that are unique to humans, an interesting observation given the fact that PE is confined to our species.
We reasoned that the differentially expressed genes might play autocrine roles in CTB functions that go awry in PE. Given our discovery that these cells undergo a vascular-like transformation as they invade the uterine wall (3), we focused our mechanistic analyses on SEMA3B, which has been proposed as an angiogenesis inhibitor. The results suggested that this molecule is a major driver of the CTB aberrations in sPE. The endpoints we analyzed included CTB invasion, which was inhibited, and apoptosis, which was increased. We confirmed the antiangiogenic properties of this molecule and described the signaling pathways it engages, a combination of novel and known mechanisms that are also dysregulated in sPE. These data extended the concept that antiangiogenic factors contribute to the maternal signs of PE (34) by showing that they also play an important role in the observed CTB phenotypic alterations that underlie the etiology of this syndrome. We previously proposed this idea in relationship to dysregulated CTB production of VEGF family members in sPE (6). In support of this concept, altering maternal levels of adrenomedullin also affects placentation (16).
Other CTB genes that were upregulated in sPE included growth hormone 2, a placental isoform, which increases the invasiveness of primary CTBs in vitro (35). In contrast, KISS-1 inhibits trophoblast (and tumor cell) invasion (36). The product of the HOP homeobox gene (HOPX), which marks intestinal epithelial stem cells (37), interacts with HDAC2 to enable GATA4 deacetylation, thereby inhibiting proliferation of embryonic cardiomyocytes (38). We described human trophoblast progenitor expression of GATA4 (39), which indicates that it might be possible that this same relationship regulates placental growth. In mice, deletion of Hopx expands the trophoblast giant cell population, with a commensurate reduction in spongiotrophoblasts (40). We confirmed a sPE-associated upregulation of HOPX expression in CTBs (our unpublished observations). Frizzle-related proteins, a component of the sPE gene signature, have actions that are often context dependent. They usually act as Wnt inhibitors, which could also negatively regulate β-catenin, a downstream target of SEMA3B signaling (41). Placenta-specific-1 is unique to this organ (42). Its function(s) is as yet unknown, but it is overexpressed in several pregnancy complications (43). CTB production of numerous isoforms of pregnancy-specific β-1 glycoprotein (PSG-1) was also upregulated. Although their functions are not well understood, they appear to have vasculogenic (44, 45) and immune properties (46, 47). Glypican 3, proposed as a receptor for some PSG family members (48), was also upregulated. ADAM-12 is involved in STB formation (49) and promotes CTB invasion (50). Thus, as a group, the functions of many of these molecules were relevant to the impact of sPE on the placenta. Furthermore, many of them are expressed by both CTBs and STBs. Since placental hypoperfusion is associated with increased numbers of giant cells (51), the dysregulated molecules could bias the cells toward fusion in vivo.
Additionally, the microarray analyses produced several surprising results. For one, nearly all of the CTB genes that were dysregulated in sPE reverted to control levels over 48 hours of culture. This unexpected finding was not due to apoptosis, because we failed to observe upregulation of genes involved in this process (Supplemental Figure 3). This is in contrast to our previous report that sPE is associated with many TUNEL-positive invasive CTBs in situ (13). Thus, we concluded that the in vivo environment, rather than intrinsic CTB defects, was involved in the observed gene dysregulation, which resolved in vitro and progressed to apoptosis in situ. Given the importance of stromal factors in influencing the behavior of epithelial cells (52), it may be that signals from chorionic villous fibroblasts or decidual cells impede invasion in PE. As to the possible involvement of other maternal factors, metabolic syndrome (53), vascular disease (54), and/or advanced maternal age (55) increase PE risk. As to fetal factors, trisomy 21 is associated with an increased incidence of PE, and some of the same aberrations in CTB invasion/differentiation are observed in these pregnancies (56).
Other findings suggest that maternal and placental factors work in concert. For example, we showed that physiological hypoxia regulates the balance between CTB replication and differentiation (57). In support of a role for oxygen tension, by 48 hours, COS-1 cells and second trimester chorionic villi upregulated SEMA3B in 2% vs. 20% O2 (Supplemental Figure 4). This finding suggested that some of the changes in gene expression that were observed in sPE could be due to reduced placental perfusion, which might explain the in vitro “rescue” that we observed. However, given the number of hypoxia-responsive elements in the genome, it was impossible to determine whether genes with this promoter sequence were overrepresented in the sPE signature. Additionally, querying the Ingenuity Knowledge Database failed to identify any pathways that linked the dysregulated molecules, suggesting a complex etiology that will be interesting to unravel. For example, aberrations in the dialogue between trophoblasts and decidua that are initiated at implantation and required for normal CTB invasion could begin the process that culminates in PE (58).
Another surprising finding was that CTBs from the severe forms of PE have common gene signatures. Since we were assaying the same CTB preparation at four 12-hour intervals, which yielded highly statistically significant data, we had the opportunity to compare sPE with other variants of the syndrome. For this purpose, we included superimposed hypertension (1 case) and HELLP syndrome (2 cases). We also added IUGR as a variable. Despite the different maternal and fetal manifestations, we found essentially the same pattern of CTB gene dysregulation in all cases. We took this as preliminary evidence that the various forms of sPE diverged at the level of maternal (and fetal) responses, a theory that will need further validation given the low number of samples that we analyzed from the sPE variants. Nevertheless, our findings are possible evidence of individual differences in the mechanisms that lead to the signs of PE. Although it is not clear what these are, they could include the same risk factors (discussed above) that predispose women to develop this pregnancy complication. However, why PE is more common in nulliparous women remains enigmatic (59). One explanation could be that CTB remodeling of the uterine stroma and/or spiral arterioles becomes progressively easier with each subsequent pregnancy.
Our data demonstrating that sPE-associated aberrations in CTB gene expression are reversible bolster the rationale for developing therapeutic interventions and biomarkers for gauging their utility. One strategy is to target the CTB population that we studied. A small molecule inhibitor that blocks binding of SEMA3A to NRP-1 has been described (60), raising the possibility that a similar compound could be developed for disrupting SEMA3B actions in sPE. Of note, a portion of the molecules that we identified are dysregulated in this pregnancy complication, e.g., the particular combination of PSGs whose expression was upregulated, could be useful biomarkers for predicting elevated risk and/or assessing therapeutic efficacy in women. Another strategy would be to target maternal responses. However, our data also suggest that potential therapeutics may need to be tailored to specific patient populations according to the signs of PE, which are not consistent despite a common set of CTB defects. Finally, if faulty decidual signals play a role in restraining CTB invasion, they could also be therapeutic targets.
In summary, our data suggest new concepts or reinforce proposed mechanisms that suggest a possible theory regarding the pathophysiology of PE. First, in vivo signals drive CTB gene dysregulation. Cells that were cultured from affected placentas normalized their gene expression patterns over 48 hours. These data also suggested the second concept, i.e., CTB phenotypic alterations in sPE are reversible and recovery is possible. Third, our data suggest that the autocrine actions of the dysregulated molecules contribute to the CTB functional defects that are the hallmarks of this syndrome — shallow invasion and apoptosis that are associated with deficits in particular signaling pathways. In this context, we identified previously unknown relevant actions of molecules, including SEMA3B, an angiogenesis inhibitor. Fourth, unified CTB defects manifest as diverse signs in different patients. Thus, our data suggested that individual differences in maternal responses are driving the clinical presentation of the various forms of sPE. In this regard, we also noted disparate fetal responses in terms of growth effects. Together, our findings provide an important rationale and framework for pursuing treatments, a research area that is usually relegated to the back burner because of questions regarding feasibility, which our data support.
Methods
Antibodies and chemicals.
The sources and concentrations used are summarized in Supplemental Table 2.
Tissue collection, CTB, and RNA isolation.
Pregnancy complications were diagnosed using published criteria (18, 19). Biopsies were either fixed in paraformaldehyde and frozen in OCT or fixed in formalin and embedded in paraffin. The remaining tissue was used for CTB isolation as described previously (61), and the cells were cultured up to 48 hours. RNA was purified using RNeasy Plus Kits (QIAGEN) immediately after the cells were isolated (0 hour) and at 12, 24, and 48 hours after culture. RNA concentration/quality was initially assessed on a Nanodrop spectrophotometer (Thermo Scientific) followed by evaluation using an Agilent RNA 6000 Nano LabChip Kit (Agilent Technologies) and an Agilent Bioanalyzer 2100 System. Samples with a RIN number >9 were used in the microarray experiments.
Global transcriptional profiling and data analysis.
Microarray analyses were performed using the GeneChip Human Genome U133 Plus 2.0 array (Affymetrix). Sample processing and hybridization was accomplished by using the protocols devised by the UCSF Gladstone (NHLBI) Genomics Core Facility as previously described (62). The raw image data from the microarrays were analyzed for quality by using RLE-NUSE T2 plots, and the data were normalized using Robust Multi-array Average (RMAExpress v.1.05). Significant differential expression between groups was determined using the Bioconductor (v.2.11) packages limma (v.3.14) and maSigPro (v.1.12) within the software environment R (v.2.15). Then differentially expressed genes were determined by statistical analysis of log odds ratio (B > 0) and absolute fold change of ≥2. maSigPro was used to evaluate gene expression over time (Benjamini-Hochberg P ≤ 0.05) (63). Microarray data were deposited in the Gene Expression Omnibus (accession no. GSE40182).
TaqMan qPCR.
cDNA libraries were prepared with 500 ng RNA using the iScript Kit (Bio-Rad) and diluted 20 fold in water. TaqMan qPCR reactions were carried out in triplicate. Differences among target expression levels were estimated by the ΔΔCT method with normalization to GAPDH. Differences between means were assessed using a 2-tailed Student’s t test (P = 0.05), assuming unequal variance. The values shown are the mean ± SD.
Multiple tissue expression array.
The BD MTE Multiple Tissue Expression Human Array 3, which contained poly A+ RNAs from 67 different human tissues, 8 cancer cell lines, and 8 controls, was from BD Biosciences Clontech. A 32P-labeled SEMA3B antisense probe was prepared using 32P-UTP and RNA polymerase. Hybridization to the MTE Array was carried out in BD ExpressHyb solution according to the manufacturer’s instructions. Signals were detected by exposing the membranes to Kodak BioMax MS film.
Plasmid constructs.
The cDNA fragment encoding SEMA3B, nt 585–1,464, was amplified using a PCR Master Mix Kit (Promega). To obtain the construct for the Northern/in situ hybridization probes, the cDNA fragment was subcloned into a pCRII-TOPO vector (Invitrogen). The cDNA encoding full-length SEMA3B was subcloned into the pcDNA3.1 directional TOPO plasmid (Invitrogen) or pET23(+) to obtain pcDNA3.1-SEMA3B and pET23(+)-SEMA3B constructs tagged with V5 and histidine repeats, respectively. All constructs were confirmed by DNA sequencing.
Northern and in situ hybridization, immunolocalization, and immunoblotting.
Northern and in situ hybridization (4), immunolocalization (6), and immunoblotting (6) were performed as previously described. In every case, at least 3 different placentas or CTB preparations were analyzed per experimental group.
Preparation of recombinant SEMA3B.
COS-1 cells were transfected with the full-length pcDNA3.1-SEMA3B construct or, as a control, an empty pcDNA3.1 plasmid. Following the manufacturer’s instructions, transfections were performed in TransFast Reagent (Promega Corp.). After 48 hours, the cells were transferred to culture medium containing 500 μg/ml geneticin to select for those expressing the plasmid (Invitrogen). Gene expression was confirmed by Northern hybridization. Protein expression and secretion were assessed by immunoblotting for V5. BL-21 E. coli cells were transformed with the pET23(+)-SEMA3B construct. His-tagged proteins were purified on Ni-NTA agarose according to the manufacturer’s instructions (QIAGEN). The identity of the purified protein was confirmed by mass spectrometry (QqTOF) (QSTAR Elite mass spectrometer; AB Sciex).
CTB invasion and apoptosis assays.
The assays were performed as previously described (6). CTBs isolated from first or second trimester placentas were plated in Transwell inserts (6.5 mm; Costar Corp.) on Matrigel-coated polycarbonate filters (8-μm pores). After 36 hours, the Transwell filters were divided in half. One part was stained with an anti-cytokeratin antibody (7D3), which enabled the counting of the number of CTBs that reached the filter’s underside. The other half was labeled using a TUNEL Kit to determine the number of CTBs undergoing apoptosis (R&D Systems). Three filters were plated per test variable, and the entire experiment was performed 6 times. Data were expressed as percentage of control; the statistical significance of the data was determined by Student’s t test.
Culture of uterine vein microvascular cells, tracking migration, and chorioallantoic membrane angiogenesis assay.
The cells were obtained from BioWhittaker Inc. They were cultured as described previously (64). Migration was analyzed using methods that we previously published (4). Endothelial apoptosis was estimated as described above for CTBs. The chick chorioallantoic membrane angiogenesis assay was performed as we previously described (65). Each experiment was performed at least 3 times.
PI3K activity assay.
Isolated second trimester CTBs were cultured for 45 minutes in medium containing SEMA3B, VEGF165, wortmannin, or CD6-Fc. Lysates were prepared using a buffer of 1% NP-40, 20 mM Tris, pH 7.4, 137 mM NaCl, 1 mM CaCl2, 1 mM MgCl2, and 1 mM NaVO3, which contained a proteinase inhibitor cocktail (57). The PI3K activity assay was performed according to the manufacturer’s instructions (Echelon Biosciences Inc.). Briefly, precleared lysates were incubated with anti-PI3K for 2 hours, and then the immunocomplexes were collected on protein A-Sepharose beads. After thoroughly washing, the PI(3,4)P2 substrate was added. The reaction was initiated by the addition of 5 mM HEPES, pH 7.0, 10 mM MgCl2, and 25 mM ATP and terminated by the addition of 5 mM EDTA. Then, the reaction mixtures were incubated with anti-PIP3 for 2 hours. The PIP3 signals were detected by the addition of 3,4,5-trimethoxy benzaldehyde and quantified by detection at 450 nm by using an absorbance plate reader. Each sample was run in duplicate. The experiment was performed 6 times. Data were expressed as percentage of control.
Statistics.
Analysis of the microarray and qPCR data is described above. CTB invasion, CTB/endothelial apoptosis, endothelial migration, PI3K activity, and the clinical data were analyzed by using a 2-tailed Student’s t test. P values of less than 0.05 were considered significant.
Study approval.
The UCSF Institutional Review Board approved this study. Informed consent was obtained from all tissue donors. Placentas were collected immediately following pregnancy terminations, after delivery due to a complication (nPTL or a form of sPE), or at the conclusion of normal pregnancy.
Supplementary Material
Acknowledgments
We are grateful to Jean Perry, Julie de la Garza, and Brittni Johnson for patient recruitment. We also thank Mari-Paule Thiet, the UCSF Maternal-Fetal Medicine Division and The Women’s Option Center at San Francisco General Hospital, for invaluable help in obtaining the placental samples that made this study possible. Finally, we are grateful to the study participants. This work was supported by NIH grant HD030367 and the UCSF Department of Obstetrics, Gynecology and Reproductive Sciences.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2013;123(7):2862–2872. doi:10.1172/JCI66966.
Fei Wang’s present address is: Department of Cell and Developmental Biology, School of Molecular and Cellular Biology, University of Illinois at Urbana-Champaign, Urbana, Illinois, USA.
See the related Attending physician beginning on page 2775.
References
- 1.Maltepe E, Bakardjiev AI, Fisher SJ. The placenta: transcriptional, epigenetic, and physiological integration during development. J Clin Invest. 2010;120(4):1016–1025. doi: 10.1172/JCI41211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pijnenborg R, Vercruysse L, Hanssens M. The uterine spiral arteries in human pregnancy: facts and controversies. Placenta. 2006;27(9–10):939–958. doi: 10.1016/j.placenta.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 3.Zhou Y, et al. Human cytotrophoblasts adopt a vascular phenotype as they differentiate. A strategy for successful endovascular invasion? J Clin Invest. 1997;99(9):2139–2151. doi: 10.1172/JCI119387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Red-Horse K, Kapidzic M, Zhou Y, Feng KT, Singh H, Fisher SJ. EPHB4 regulates chemokine-evoked trophoblast responses: a mechanism for incorporating the human placenta into the maternal circulation. Development. 2005;132(18):4097–4106. doi: 10.1242/dev.01971. [DOI] [PubMed] [Google Scholar]
- 5.Hunkapiller NM, et al. A role for Notch signaling in trophoblast endovascular invasion and in the pathogenesis of pre-eclampsia. Development. 2011;138(14):2987–2998. doi: 10.1242/dev.066589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou Y, et al. Vascular endothelial growth factor ligands and receptors that regulate human cytotrophoblast survival are dysregulated in severe preeclampsia and hemolysis, elevated liver enzymes, and low platelets syndrome. Am J Pathol. 2002;160(4):1405–1423. doi: 10.1016/S0002-9440(10)62567-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Redman CW, Sargent IL. Latest advances in understanding preeclampsia. Science. 2005;308(5728):1592–1594. doi: 10.1126/science.1111726. [DOI] [PubMed] [Google Scholar]
- 8.Gebb J, Dar P. Colour Doppler ultrasound of spiral artery blood flow in the prediction of pre-eclampsia and intrauterine growth restriction. Best Pract Res Clin Obstet Gynaecol. 2011;25(3):355–366. doi: 10.1016/j.bpobgyn.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 9.Naicker T, Khedun SM, Moodley J, Pijnenborg R. Quantitative analysis of trophoblast invasion in preeclampsia. Acta Obstet Gynecol Scand. 2003;82(8):722–729. doi: 10.1034/j.1600-0412.2003.00220.x. [DOI] [PubMed] [Google Scholar]
- 10.Brosens IA, Robertson WB, Dixon HG. The role of the spiral arteries in the pathogenesis of preeclampsia. Obstet Gynecol Annu. 1972;1:177–191. [PubMed] [Google Scholar]
- 11.Zhou Y, Damsky CH, Chiu K, Roberts JM, Fisher SJ. Preeclampsia is associated with abnormal expression of adhesion molecules by invasive cytotrophoblasts. J Clin Invest. 1993;91(3):950–960. doi: 10.1172/JCI116316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou Y, Damsky CH, Fisher SJ. Preeclampsia is associated with failure of human cytotrophoblasts to mimic a vascular adhesion phenotype. One cause of defective endovascular invasion in this syndrome? J Clin Invest. 1997;99(9):2152–2164. doi: 10.1172/JCI119388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DiFederico E, Genbacev O, Fisher SJ. Preeclampsia is associated with widespread apoptosis of placental cytotrophoblasts within the uterine wall. Am J Pathol. 1999;155(1):293–301. doi: 10.1016/S0002-9440(10)65123-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maynard SE, et al. Excess placental soluble fms-like tyrosine kinase 1 (sFlt1) may contribute to endothelial dysfunction, hypertension, and proteinuria in preeclampsia. J Clin Invest. 2003;111(5):649–658. doi: 10.1172/JCI17189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Venkatesha S, et al. Soluble endoglin contributes to the pathogenesis of preeclampsia. Nat Med. 2006;12(6):642–649. doi: 10.1038/nm1429. [DOI] [PubMed] [Google Scholar]
- 16.Li M, Yee D, Magnuson TR, Smithies O, Caron KM. Reduced maternal expression of adrenomedullin disrupts fertility, placentation, and fetal growth in mice. J Clin Invest. 2006;116(10):2653–2662. doi: 10.1172/JCI28462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou Y, et al. Comparative analysis of maternal-fetal interface in preeclampsia and preterm labor. Cell Tissue Res. 2007;329(3):559–569. doi: 10.1007/s00441-007-0428-0. [DOI] [PubMed] [Google Scholar]
- 18.ACOG Practice Bulletin Diagnosis and management of preeclampsia and eclampsia. Number. 2002;77:67–75. [PubMed] [Google Scholar]
- 19.Haram K, Svendsen E, Abildgaard U. The HELLP syndrome: clinical issues and management. A Review. BMC Pregnancy Childbirth. 2009;9:8. doi: 10.1186/1471-2393-9-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mittal P, et al. Placental growth hormone is increased in the maternal and fetal serum of patients with preeclampsia. J Matern Fetal Neonatal Med. 2007;20(9):651–659. doi: 10.1080/14767050701463571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Winn VD, et al. Severe preeclampsia-related changes in gene expression at the maternal-fetal interface include sialic acid-binding immunoglobulin-like lectin-6 and pappalysin-2. Endocrinology. 2009;150(1):452–462. doi: 10.1210/en.2008-0990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang H, Long Q, Ling L, Gao A, Li H, Lin Q. Elevated expression of KiSS-1 in placenta of preeclampsia and its effect on trophoblast. Reprod Biol. 2011;11(2):99–115. doi: 10.1016/S1642-431X(12)60048-5. [DOI] [PubMed] [Google Scholar]
- 23.Gack S, et al. Preeclampsia: increased expression of soluble ADAM 12. J Mol Med (Berl). 2005;83(11):887–896. doi: 10.1007/s00109-005-0714-9. [DOI] [PubMed] [Google Scholar]
- 24. Kolodkin AL, Tessier-Lavigne M. Mechanisms and molecules of neuronal wiring: a primer. Cold Spring Harb Perspect Biol. 2011. 3 (6).pii: a001727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Castro-Rivera E, Ran S, Brekken RA, Minna JD. Semaphorin 3B inhibits the phosphatidylinositol 3-kinase/Akt pathway through neuropilin-1 in lung and breast cancer cells. Cancer Res. 2008;68(20):8295–8303. doi: 10.1158/0008-5472.CAN-07-6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dayanir V, Meyer RD, Lashkari K, Rahimi N. Identification of tyrosine residues in vascular endothelial growth factor receptor-2/FLK-1 involved in activation of phosphatidylinositol 3-kinase and cell proliferation. J Biol Chem. 2001;276(21):17686–17692. doi: 10.1074/jbc.M009128200. [DOI] [PubMed] [Google Scholar]
- 27.Datta SR, et al. Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. Cell. 1997;91(2):231–241. doi: 10.1016/S0092-8674(00)80405-5. [DOI] [PubMed] [Google Scholar]
- 28.Kandel ES, Hay N. The regulation and activities of the multifunctional serine/threonine kinase Akt/PKB. Exp Cell Res. 1999;253(1):210–229. doi: 10.1006/excr.1999.4690. [DOI] [PubMed] [Google Scholar]
- 29.Cross DA, Alessi DR, Cohen P, Andjelkovich M, Hemmings BA. Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature. 1995;378(6559):785–789. doi: 10.1038/378785a0. [DOI] [PubMed] [Google Scholar]
- 30.Shaw M, Cohen P, Alessi DR. Further evidence that the inhibition of glycogen synthase kinase-3beta by IGF-1 is mediated by PDK1/PKB-induced phosphorylation of Ser-9 and not by dephosphorylation of Tyr-216. FEBS Lett. 1997;416(3):307–311. doi: 10.1016/S0014-5793(97)01235-0. [DOI] [PubMed] [Google Scholar]
- 31.Doble BW, Woodgett JR. GSK-3: tricks of the trade for a multi-tasking kinase. J Cell Sci. 2003;116(pt 7):1175–1186. doi: 10.1242/jcs.00384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zucman-Rossi J, et al. Differential effects of inactivated Axin1 and activated beta-catenin mutations in human hepatocellular carcinomas. Oncogene. 2007;26(5):774–780. doi: 10.1038/sj.onc.1209824. [DOI] [PubMed] [Google Scholar]
- 33.Daugherty RL, et al. Phospho-regulation of Beta-catenin adhesion and signaling functions. Physiology (Bethesda). 2007;22:303–309. doi: 10.1152/physiol.00020.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Powe CE, Levine RJ, Karumanchi SA. Preeclampsia, a disease of the maternal endothelium: the role of antiangiogenic factors and implications for later cardiovascular disease. Circulation. 2011;123(24):2856–2869. doi: 10.1161/CIRCULATIONAHA.109.853127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lacroix MC, et al. Stimulation of human trophoblast invasion by placental growth hormone. Endocrinology. 2005;146(5):2434–2444. doi: 10.1210/en.2004-1550. [DOI] [PubMed] [Google Scholar]
- 36.Hiden U, Bilban M, Knofler M, Desoye G. Kisspeptins and the placenta: regulation of trophoblast invasion. Rev Endocr Metab Disord. 2007;8(1):31–39. doi: 10.1007/s11154-007-9030-8. [DOI] [PubMed] [Google Scholar]
- 37.Takeda N, Jain R, LeBoeuf MR, Wang Q, Lu MM, Epstein JA. Interconversion between intestinal stem cell populations in distinct niches. Science. 2011;334(6061):1420–1424. doi: 10.1126/science.1213214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trivedi CM, et al. Hopx and Hdac2 interact to modulate Gata4 acetylation and embryonic cardiac myocyte proliferation. Dev Cell. 2010;19(3):450–459. doi: 10.1016/j.devcel.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Genbacev O, et al. Establishment of human trophoblast progenitor cell lines from the chorion. Stem Cells. 2011;29(9):1427–1436. doi: 10.1002/stem.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Asanoma K, et al. HOP/NECC1, a novel regulator of mouse trophoblast differentiation. J Biol Chem. 2007;282(33):24065–24074. doi: 10.1074/jbc.M701380200. [DOI] [PubMed] [Google Scholar]
- 41.Mii Y, Taira M. Secreted Wnt “inhibitors” are not just inhibitors: regulation of extracellular Wnt by secreted Frizzled-related proteins. Dev Growth Differ. 2011;53(8):911–923. doi: 10.1111/j.1440-169X.2011.01299.x. [DOI] [PubMed] [Google Scholar]
- 42.Rawn SM, Cross JC. The evolution, regulation, and function of placenta-specific genes. Annu Rev Cell Dev Biol. 2008;24:159–181. doi: 10.1146/annurev.cellbio.24.110707.175418. [DOI] [PubMed] [Google Scholar]
- 43.Fant M, Farina A, Nagaraja R, Schlessinger D. PLAC1 (Placenta-specific 1): a novel, X-linked gene with roles in reproductive and cancer biology. Prenat Diagn. 2010;30(6):497–502. doi: 10.1002/pd.2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ha CT, et al. Human pregnancy specific beta-1-glycoprotein 1 (PSG1) has a potential role in placental vascular morphogenesis. Biol Reprod. 2010;83(1):27–35. doi: 10.1095/biolreprod.109.082412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lisboa FA, et al. Pregnancy-specific glycoprotein 1 induces endothelial tubulogenesis through interaction with cell surface proteoglycans. J Biol Chem. 2011;286(9):7577–7586. doi: 10.1074/jbc.M110.161810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Motran CC, Diaz FL, Gruppi A, Slavin D, Chatton B, Bocco JL. Human pregnancy-specific glycoprotein 1a (PSG1a) induces alternative activation in human and mouse monocytes and suppresses the accessory cell-dependent T cell proliferation. J Leukoc Biol. 2002;72(3):512–521. [PubMed] [Google Scholar]
- 47.Snyder SK, et al. Pregnancy-specific glycoproteins function as immunomodulators by inducing secretion of IL-10, IL-6 and TGF-beta1 by human monocytes. Am J Reprod Immunol. 2001;45(4):205–216. doi: 10.1111/j.8755-8920.2001.450403.x. [DOI] [PubMed] [Google Scholar]
- 48.Sulkowski GN, Warren J, Ha CT, Dveksler GS. Characterization of receptors for murine pregnancy specific glycoproteins 17 and 23. Placenta. 2011;32(8):603–610. doi: 10.1016/j.placenta.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huppertz B, Bartz C, Kokozidou M. Trophoblast fusion: fusogenic proteins, syncytins and ADAMs, and other prerequisites for syncytial fusion. Micron. 2006;37(6):509–517. doi: 10.1016/j.micron.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 50.Beristain AG, Zhu H, Leung PC. Regulated expression of ADAMTS-12 in human trophoblastic cells: a role for ADAMTS-12 in epithelial cell invasion? PLoS One. 2011;6(4):e18473. doi: 10.1371/journal.pone.0018473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Redline RW, et al. Maternal vascular underperfusion: nosology and reproducibility of placental reaction patterns. Pediatr Dev Pathol. 2004;7(3):237–249. doi: 10.1007/s10024-003-8083-2. [DOI] [PubMed] [Google Scholar]
- 52.Glaire MA, El-Omar EM, Wang TC, Worthley DL. The mesenchyme in malignancy: A partner in the initiation, progression and dissemination of cancer. Pharmacol Ther. 2012;136(2):131–141. doi: 10.1016/j.pharmthera.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 53.Hubel CA, Roberts JM. Chesley’s Hypertensive Disorders in Pregnancy. 2009. Metabolic syndrome and preeclampsia. In: Lindheimer MD, Roberts JM, Cunningham FG, eds. pp. 105–128. 3rd ed. Boston, Massachusetts, USA: Academic Press; [Google Scholar]
- 54.Harville EW, Viikari JS, Raitakari OT. Preconception cardiovascular risk factors and pregnancy outcome. Epidemiology. 2011;22(5):724–730. doi: 10.1097/EDE.0b013e318225c960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chibber R. Child-bearing beyond age 50: pregnancy outcome in 59 cases “a concern?”. Arch Gynecol Obstet. 2005;271(3):189–194. doi: 10.1007/s00404-003-0577-7. [DOI] [PubMed] [Google Scholar]
- 56.Wright A, et al. Trisomy 21 is associated with variable defects in cytotrophoblast differentiation along the invasive pathway. Am J Med Genet A. 2004;130A(4):354–364. doi: 10.1002/ajmg.a.30254. [DOI] [PubMed] [Google Scholar]
- 57.Genbacev O, Zhou Y, Ludlow JW, Fisher SJ. Regulation of human placental development by oxygen tension. Science. 1997;277(5332):1669–1672. doi: 10.1126/science.277.5332.1669. [DOI] [PubMed] [Google Scholar]
- 58.Dey SK. How we are born. J Clin Invest. 2010;120(4):952–955. doi: 10.1172/JCI42380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.George EM, Granger JP. Mechanisms and potential therapies for preeclampsia. Curr Hypertens Rep. 2011;13(4):269–275. doi: 10.1007/s11906-011-0204-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kikuchi K, et al. In vitro and in vivo characterization of a novel semaphorin 3A inhibitor, SM-216289 or xanthofulvin. J Biol Chem. 2003;278(44):42985–42991. doi: 10.1074/jbc.M302395200. [DOI] [PubMed] [Google Scholar]
- 61.Hunkapiller NM, Fisher SJ. Chapter 12. Placental remodeling of the uterine vasculature. Methods Enzymol. 2008;445:281–302. doi: 10.1016/S0076-6879(08)03012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Winn VD, et al. Gene expression profiling of the human maternal-fetal interface reveals dramatic changes between midgestation and term. Endocrinology. 2007;148(3):1059–1079. doi: 10.1210/en.2006-0683. [DOI] [PubMed] [Google Scholar]
- 63.Conesa A, Nueda MJ, Ferrer A, Talon M. maSigPro: a method to identify significantly differential expression profiles in time-course microarray experiments. Bioinformatics. 2006;22(9):1096–1102. doi: 10.1093/bioinformatics/btl056. [DOI] [PubMed] [Google Scholar]
- 64.Zhou Y, Bellingard V, Feng KT, McMaster M, Fisher SJ. Human cytotrophoblasts promote endothelial survival and vascular remodeling through secretion of Ang2, PlGF, and VEGF-C. Dev Biol. 2003;263(1):114–125. doi: 10.1016/S0012-1606(03)00449-4. [DOI] [PubMed] [Google Scholar]
- 65.Damsky CH, Fitzgerald ML, Fisher SJ. Distribution patterns of extracellular matrix components and adhesion receptors are intricately modulated during first trimester cytotrophoblast differentiation along the invasive pathway, in vivo. J Clin Invest. 1992;89(1):210–222. doi: 10.1172/JCI115565. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.