Abstract
Relatively, few species have been able to colonize extremely cold alpine environments. We investigate the role played by the cushion life form in the evolution of climatic niches in the plant genus Androsace s.l., which spreads across the mountain ranges of the Northern Hemisphere. Using robust methods that account for phylogenetic uncertainty, intraspecific variability of climatic requirements and different life-history evolution scenarios, we show that climatic niches of Androsace s.l. exhibit low phylogenetic signal and that they evolved relatively recently and punctually. Models of niche evolution fitted onto phylogenies show that the cushion life form has been a key innovation providing the opportunity to occupy extremely cold environments, thus contributing to rapid climatic niche diversification in the genus Androsace s.l. We then propose a plausible scenario for the adaptation of plants to alpine habitats.
Keywords: Alpine plants, climatic niche, key innovation, niche conservatism, phylogenetic signal, phylogenetic uncertainty
The evolutionary mechanisms that drive species’ ranges have fascinated evolutionists and biogeographers since Darwin (1859). One key element lying at the heart of these questions is the set of ecological conditions required for a given species to maintain viable populations (i.e., the species’ ecological niche, Grinnell 1917; Hutchinson 1957). Among the different niche dimensions, climatic niches are particularly interesting in the current context of climate change, because their evolutionary stasis (i.e., niche conservatism) combined with insufficient migration capacities (Loarie et al. 2009), could lead to disproportionate species loss in certain clades (Mace et al. 2003; Parmesan 2006; Thuiller et al. 2011). Recent literature emphasizes that there is an urgent need to test whether niche conservatism can be considered as a general principle (Wiens et al. 2010), and to determine the rates and drivers of climatic niche evolution (Lavergne et al. 2010a).
Despite the current debate on the prevalence of niche conservatism in nature (Losos 2008a; Wiens et al. 2010), some general principles have been identified. The most common view is that high dispersal rates in a species enable habitat selection, leading to stabilizing selection on key ecological traits and hence niche conservatism. Donoghue (2008) posited that it might be easier and quicker for species to migrate than to evolve when exposed to environmental change. In contrast, situations suited for rapid niche evolution often involve dispersal limitations. Ackerly (2003) pointed out that isolation on “environmental islands” subject to environmental change might expose species to strong selection pressures, resulting in punctual and abrupt niche shifts. The literature on adaptive radiations contains numerous examples of rapid niche evolution in insular settings (e.g., Witter and Carr 1988; Pinto et al. 2008). Most of these cases involve the emergence of a key innovation, that is, a trait that allows a species to interact with the environment in a fundamentally different way and provides the stimulus for niche evolution (Miller 1949; Losos and Mahler 2010; we do not use in this article the recent definition of a key innovation as a trait enhancing diversification rates).
A commonly used approach for testing niche conservatism has been the measurement of “phylogenetic signal.” This statistical pattern measures “the tendency for related species to resemble each other more than if they were taken at random from a phylogenetic tree” (Blomberg and Garland 2002), a strong phylogenetic signal in niche-related traits being usually interpreted as evidence for niche conservatism (Losos 2008a). However, although phylogenetic signal describes a statistical pattern of trait autocorrelation across phylogenies, it is not useful for inferring the rate (amount of change per unit of time), the tempo (early vs. late) and the mode (punctual vs. gradual) of niche evolution (Revell et al. 2008; Lavergne et al. 2010a). Recently, Evans et al. (2009) investigated the tempo (i.e., early vs. late) of climatic niche diversification in a group of plants (Oenothera spp. [Onagraceae], sections Anogra and Kleinia) and showed that comparing niche evolutionary patterns to the ones expected under a null model of Brownian evolution can reveal interesting insights into the processes driving niche evolution (see also Yesson and Culham 2006a,b). Surprisingly, little attention has been paid to the biological traits potentially driving niche evolution (but see Luxbacher and Knouft 2009; Edwards and Smith 2010). Indeed, most of the studies on climatic niche conservatism have detected patterns of niche evolution and in some cases have discussed their consequences, but have rarely tackled the processes that are directly involved (Wiens et al. 2010).
Alpine species are well suited to the study of niche evolution because they have experienced major climatic fluctuations in the past (Zachos et al. 2008), and may exhibit key phenotypic innovations that have allowed the colonization of alpine niches following mountain uplifts. Moreover, the mountain ranges they occupy make their distribution highly fragmented and their survival particularly vulnerable to climate change (Randin et al. 2009). Here, we focus on the Androsace (sensu Martins et al. 2003) genus (Primulaceae), a group of about 110 species (according to the International Plant Names Index, http://www.ipni.org) and distributed among the temperate and cold regions of the Northern Hemisphere, many of them being endemic to certain mountain ranges. It is suspected that the evolutionary history of Androsace has been primarily shaped by mountains uplifts and past climate fluctuations, as already detected in some groups in which floras have been fragmented by the emergence of high-altitude island-like habitats (e.g., Hughes and Eastwood 2006). Interestingly, species within this genus harbor a variety of life forms, such as annual and perennial herbaceous, but also long-lived cushions, which have long been thought to be adapted to harsh alpine conditions (Körner 1999). However, the link between life form and particular climatic niches has never been quantitatively tested.
This leads us to address the following questions: (1) is there any trace of past rapid climatic niche evolution in Androsace, as expected given the fragmentation of the high-altitude habitats they occupy? (2) Is the cushion life-form ancestral in Androsace or is it a derived character? And if derived, when and how many times has it evolved? (3) Is the cushion life form a key innovation (sensu Miller 1949) that triggered a significant shift toward alpine niches? To answer these questions and understand the evolutionary history of climatic niches and life forms in the Androsace genus, we developed a comprehensive phylo-climatic modeling approach. We first built the most complete phylogeny of the Androsace genus to date. Then, we used robust methods to estimate species’ climatic niches and compared them in the phylogenetic framework while accounting for intraspecific niche variability, and uncertainty in phylogenetic inference and ancestral traits reconstructions.
Material and Methods
STUDY GROUP AND TAXON SAMPLING
Recent phylogenetic studies (Martins et al. 2003; Schneeweiss et al. 2004) have shown that the former Androsace L., Vitaliana Sesl., Douglasia Lindl., and Pomatosace Maxim. genera form a monophyletic group (hereafter referred to as the Androsace genus). The hotspot of species richness for Androsace occurs in the Eastern Himalaya, as for Primulaceae in general, suggesting that this clade is of Asian origin. All Androsace species except Vitaliana primuliflora are characterized by white or pink homostylous flowers that have a short corolla tube and are relatively large compared to plant size (Ruffier-Lanche 1964). They are mainly pollinated by Hymenoptera and have low competition abilities. The main differences between species of the genus are their contrasted soil ecologies (e.g., Androsace alpina is described as mainly silicicolous and A. helvetica as mainly calcicolous, Lauber and Wagner 2007), their climatic requirements (from dry steppes to mountain tops) and their different life forms, ranging from annual species to cushions with high individual longevity (see illustrations on Fig. 1).
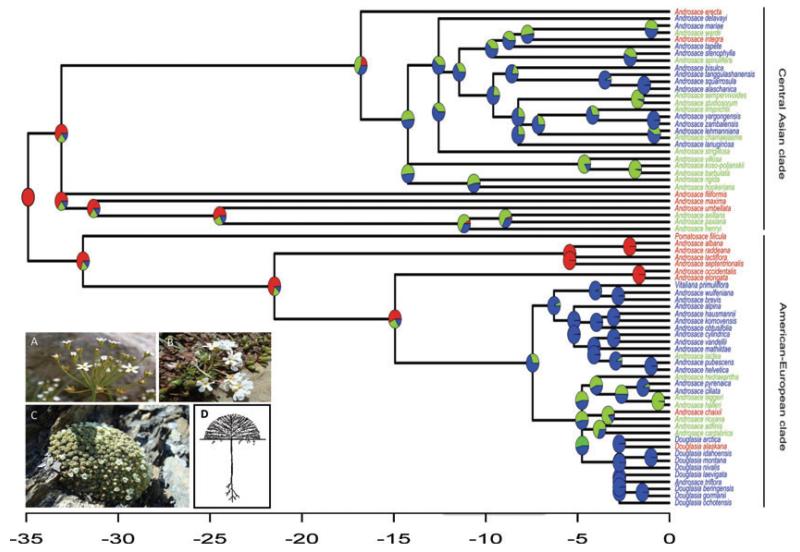
Figure 1.
Evolution of life forms in the Androsace genus. The tree is the consensus tree for 71 species of the genus, with the time scale given in million years. Two main clades segregate from the root. Each ancestral node is colored according to the marginal likelihoods of the different life forms in the NEJ-root model (red = short-lived; green = perennial; blue = cushion). Tips are colored the same way. The cushion life form appears independently in the two main clades. Photographs show representatives of the different life forms. (A) A. septentrionalis, short-lived (annual, photo: S. Aubert/SAJF). (B) A. adfinis, perennial (photo: F.Boucher/LECA). (C) A. helvetica, cushion (photo: F. Boucher/LECA). (D) Schematic representation of the cushion morphology, with its dense canopy formed by radial ramifications (illustration in R. Ruffier-Lanche 1964).
The phylogenetic trees we built contain 77 taxa (see Appendix S1). Subspecies were first collapsed together (always collapsed to the type subspecies) as the occurrence data we used did not distinguish between subspecies (71 species left). All the species were used to estimate ancestral life forms. Finally, 19 species were pruned from the trees and removed from the analyses on climatic niches, as there was little knowledge about their current distribution (fewer than five known occurrences, see list in Appendix S1).
PHYLOGENETIC INFERENCE
All ITS and trnL-F sequences available in Genbank for Androsace, Douglasia, Potamosace, and Vitaliana plus three outgroups (Soldanella alpina L., Trientalis borealis Raf., and T. europaea L.) were retrieved (accession numbers available in Appendix S1). Both regions were available for 61 ingroup species (66 taxa including subspecies), and only one region was available for 11 additional species, thus covering around 65% (72 out of ca. 110) of the Androsace species. Those sequences correspond to the following studies: Dixon et al. (2009), Martins et al. (2003), Mast et al. (2006), Schneeweiss et al. (2004), Schönswetter and Schneeweiss (2009), Wang et al. (2004). When more than one sequence was available for each taxa and region, a consensus sequence was created with Bioedit (Hall 1999). For each region, sequences were aligned with ClustalW2 (Larkin et al. 2007), Kalign (Lassmann and Sonnhammer 2005), MAFFT (Katoh et al. 2005), and Muscle (Edgar 2004). The best alignment was chosen with the multiple overlap score of MUMSA (Lassmann and Sonnhammer 2006). MUMSA is a tool for automatic assessment of alignment quality that provides two indices: the average overlap score, which indicates whether the sequences cumulated for a region are too divergent to be aligned consistently; and the multiple overlap score, which identifies the most consistent alignment when comparing different alignments for a same set of sequences. For both regions, the best alignment was the one produced by MAFFT. We removed ambiguous sites from the alignments matrices with trimAl using the heuristic algorithm—automated1 (Capella-Gutierrez et al. 2009), which uses gaps and similarities distribution to determine the thresholds for trimming the poorly aligned sites of an alignment. We determined the best-fitting model of evolution for each region with the Akaike Information Criterion (AIC) as implemented in MrModeltest version 2 (Nylander 2004). Both regions were concatenated with FASconCAT (Kuck and Meusemann 2010). Phylogenetic analyses were conducted for the combined dataset with MrBayes 3.1.2. (Ronquist and Huelsenbeck 2003) with partitioned model parameters for each region. Two independent analyses were run with 20 million generations sampling one of every 100 trees. Run convergences were checked with AWTY (Nylander et al. 2008), a tool for graphical exploration of convergence in rates of posterior split probabilities and branch lengths. The first 25% of trees were eliminated in the burn-in phase. The combined matrix and the 50% majority-rule consensus phylogenetic tree were deposited in Treebase; study number TB2:S11159 (http://www.treebase.org).
Dating analyses were performed with PAML (Yang 1997) and Multidivtime packages (Kishino et al. 2001; Thorne and Kishino 2002), which provide a mean age and a 95% confidence interval (95% CI) for each node. This was applied to 100 randomly selected trees from the posterior distribution of Bayesian analyses to take into account phylogenetic uncertainty in subsequent analyses. To calibrate the trees, due to the lack of a suitable fossil record for Androsace, a minimum and maximum age constraint (34.5–35.3 million years ago [Mya]) was applied to the Androsace crown node based on the results (95% CI limits) obtained in Yesson et al. (2009), which produced a dated generic level molecular phylogeny of Primulaceae and Myrsinaceae. A maximum age of 45.3 Mya was applied to the root node, which corresponds to the upper limit of the 95% CI (Yesson et al. 2009) for the split between Primulaceae and Myrsinaceae (to which Trientalis belongs). We used the ages reported by Yesson et al. (2009) and not those of Schneeweiss et al. (2004) because we consider the data of the former are more accurate. Schneeweiss et al. (2004) dating was indeed based on divergence estimates of Maesaceae, Theophrastaceae, Primulaceae, and Myrsinaceae obtained by Wikström et al. (2001) in a study including only two genera of Primulaceae, whereas Yesson et al. (2009) constructed expressly a phylogeny of Myrsinaceae (18 genera included) and Primulaceae to obtain a secondary age estimate for the genera of Primulaceae (13 genera sampled).
For the following analyses, each of the 100 trees was pruned to keep only 62 (for life-form reconstructions) or 51 tips (for all calculations including climatic niches) using PAUP* 4.0b10 (Swofford 2002).
LOCATION
Occurrence data for 51 species of Androsace (see list in Appendix S1) were extracted from the Global Biodiversity Information Facility database (GBIF, http://www.gbif.org). Occurrences for 13 species from the Alps obtained from two French National Botanical Conservatories (CBNA, http://www.cbna05.com, and CBNMED, http://www.cbnmed.fr) and from the CRSF Swiss Floristics Network (http://www.crsf.ch) were also added to the dataset. After deleting all points with coordinate precisions lower than 0.01 arc-degree, more than 7000 points remained almost equally distributed between the two different data sources. European species were generally more represented in the data than Central Asian and American ones (see number of occurrences for each species in Appendix S1). As no additional sources of distribution data are available for the Asian part of the study, and because our results were robust to the low number of occurrence for some species (Appendix S2), this data asymmetry is not likely to generate any bias in our study but should only reduce the statistical power of interspecific comparisons of niche characteristics
NICHE SEPARATION
To separate species’ niches in a multidimensional space, we used an ordination method called the “outlying mean index” (OMI, Dolédec et al. 2000), which measures the distance between a species’ niche and the mean conditions of the sampling area (in this case, all locations where Androsace species are present). Unlike other ordination techniques, the OMI makes no assumption about the shape of the species’ response curve to environmental gradients and gives equal weight to all sites regardless of their species richness (Thuiller et al. 2004). The latter characteristic was particularly appreciable in our case, given the low number of observed occurrences in Central Asia.
Global climate across the Northern Hemisphere was represented by the 19 “Bioclim” variables from the WORLDCLIM database (Hijmans et al. 2005, http://worldclim.org) for the baseline period (1950-2000). These variables represented a range of metrics (mean value, variance, and extremes) on global temperature and precipitations. The topographic heterogeneity of the mountain ranges where Androsace mostly occurs and the average precision of the occurrence data led us to choose a moderate resolution of 2.5′. We also added a critical variable for plant physiology, namely the ratio of actual over potential evapotranspiration (aetpet, see Thuiller et al. 2005 for more details). The choice of including only climatic variables into the niche estimation was motivated by the global scale of our study, at which it is known that primary determinants of species distributions are climatic (Woodward 1990, 1992). Furthermore, in the special case of Androsace, which share similar biotic interactions and occur in relatively cold and dry environments, abiotic variables might be more important than biotic ones for setting niche boundaries (Körner 1999). Although Androsace species show contrasting soil ecologies, and even if substrate type strongly influences alpine plant distributions (Alvarez et al. 2009), we did not include soil preferences in our study due to the absence of data with a sufficient global scale resolution.
INTEGRATING MULTIPLE SOURCES OF UNCERTAINTY
In this study, we developed an original workflow to account for multiple sources of uncertainty (Appendix S4).
To date, few studies of niche evolution have explicitly incorporated phylogenetic uncertainty through the use of multiple phylogenetic trees (but see Edwards and Smith 2010). Here, all subsequent analyses were carried out on a set of 100 phylogenetic trees sampled randomly from the stationary phase of Bayesian analyses.
Another possible source of error is due to the fact that most comparative studies often consider only the mean value of a character for each species (Losos 2008a; Kozak and Wiens 2010). Indeed, evolutionary biologists since Darwin (1859) have known about intraspecific variability in niche-related traits, as it is the basis of lineage differentiation and speciation. It can sometimes be higher than interspecific variation among closely related species (Felsenstein 2008; Albert et al. 2010) and can inflate type I errors in phylogenetic tests (Harmon and Losos 2005). Still, niches of species are almost always represented by their mean values in phylogenetic studies (e.g., Luxbacher and Knouft 2009; Kozak and Wiens 2010; but see Evans et al. 2009). Here, we propose a way of incorporating intraspecific niche variability in studies of niche evolution. Instead of retaining the mean niche position and niche breadth over the most important axes, we extracted the scores of all occurrence points on the first two axes yielded by the OMI. For each species, we obtained a bidimensional distribution that can be interpreted as a projection of the climatic niche in the environmental space, maximizing niche differentiation across the whole study group. We used these distributions to resample species’ positions along climatic gradients: at each resampling step one, “niche value” on each gradient was randomly selected for each species from its niche distribution on the OMI axes. These values were then used in the phylogenetic analyses. This allowed us to overcome the biases induced by intraspecific niche variability (see the comparison with results without resampling in Appendix S3).
Finally, ancestral state estimation is a step that is traditionally known to generate major uncertainties (Pagel 1997; Losos 1999). After inferring the best model of life-form evolution (see below), we used joint likelihoods of the different life forms for every node in the tree to generate 100 ancestral state reconstructions per tree.
The methods presented below are detailed for one given tree and one set of characters (niche values for extant species and ancestral life forms), but by pooling the results obtained for all resamples of phylogenetic trees, climatic niche positions, and ancestral life forms, we obtained the distributions of all estimated parameters and AIC scores of alternative models. Given that they were not normal, these distributions were always compared using pairwise Wilcoxon signed-rank tests to evaluate if their means were statistically different. All the following analyses have been performed using the R software (R Development Core Team 2011) version 2.12.0 with the ade4 (Dray and Dufour 2007), ape (Paradis et al. 2004), geiger (Harmon et al. 2008), picante (Kembel et al. 2009), and diversitree (FitzJohn et al. 2009) packages.
EVOLUTIONARY HISTORY OF THE CLIMATIC NICHE
As the fragmentation of habitats occupied by Androsace could have stimulated niche evolution, we first measured the amount of phylogenetic signal in the climatic niches using two statistics: Blomberg’s K (Blomberg et al. 2003; Kembel 2009) and Pagel’s λ (Pagel 1999; Freckleton et al. 2002), which differ in their statistical implementation and parameter testing procedures. Using both of them, we prevent ourselves from drawing conclusions from a single method, the potential pitfalls of which have been pointed out (Freckleton et al. 2002; Revell et al. 2008). K compares the distribution of independent contrasts (Felsenstein 1985) to that expected under a Brownian motion (BM) model of trait evolution. Values of K close to 0 imply no signal and values closer or higher than 1 indicate a signal close or greater than expected under a BM model of evolution. K’s significance is assessed by data randomization. Pagel’s λ is a multiplicative parameter affecting the covariances of characters between different tips of the tree, using a generalized least square optimization. It ranges from 0 to 1, indicating no signal or a signal equivalent to the one expected under BM, respectively. Its significance can be assessed by a likelihood ratio comparison of nested models with particular values (i.e., 0 or 1).
Given the low amount of phylogenetic signal in climatic niches of Androsace (see Results), we went a step further and tried to determine how and when niche evolution took place. For this we estimated the tempo and the mode of climatic niche evolution. The tempo describes whether a character has evolved early (close to the root) or late (close to the tips) in a given phylogeny; the mode of evolution discriminates between gradual (i.e., changes proportional to branch length) and punctual evolution. This was first performed by computing Pagel’s δ and κ parameters, which, respectively, measure the tempo and mode of evolution (Pagel 1999; Verdú 2006).
To get a precise view of how niches diversified over time during clade growth and to be able to compare it with the timing of the appearance of the cushion life form, the tempo of niche evolution was also assessed through a disparity analysis (Harmon et al. 2003). Disparity within a clade is calculated here from average pairwise Euclidean distance between species on the two niche axes, and standardized by the disparity of the whole genus. Then, for each speciation event in the phylogeny, the mean disparity is calculated as the average of the disparities of the clades whose ancestral lineage was present at the time of the speciation. By plotting the average subclade disparity against evolutionary time, we obtain a disparity-through-time (DTT) plot, which ranges between 1 (all disparity still to be built) and 0 (all disparity has been accumulated). Observed DTT is then compared to the one expected under a null model of evolution, that is, to BM simulations (10 simulations for each observed DTT). To quantify the results of the disparity analysis, we computed the morphological disparity index (MDI), which compares observed disparity to the one expected under BM (Harmon et al. 2003). Positive values of MDI indicate that disparity is relatively distributed within clades and hence that the trait being studied has evolved relatively recently; alternatively, negative values of MDI are interpreted as evidence of disparity being mainly between clades and of early evolution of the trait (Evans et al. 2009).
EVOLUTION OF LIFE FORMS
Information on species life forms was gathered from the following sources: Flora Helvetica (Lauber and Wagner 2007), Flora Europeae (Tutin et al. 1964+), Flora of China (Hu and Kelso 2007), and Flora of North America (Cholewa and Kelso 2009). Life forms were classified into the three following groups: “short-lived” species for annuals and biannuals, “perennials” for herbaceous perennials, and “cushion” species for long-lived species forming dense mats or cushions, generally displaying woody structures. Using our newly developed phylogeny and by distinguishing cushions from other perennials species, we are extending a previous work on the evolution of life form in Androsace that had been performed on a smaller phylogeny (Schneeweiss et al. 2004).
To determine the ancestral life form of the Androsace genus and the number of times the cushion life form has evolved, we estimated ancestral life forms over our set of phylogenetic trees using ancestral state reconstructions following the Markov (Mk) model (Lewis 2001) as implemented by Fitzjohn et al. (2009). This was done by fitting and comparing six possible discrete Markov models: a model with all transition rates being different (ARD), the same model but with the tree root constrained to be short-lived (ARD-root), a model with equal transition rates (ER), the same with a short-lived ancestor (ER-root), a model with different rates and transitions between short-lived species and cushions set to zero (NEJ for “No Evolutionary Jumps”) and the same with a short-lived ancestor (NEJ-root). Models were compared using AIC. The possibility of having a short-lived ancestor of the genus is highly motivated by the fact that the cushion life form is a derived character in most angiosperm genera (S. Aubert, unpubl. data). Following model selection, we used the best model to run evolutionary simulations of life-form evolution. This was done by drawing histories of life forms from the joint likelihood of each life form on every node of the phylogeny (100 character histories per tree, following Fitzjohn et al. 2009)
RELATION BETWEEN LIFE FORM AND NICHE EVOLUTION
To investigate the role played by life-form transitions in climatic niche evolution and see whether the cushion life form is a key innovation in this genus, we fitted different models of niche evolution to the data, covering a broad range of evolutionary scenarios. We compared a BM model of evolution with various Orstein–Ulhenbeck models (OU, Butler and King 2004). BM models the random walk of niche values; it has been introduced as a model for genetic drift but can also be interpreted as a model of selection in a fluctuating environment, like the one that ancestors of Androsace probably experienced. The OU model includes both a random walk and a constraint term. It has been designed to model selection of a continuous character along a phylogeny (Butler and King 2004), with one or several selective optima depending on a priori hypotheses (e.g., Lavergne et al. 2010b). A good fit of an OU model is often interpreted as evidence of evolutionary constraints acting on the character under study. Here, we use OU models to model habitat selection along climatic gradients depending on different possible selective scenarios. The first OU model has a single optimum, common to all current and ancestral species (OU1). In this case, the model assumes that there is a single optimal niche for all the species of the group. The second model (OU2) has two optima depending on well-delimitated clades (Central Asian vs. American–European, see Results), which would imply that different climatic optima have evolved into separate geographic regions, for example due to different specific genetic adaptations of the ancestors of each clade or to climatic differences between the two regions placing different constraints on niche evolution. The last model (OU3) is a model with three optima depending on life form and parameterized with the ancestral reconstructions performed as described above. This last model makes the hypothesis that the three different life forms are suited to different climatic conditions and could indicate that they selected for different optimal climatic tolerances.
Results
PHYLOGENETIC INFERENCE
To date, the phylogeny obtained in this study is the most complete for Androsace, including 77 taxa and 72 recognized species of the genus. The topology of the 50% majority-rule consensus tree (Appendix S5) is totally congruent with the results obtained by two complementary works that explored the phylogenetic relationships within Androsace and allied genera (Schneeweiss et al. 2004; Wang et al. 2004). Clade supports are, in general, equal or higher to those obtained in these two studies. The ingroup species are divided into two well-supported clades: one constituted by species distributed mainly in Central Asia, the other formed mainly by American and/or European species, and including the nested genera Douglasia, Vitaliana, and Pomatosace (Fig. 1).
Divergence time estimation analyses yielded older age estimates for all nodes (especially deeper ones) than those obtained in previous studies (Schneeweiss et al. 2004; Wang et al. 2004). This is probably due mainly to calibration based on the updated phylogeny of Primulaceae made by Yesson et al. (2009), who obtained a date of 35.4–34.7 Mya instead of the 23 Mya reported by Schneeweiss et al. (2004). The split between the two main clades of Androsace s. l. would have occurred 34.9 Mya (95% CI: 34.5–35.3 Mya). The genus Pomatosace, sister to the European–American clade, would have diverged 31.9 Mya (95% CI: 28.8–34.2 Mya). In contrast, the two other genera nested within Androsace, that is, Vitaliana and Douglasia, are much more recent: Vitaliana would have appeared 1.8 Mya (95% CI: 1.3–8.5 Mya), and the ancestor of Douglasia and A. triflora Adans. would have diversified 1.5 Mya (95% CI: 0.8–6.6 Mya). Node ages and 95% CI are reported in Appendix S6.
NICHE SEPARATION
The first two axes of the OMI analysis represented 77% of the total inertia of the points included in the study. The first axis (50% of total inertia) correlated negatively with seven (out of eight) precipitation variables, and most strongly with precipitation of the driest quarter and precipitation of the driest month. This first gradient will therefore be referred to hereafter as the “moisture gradient.” The second gradient (27% of total inertia) correlated negatively with seven (out of 11) temperature variables and above all with the mean temperature of the coldest quarter, minimal temperature of the coldest month, and mean annual temperature. This second gradient will be referred to hereafter as the “temperature gradient.” Although most Androsace species could be described as arctic-alpine or alpine species, niche separation was, however, quite significant along these two climatic axis, suggesting that strong niche diversification has occurred in the genus (species scores on the OMI axes shown in Fig. 2).
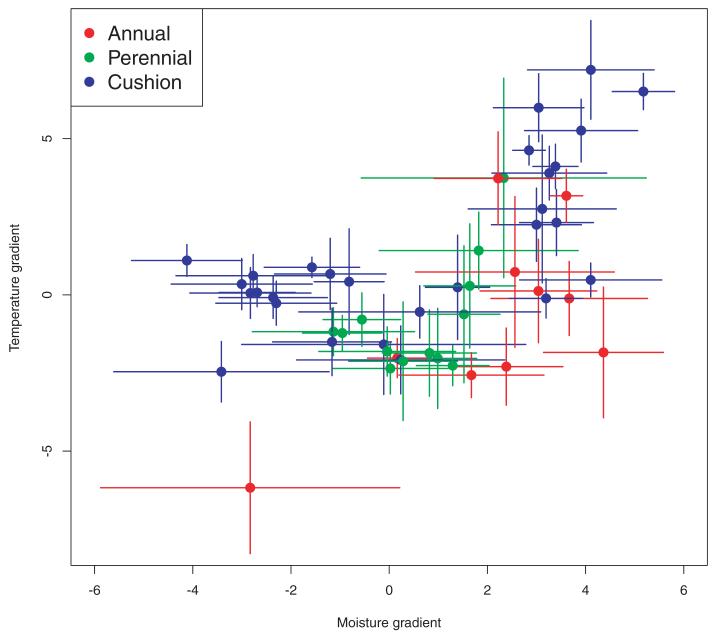
Figure 2.
Climatic niche separation. Ordination plot of mean OMI scores, with standard deviation bars, for 51 species of the Androsace genus. Moisture increases and temperature decreases when moving toward higher positive values. Species are colored according to their life forms. Cushions occupy a larger part of the climatic space than other life forms.
EVOLUTIONARY HISTORY OF THE NICHE
Although our two estimates of phylogenetic signal were not always convergent, the common pattern was a moderate phylogenetic signal for the niche distributions along the two climatic axes for all sampled phylogenetic trees (Table 1). However, K was never estimated to be greater than 1 and lambda was never close to 1, suggesting a phylogenetic signal lower than the one expected under BM.
Table 1. Estimated indices of phylogenetic signal, tempo, and mode of evolution. For each index used, the mean (± standard deviation) of all values obtained over the 10,000 resamples is presented for each OMI axis. The two exponents indicate the percentage of P-values ≤ 0.05 in the tests against the particular values of 0 and 1, respectively (K is only tested against 0). Note that δ stops at 2.99 due to its calculation in the geiger package in R.
| K | λ | δ | κ | |
|---|---|---|---|---|
| OMI 1 | 0.29 (±0.08)85− | 0.63 (±0.21)83 92 | 2.99 (±0.02)100 100 | 0.21 (±0.19)100 100 |
| OMI 2 | 0.29 (±0.09)72− | 0.64 (±0.20)68 90 | 2.99 (±0.05)100 100 | 0.16 (±0.16)100 100 |
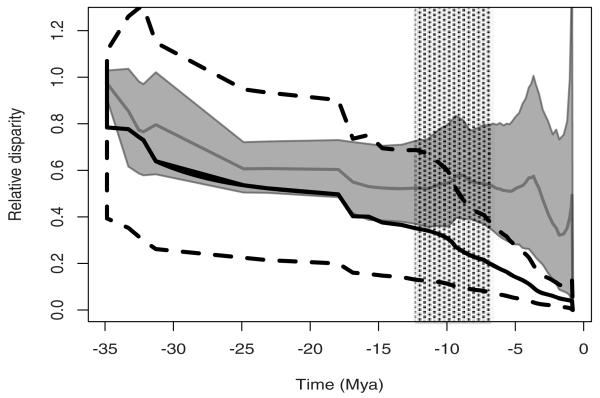
On the two environmental axes, δ was always greater than 1 (δ = 2.999 for 99.8% of the resamples on axis 1 and for 99.4% on axis 2, Table 1), thus clearly confirming the expected late diversification of climatic niches. Estimated κ values tend to be quite small for the two axes of the climatic niche (Table 1), indicating that niches have tended to evolve in a punctual way, that is, quite independently of branch lengths. Disparity plots revealed that climatic niches have evolved in a way that is not discernable from BM for most of the evolutionary history of Androsace (Fig. 3). This was followed by a fast and marked increase of the niche disparity of climatic niches occurring relatively recently in evolutionary time, with this burst of niche disparification starting approximately around 10 Mya. The MDI obtained across all the resamples was recurrently positive (mean = 0.182, SD = 0.075), suggesting that niche disparity was mostly distributed within subclades and thus confirming the late evolution of climatic niches.
Figure 3.
Relative disparity through time (DTT) for the climatic niche in Androsace. In black, the bold and dashed lines indicate, respectively, the mean and the 5% and 95% quantiles of the Brownian Motion simulations (10 simulations for each resampling step). The bold line in gray and the shaded area indicate the mean and 95% envelope of the observed disparities in the 10,000 resamples taken from niches and trees. As trees have different branching times, all curves are plotted against the mean time of each speciation event across the 100 trees. The dashed area indicates the approximate period between the two appearances of the cushion life form, probably in Asia (−12.5 ± 2.8 Mya) and in Europe (−7.4 ± 2.6 Mya), which seems to coincide with the increase in disparity.
EVOLUTION OF LIFE FORM
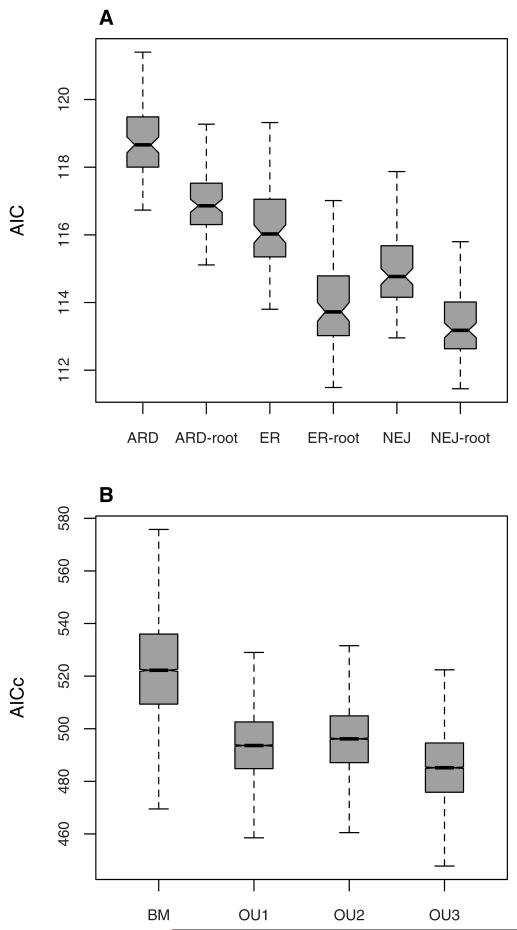
The six different discrete Markov models of life-form evolution yielded significantly different AIC values, with the model with the lowest AIC distribution being NEJ-root (Fig. 4A), that is a model with all the rates being different, forbidden transitions between short-lived species and cushions, and the ancestor forced to be short-lived. Rates of transition between perennials and cushions were on average about 10 times higher than rates for the transitions between short-lived species and perennials (qshortlived→perennial = 0.023 Mya−1, qperennial→shortlived = 0.0424 Mya−1, qperennial→cushion = 0.541 Mya−1, qcushion→perennial = 0.268 Mya−1), suggesting that shifts between the short-lived and the perennial life form seldom occurred in the genus. Our results mainly agree with those of Schneeweiss et al. (2004). Reconstructions showed that the cushion life form appeared independently in the two main clades. Hence, we can affirm that it is a homoplasy shared by some Himalayan and some Western (European and North American) species. Only three reversals from perennials toward short-lived species were inferred, whereas Schneeweiss et al. (2004) found at least four. This new result is due to the improvement of the phylogeny, which includes more Central Asian species and thus helps to resolve uncertainties in this clade. We used ancestral life-form reconstructions made using the NEJ-root model for fitting the OU3 model in the following section (marginal likelihoods of the three ancestral states at each node of the phylogeny are shown on Fig. 1).
Figure 4.
Evolutionary models comparisons. (A) AIC distributions of the six Mk models fitted on the 100 trees with 62 species. (B) AICc distributions of the four niche evolution models, fitted for 51 species on 10,000 resamples from trees, niches, and ancestral life forms where necessary. All AIC and AICc distributions are significantly different according to Wilcoxon signed-rank tests (all P-values < 0.01).
INFLUENCE OF LIFE FORM ON NICHE EVOLUTION
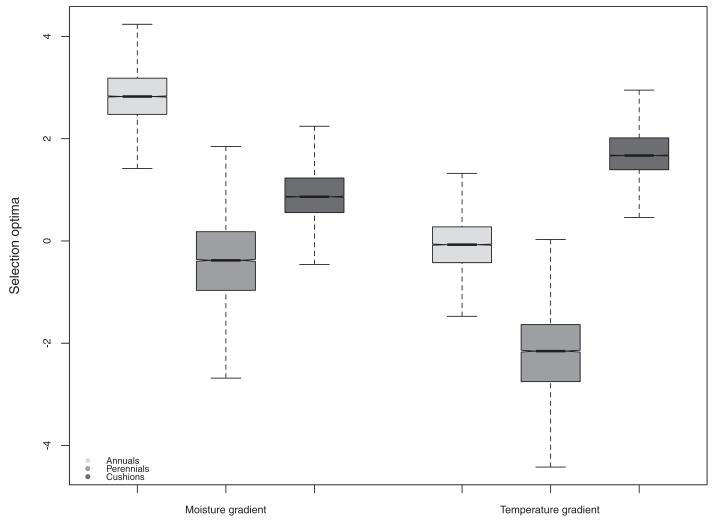
Different evolutionary scenarios yielded extremely variable AICc distributions, with BM generally yielding the highest AICc values (mean = 523, SD = 20). OU3 had the lowest AICc values (mean = 485, SD = 15), and was therefore the model that best described niche evolution among the models we compared (Fig. 4B). Climatic optima estimated for different life forms and across all trees, drawn niche values and ancestral state reconstructions showed that short-lived species displayed the driest climatic optimum, and that the coldest temperature optimum was for cushion species (Fig. 5).
Figure 5.
Estimation of the niche evolutionary optima for the OU3 model on the two main environmental gradients, for the three different life forms. All optima distributions are significantly different according to Wilcoxon signed-rank tests (all P-values < 0.01). The driest niches have been selected in short-lived species and the coldest in cushions.
Discussion
One of the main arguments supporting the importance of niche conservatism is that most clades generally occupy only one or a few given biomes (e.g., Crisp et al. 2009 for plants). This observation leads to the conclusion that adaptations to new climates have often been difficult (Donoghue 2008), carrying important implications for the future distribution of species in the face of climate change. However, a few biological groups have managed to colonize extreme environments over relatively short timescales, thus challenging common wisdom about niche conservatism. The macroevolutionary study of such groups is therefore necessary to reveal the general mechanisms that enable adaptation to changing environments. Our study of niche and trait evolution in Androsace provides one of these much-needed examples.
NICHE LABILITY IN ANDROSACE
This study reveals significant evolutionary lability of climatic niches in the Androsace genus, as indicated by a weak phylogenetic signal and strong interspecific disparity in climatic optima along temperature and moisture gradients. Although most Androsace species tend to occur in relatively cold environments, niche separation illustrates clear differences between their climatic requirements. For example, the mean annual temperature in the genus ranges from −10.4°C for Douglasia ochotensis to 18.4°C for A. umbellata, European species occupying slightly wetter and warmer environments than American or Central Asian ones (Appendix S1). Dimensions of niche differentiation vary between the two main Androsace clades. In the Central Asian clade, species’ niches are primarily distinguished along the temperature gradient. For example, A. umbellata, found in New Guinea, has the warmest niche position and shows almost no niche overlap with other Androsace species. Within the American-European clade, niche differentiation is mainly driven by the moisture gradient, which is due to the Rocky Mountains or Alaska regions being relatively drier than European mountain ranges. Phylogenetic comparative analyses of climatic niches in the Androsace genus reveal the reasons for this lability. Indeed, the recent diversification of species’ niches (high values of δ and positive MDI) through punctual evolution (low values of κ) is likely to have erased most of the phylogenetic legacy in species environmental requirements, resulting in low phylogenetic signal. We can conclude that the climatic niches of Androsace have not been conserved through evolutionary time but instead underwent a late and rapid radiation.
These findings are in agreement with the prediction that isolation of lineages leads to niche evolution (Ackerly 2003; Donoghue 2008). Indeed, the orogeny of the Alpide belt (the mountain range that extends along the southern margin of Eurasia, from the Pyrenees to New Guinea) started around 40 Mya (Nikonov 1988) and proceeded at high rates until the present day, leading to the sharp topography we observe today (Hergarten et al. 2010). Ancestral lineages of Androsace probably rode this mountain uplift, thus becoming progressively isolated from each other by eroded valleys (Kuhlemann 2007) and lowlands. The lack of zoochory opportunities for plants that do not produce fleshy fruits and the absence of seed adaptation to long distance dispersal (as noted in A. alpina by Schonswetter et al. 2003) may have increased this geographic isolation. In addition, Cenozoic Era climatic and glacial oscillations (Zachos et al. 2008) are likely to have alternately opened pathways between populations and created environmental barriers between favorable regions. The punctual evolution of the climatic niche we observed in Androsace could be the result of colonization events enabled by the opening of these pathways during colder periods, followed by speciation and rapid adaptation after isolation. Numerous cases of rapid niche disparification in plant lineages following colonization of island-like habitats have already been documented. Perhaps the most striking cases are those of the Hawaiian silverswords (Witter and Carr 1988 Evolution) and of the Andean species of the genus Lupinus (Hughes et al. 2006), both displaying tremendous variation in life form (from weeds of a few centimeters to trees) and habitat. The case of Androsace is however different because it has less variation in life form and habitat but higher variation in climatic tolerances.
THE RISE OF THE CUSHION LIFE FORM
Interestingly, the diversification of climatic niches observed in Androsace has been enhanced by life-form evolution. Short-lived Androsace tend to occupy the driest niches (e.g., dry steppes and plains), which is consistent with previous observations that seed dormancy of annual plants prevents local extinction during drought periods (Körner 1999), even in dry alpine regions (e.g., the central Chilean Andes, Arroyo et al. 1999). The fact that the coldest environments are occupied by cushion Androsace corroborates eco-physiological evidence that the cushion’s dense canopy acts as a temperature variation buffer by elevating the temperature inside the plant (Körner 1999; Larcher et al. 2010), thus enabling tolerance to colder conditions.
Our study reveals that the ancestor of Androsace was a short-lived species, probably therefore inhabiting cold steppes in Eurasia. The cushion life form then appeared independently in the two main clades, as they were already geographically isolated in Asia and Europe (Schneeweiss et al. 2004). These two events occurred roughly around 12.5 Mya in Asia and 7.4 Mya in Europe (see Fig. 1, nodes 24 and 57 are shown in Appendix S5 and dated in Appendix S6). Similar cases of repeated evolution of a morphological trait in a clade have already been studied and can be explained by similar selection pressures in geographically isolated environments (Wiens et al. 2006). Even if our large-scale approach cannot directly demonstrate it, the convergent evolution of the cushion life form in the Central Asian and American–European clades could be attributed to strong selection pressures toward cold tolerance resulting from the rapid rise of the Alpide belt in the Miocene and to similar biotic contexts (i.e., temperate biomes of the Northern Hemisphere, Fine and Ree 2006).
THE CUSHION LIFE FORM: A KEY INNOVATION
Although a macroevolutionary study such as ours does not enable the investigation of the precise mechanisms by which plant morphological adaptations permit particular climatic tolerances, we can suggest from our results that the evolution toward harsh alpine niches has been enabled by the emergence of the cushion life form. Indeed, the fact that a model with several selective optima (OU3) has a better fit than a model of drift (BM) model indicates that different climatic constraints have acted on the different life forms. This result is even stronger when we note that the ancestors of Androsace have probably experienced a fluctuating environment, a situation ideally modeled by BM. Therefore, the selection toward optimal niches in each life form has been strong enough to overcome the effects of climate fluctuations. We can consequently affirm that the cushion life form is a key innovation in the genus Androsace, leading to a significant change in the fundamental niche of the species that possessed it and allowing them to colonize the cold habitats created by the alpide orogeny. Key innovations are known to be major sources of ecological opportunity and are often associated with increased niche disparity (Losos and Mahler 2010). Even if our study does not specifically address this question, it can provide a qualitative answer to it. Indeed, the fast increase of niche disparity found here seems to coincide with the emergence of the cushion life form (Fig. 3), even if we did not specifically tested that this increase was mainly due to clades possessing the cushion life form. Moreover, cushions seem to have a broader occupancy of the climatic space than other life forms (Fig. 2). Further work is still needed to see whether cushions also stimulated diversification rates, a consequence that would be expected from such a key innovation (Glor 2010) and that has already been observed in many lineages that colonized the Andes (Donoghue 2008).
A GENERAL SCENARIO FOR THE EVOLUTION OF ALPINE PLANTS?
Putting together our results gives a clear picture of the evolution of cold tolerance in Androsace, which can be summarized in the following scenario. Ancestors of the genus, probably already adapted to the temperate conditions of cold steppes, rode the rising mountain chain of the Alpide belt. The increasing isolation of these new “continental islands” combined with poor dispersal ability prevented species from migrating back to their optimal habitats and instead forced them to adapt in situ to survive. Under the strong selection pressures for increased cold tolerance that these species experienced, the cushion life form evolved as a key morphological innovation, enabling the occupation of novel alpine habitats.
This scenario is strikingly similar to the one inferred for the evolution of Espeletia (Asteraceae) by Monasterio and Sarmiento (1991). This Andean plant genus originated as a rainforest tree and subsequently colonized the high-altitude paramo habitats created by the Andean orogeny due to the evolution of the pachycaulous life form. The similar environmental conditions and orogenic history along with the presence of the same pachycaulous morphology in several genera of tropical-alpine plants such as Puya in the Andes and Dendrosenecio and Lobelia in East Africa suggest that their evolutionary history could have been similar. Despite differences between climatic conditions in tropical highlands and cold areas of the Northern Hemisphere (Körner 2007), the scenario proposed above may quite effectively describe the general stages for the evolution of adaptation to alpine habitats in plants. Several plant genera distributed in the Holarctic ecozone and that contain species bearing the cushion life form such as Saxifraga, Draba, or Silene would be preferred candidates to test the generality of this scenario.
ASSUMPTIONS AND POSSIBLE SOURCES OF ERROR
Our study potentially presents some limitations. First, there is a strong disequilibrium between Central Asian and American–European species, the former being sampled a lot less (in terms of the occurrence points as well as for the phylogeny). However, the analysis conducted on a subset of species with at least 15 points (a number that has been demonstrated to be large enough for a reliable niche estimation, Stockwell and Peterson 2002) shows the robustness of our results (Appendix S2) and the low fit of the OU2 model compared to OU3 also indicates that climatic differences between clades are not as important as differences explained by biological traits. Second, we used climate data obtained by large-scale interpolations and thus containing potential errors. It is hoped that the resampling procedure we applied on the niche distributions helped to alleviate the effects of these errors. We must also recognize that as in all attempts to incorporate climatic variables in studies of macroevolution, we cannot rule out that climatic variables were differently correlated in the past. As already mentioned, soil ecology is known to be an important driver of alpine plants distributions (Alvarez et al. 2009) and could be a confounding factor in our work. This variable was anyway not available at such a large geographic scale and we believe moreover that this large scale prevents soil type from being totally confounded with one climatic variable. Lastly, it is important to stress that microevolutionary experiments on the selective advantage of different life forms in different environmental conditions would really help to corroborate our speculation concerning the parallel evolution of cushions and alpine niches.
Conclusion
In a recent debate on the niche conservatism paradigm, some authors (e.g., Losos 2008b) suggested that more studies on different groups were necessary to assess the generality of this phenomenon. Moving away from the extensively studied examples of groups of reptiles and amphibians, we provide, through comprehensive analysis, a robust and clear answer for a genus of arctic-alpine plants in which niche lability prevails. As noted by some authors (e.g., Wiens 2008; Cooper et al. 2010), raw estimations of phylogenetic signal are of little interest when attempting to understand niche evolution. Including simple measures such as the tempo and mode of evolution of the niche, as well as a biological trait, enables us to better understand niche evolution in a particular group, making the link between ecology and evolution clearer. In doing so, we showed how occupation of alpine niches in Androsace has been triggered by the emergence of a key innovation: the cushion life form.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to all the people who contributed to GBIF, and to the botanists from the CBNA, the CBNMED, and the CRSF. We also thank M. Alfaro, L. Sack, and two anonymous reviewers for constructive criticism and advice on this work, and R. Fitzjohn for help with functions of the “diversitree” package. L. Gallien provided useful feedback on the focus of the study. Thanks also to Version Originale for checking and correcting the English in this article. This work was funded by the French “Agence Nationale de la Recherche” with the EVORANGE (ANR-09-PEXT-011) project, and by the European Commission’s FP6 ECOCHANGE project (Contract No. 066866 GOCE). The grant to FB was provided by the Ecole Polytechnique, Saclay (AMX 2010-2013). NA was funded by the Swiss National Science Foundation (Ambizione fellowship PZ00P3_126624). CR was supported by a grant from the Fundación Ramón Areces.
Footnotes
The following supporting information is available for this article:
Appendix S1. List of all taxa included in the study, showing all GenBank IDs.
Appendix S2. Sensitivity analysis.
Appendix S3. Utility of the resampling procedure.
Appendix S4. Overall workflow of the comparative study.
Appendix S5. Chronogram based on Bayesian 50% majority-rule consensus tree.
Appendix S6. Node ages estimated for the Bayesian 50% majority-rule consensus tree, standard deviation (SD), and 95% confidence interval (95% CI) for each node.
Supporting Information may be found in the online version of this article.
Please note: Wiley-Blackwell is not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
LITERATURE CITED
- Ackerly DD. Community assembly, niche conservatism, and adaptive evolution in changing environments. Int. J. Plant Sci. 2003;164:S165–S184. [Google Scholar]
- Albert CH, Thuiller W, Yoccoz NG, Douzet R, Aubert S, Lavorel S. A multi-trait approach reveals the structure and the relative importance of intra- vs. interspecific variability in plant traits. Funct. Ecol. 2010;24:1192–1201. [Google Scholar]
- Alvarez N, Thiel-Egenter C, Tribsch A, Holderegger R, Manel S, Schonswetter P, Taberlet P, Brodbeck S, Gaudeul M, Gielly L, et al. History or ecology? Substrate type as a major driver of patial genetic structure in Alpine plants. Ecol. Lett. 2009;12:632–640. doi: 10.1111/j.1461-0248.2009.01312.x. [DOI] [PubMed] [Google Scholar]
- Arroyo MTK, Cavieres LA, Castor C, Humana AM. Persistent soil seed bank and standing vegetation at a high alpine site in the central Chilean Andes. Oecologia. 1999;119:126–132. doi: 10.1007/s004420050768. [DOI] [PubMed] [Google Scholar]
- Blomberg SP, Garland T. Tempo and mode in evolution: phylogenetic inertia, adaptation and comparative methods. J. Evol. Biol. 2002;15:899–910. [Google Scholar]
- Blomberg SP, Garland T, Ives AR. Testing for phylogenetic signal in comparative data: behavioral traits are more labile. Evolution. 2003;57:717–745. doi: 10.1111/j.0014-3820.2003.tb00285.x. [DOI] [PubMed] [Google Scholar]
- Butler MA, King AA. Phylogenetic comparative analysis: a modeling approach for adaptive evolution. Am. Nat. 2004;164:683–695. doi: 10.1086/426002. [DOI] [PubMed] [Google Scholar]
- Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cholewa AF, Kelso S. In: Primulaceae. Flora of North America Editorial Committee, editor. Flora of North America North Mexico; New York and Oxford: 2009. 1993+. [Google Scholar]
- Cooper N, Jetz W, Freckleton RP. Phylogenetic comparative approaches for studying niche conservatism. J. Evol. Biol. 2010;23:2529–2539. doi: 10.1111/j.1420-9101.2010.02144.x. [DOI] [PubMed] [Google Scholar]
- Crisp MD, Arroyo MTK, Cook LG, Gandolfo MA, Jordan GJ, McGlone MS, Weston PH, Westoby M, Wilf P, Linder HP. Phylogenetic biome conservatism on a global scale. Nature. 2009;458:754–U90. doi: 10.1038/nature07764. [DOI] [PubMed] [Google Scholar]
- Darwin CR. On the origin of species by means of natural selection, or the preservation of favoured races in the struggle for life. John Murray; London: 1859. [PMC free article] [PubMed] [Google Scholar]
- Dixon CJ, Schonswetter P, Suda J, Wiedermann MM, Schneeweiss GM. Reciprocal Pleistocene origin and postglacial range formation of an allopolyploid and its sympatric ancestors (Androsace adfinis group, Primulaceae) Mol. Phylogenet. Evol. 2009;50:74–83. doi: 10.1016/j.ympev.2008.10.009. [DOI] [PubMed] [Google Scholar]
- Dolédec S, Chessel D, Gimaret-Carpentier C. Niche separation in community analysis: a new method. Ecology. 2000;81:2914–2927. [Google Scholar]
- Donoghue MJ. A phylogenetic perspective on the distribution of plant diversity. Proc. Natl. Acad. Sci. USA. 2008;105:11549–11555. doi: 10.1073/pnas.0801962105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dray S, Dufour AB. The ade4 package: implementing the duality diagram for ecologists. J. Stat. Soft. 2007;22:1–20. [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards EJ, Smith SA. Phylogenetic analyses reveal the shady history of C-4 grasses. Proc. Natl. Acad. Sci. USA. 2010;107:2532–2537. doi: 10.1073/pnas.0909672107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans MEK, Smith SA, Flynn RS, Donoghue MJ. Climate, niche evolution, and diversification of the “Bird-Cage” Evening Primroses (Oenothera, sections Anogra and Kleinia) Am. Nat. 2009;173:225–240. doi: 10.1086/595757. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Phylogenies and the comparative method. Am. Nat. 1985;125:1–15. [Google Scholar]
- Felsenstein J. Comparative methods with sampling error and within-species variation: contrasts revisited and revised. Am. Nat. 2008;171:713–725. doi: 10.1086/587525. [DOI] [PubMed] [Google Scholar]
- Fine PVA, Ree RH. Evidence for a time-integrated species-area effect on the latitudinal gradient in tree diversity. Am. Nat. 2006;168:796–804. doi: 10.1086/508635. [DOI] [PubMed] [Google Scholar]
- Fitzjohn RG, Maddison WP, Otto SP. Estimating trait-dependent speciation and extinction rates from incompletely resolved phylogenies. Syst. Biol. 2009;58:595–611. doi: 10.1093/sysbio/syp067. [DOI] [PubMed] [Google Scholar]
- Freckleton RP, Harvey PH, Pagel M. Phylogenetic analysis and comparative data: a test and review of evidence. Am. Nat. 2002;160:712–726. doi: 10.1086/343873. [DOI] [PubMed] [Google Scholar]
- Glor RE. Phylogenetic insights on adaptive radiation. Annu. Rev. Ecol. Evol. Syst. 2010;41:251–270. [Google Scholar]
- Grinnell J. The niche-relationships of the California Trasher. Auk. 1917;34:131–135. [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- Harmon LJ, Losos JB. The effect of intraspecific sample size on type 1 and type 11 error rates in comparative studies. Evolution. 2005;59:2705–2710. [PubMed] [Google Scholar]
- Harmon LJ, Schulte JA, Larson A, Losos JB. Tempo and mode of evolutionary radiation in iguanian lizards. Science. 2003;301:961–964. doi: 10.1126/science.1084786. [DOI] [PubMed] [Google Scholar]
- Harmon LJ, Weir JT, Brock CD, Glor RE, Challenger W. GEIGER: investigating evolutionary radiations. Bioinformatics. 2008;24:129–131. doi: 10.1093/bioinformatics/btm538. [DOI] [PubMed] [Google Scholar]
- Hergarten S, Wagner T, Stüwe K. Age and prematurity of the Alps derived from topography. Earth Planet. Sci. Lett. 2010;297:453–460. [Google Scholar]
- Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A. Very high resolution interpolated climate surfaces for global land areas. Int. J. Clim. 2005;25:1965–1978. [Google Scholar]
- Hu Q, Kelso S. In: Primulaceae. Flora of China Editorial Committee, editor. Flora of China. Science Press; Missouri Botanical Garden Press; Beijing, China: St. Louis, MO: 2007. 1994+. [Google Scholar]
- Hughes C, Eastwood R. Island radiation on a continental scale: Eeceptional rates of plant diversification after uplift of the Andes. Proc. Natl. Acad. Sci. USA. 2006;103:10334–10339. doi: 10.1073/pnas.0601928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson GE. Concluding remarks. Cold Spring Harbor Symp. Quant. Biol. 1957;22:145–159. [Google Scholar]
- Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kembel SW. Disentangling niche and neutral influences on community assembly: assessing the performance of community phylogenetic structure tests. Ecol. Lett. 2009;12:949–960. doi: 10.1111/j.1461-0248.2009.01354.x. [DOI] [PubMed] [Google Scholar]
- Kembel SW, Cowan PD, Helmus MR, Cornwell WK, Morlon H, Ackerly DD, Blomberg SP, Webb CO. Picante: R tools for integrating phylogenies and ecology. Bioinformatics. 2009;26:1463–1464. doi: 10.1093/bioinformatics/btq166. [DOI] [PubMed] [Google Scholar]
- Kishino H, Thorne JL, Bruno WJ. Performance of a divergence time estimation method under a probabilistic model of rate evolution. Mol. Biol. Evol. 2001;18:352–361. doi: 10.1093/oxfordjournals.molbev.a003811. [DOI] [PubMed] [Google Scholar]
- Körner C. Alpine plant life. Springer-Verlag; Berlin: 1999. [Google Scholar]
- Körner C. The use of ‘altitudinal’ in ecological research. Trends Ecol. Evol. 2007;22:569–574. doi: 10.1016/j.tree.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Kozak KH, Wiens JJ. Accelerated rates of climatic-niche evolution underlie rapid species diversification. Ecol. Lett. 2010;13:1378–1389. doi: 10.1111/j.1461-0248.2010.01530.x. [DOI] [PubMed] [Google Scholar]
- Kuck P, Meusemann K. FASconCAT: convenient handling of data matrices. Mol. Phylogenet. Evol. 2010;56:1115–1118. doi: 10.1016/j.ympev.2010.04.024. [DOI] [PubMed] [Google Scholar]
- Kuhlemann J. Paleogeographic and paleotopographic evolution of the Swiss and Eastern Alps since the Oligocene. Glob. Planet. Change. 2007;58:224–236. [Google Scholar]
- Larcher W, Kainmuller C, Wagner J. Survival types of high mountain plants under extreme temperatures. Flora. 2010;205:3–18. [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lassmann T, Sonnhammer EL. Kalign—an accurate and fast multiple sequence alignment algorithm. BMC Bioinform. 2005;6:298. doi: 10.1186/1471-2105-6-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassmann T, Sonnhammer EL. Kalign, Kalignvu and Mumsa: web servers for multiple sequence alignment. Nucleic Acids Res. 2006;34:W596–W599. doi: 10.1093/nar/gkl191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber K, Wagner G. Flora Helvetica: Flore illustrée de la Suisse. Belin; Paris: 2007. [Google Scholar]
- Lavergne S, Mouquet N, Thuiller W, Ronce O. Biodiversity and climate change: integrating evolutionary and ecological responses of species and communities. Annu. Rev. Ecol. Evol. Syst. 2010a;41:321–350. [Google Scholar]
- Lavergne S, Muenke NJ, Molofsky J. Genome size reduction can trigger rapid phenotypic evolution in invasive plants. Ann. Bot. 2010b;105:109–116. doi: 10.1093/aob/mcp271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis PO. A likelihood approach to estimating phylogeny from discrete morphological character data. Syst. Biol. 2001;50:913–925. doi: 10.1080/106351501753462876. [DOI] [PubMed] [Google Scholar]
- Loarie SR, Duffy PB, Hamilton H, Asner GP, Field CB, Ackerly DD. The velocity of climate change. Nature. 2009;462:1052–1055. doi: 10.1038/nature08649. [DOI] [PubMed] [Google Scholar]
- Losos JB. Uncertainty in the reconstruction of ancestral character states and limitations on the use of phylogenetic comparative methods. Anim. Behav. 1999;58:1319–1324. doi: 10.1006/anbe.1999.1261. [DOI] [PubMed] [Google Scholar]
- Losos JB. Phylogenetic niche conservatism, phylogenetic signal and the relationship between phylogenetic relatedness and ecological similarity among species. Ecol. Lett. 2008a;11:995–1003. doi: 10.1111/j.1461-0248.2008.01229.x. [DOI] [PubMed] [Google Scholar]
- Losos JB. Rejoinder to Wiens (2008). Phylogenetic niche conservatism, its occurrence and importance. Ecol. Lett. 2008b;11:1005–1007. [Google Scholar]
- Losos JB, Mahler DL. Adaptive radiation: the interaction of ecological opportunity, adaptation, and speciation. In: Bell MA, Futuyama DJ, Eanes WF, Levinton JS, editors. Evolution since Darwin: the first 150 years. Sinauer Associates.; Sunderland, MA: 2010. pp. 381–420. [Google Scholar]
- Luxbacher AM, Knouft JH. Assessing concurrent patterns of environmental niche and morphological evolution among species of horned lizards (Phrynosoma) J. Evol. Biol. 2009;22:1669–1678. doi: 10.1111/j.1420-9101.2009.01779.x. [DOI] [PubMed] [Google Scholar]
- Mace GM, Gittleman JL, Purvis A. Preserving the tree of life. Science. 2003;300:1707–1709. doi: 10.1126/science.1085510. [DOI] [PubMed] [Google Scholar]
- Martins L, Oberprieler C, Hellwig FH. A phylogenetic analysis of Primulaceae s.l. based on internal transcribed spacer (ITS) DNA sequence data. Plant Syst. Evol. 2003;237:75–85. [Google Scholar]
- Mast AR, Kelso S, Conti E. Are any primroses (Primula) primitively monomorphic? New Phytol. 2006;171:605–616. doi: 10.1111/j.1469-8137.2006.01700.x. [DOI] [PubMed] [Google Scholar]
- Miller AH. Some ecologic and morphologic considerations in the evolution of higher taxonomic categories. In: Mayr E, Schüz E, editors. Ornithologie als Biologische Wissenschaft. Carl Winter; Heidelberg: 1949. pp. 84–88. [Google Scholar]
- Monasterio M, Sarmiento L. Adaptive radiation of Espeletia in the cold Andean tropics. Trends Ecol. Evol. 1991;6:387–391. doi: 10.1016/0169-5347(91)90159-U. [DOI] [PubMed] [Google Scholar]
- Nikonov AA. The rate of uplift in the alpine mobile belt. Tectono-physics. 1988;163:267–276. [Google Scholar]
- Nylander JA. MrModeltest. Evolutionary Biology Centre, Uppsala University; 2004. Program distributed by the author. [Google Scholar]
- Nylander JAA, Wilgenbusch JC, Warren DL, Swofford DL. AWTY (are we there yet?): a system for graphical exploration of MCMC convergence in Bayesian phylogenetics. Bioinformatics. 2008;24:581–583. doi: 10.1093/bioinformatics/btm388. [DOI] [PubMed] [Google Scholar]
- Pagel M. Inferring evolutionary processes from phylogenies. Zool. Scri. 1997;26:331–348. [Google Scholar]
- Pagel M. Inferring the historical patterns of biological evolution. Nature. 1999;401:877–884. doi: 10.1038/44766. [DOI] [PubMed] [Google Scholar]
- Paradis E, Claude J, Strimmer K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- Parmesan C. Ecological and evolutionary responses to recent climate change. Annu. Rev. Ecol. Evol. Syst. 2006;37:637–669. [Google Scholar]
- Pinto G, Mahler DL, Harmon LJ, Losos JB. Testing the island effect in adaptive radiation: rates and patterns of morphological diversification in Caribbean and mainland Anolis lizards. Proc. R. Soc. Lond. B. 2008;275:2749–2757. doi: 10.1098/rspb.2008.0686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team, editor. R: a language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: [Accessed September 12, 2011]. 2011. Available at http://www.R-project.org. [Google Scholar]
- Randin C, Engler R, Normand S, Zappa M, Zimmermann NE, Pearman P, Vittoz P, Thuiller W, Guisan A. Climate change and plant distribution: local models predict high-elevation persistence. Glob. Change Biol. 2009;15:1557–1569. [Google Scholar]
- Revell LJ, Harmon LJ, Collar DC. Phylogenetic signal, evolutionary process, and rate. Syst. Biol. 2008;57:591–601. doi: 10.1080/10635150802302427. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Ruffier-Lanche R. Les plantes en coussinet. Bulletin de la Société des Amateurs de Jardins Alpins. 1964;49:3–13. [Google Scholar]
- Schneeweiss GM, Schonswetter P, Kelso S, Niklfeld H. Complex biogeographic patterns in Androsace (Primulaceae) and related genera: evidence from phylogenetic analyses of nuclear internal transcribed spacer and plastid trnL-F sequences. Syst. Biol. 2004;53:856–876. doi: 10.1080/10635150490522566. [DOI] [PubMed] [Google Scholar]
- Schonswetter P, Schneeweiss GM. Androsace komovensis sp nov., a long mistaken local endemic from the southern Balkan Peninsula with biogeographic links to the Eastern Alps. Taxon. 2009;58:544–549. [Google Scholar]
- Schönswetter P, Tribsch A, Niklfeld H. Phylogeography of the high alpine cushion plant Androsace alpina (Primulaceae) in the European Alps. Plant Biol. 2003;5:623–630. [Google Scholar]
- Stockwell DRB, Peterson AT. Effects of sample size on accuracy of sepcies distribution models. Eco. Mod. 2002;148:1–13. [Google Scholar]
- Swofford DL. PAUP* Phylogenetic Analysis using Parsimony (* and other methods), v.4.0 beta 10. Sinauer Associates; Sunderland, MA: 2002. [Google Scholar]
- Thorne JL, Kishino H. Divergence time and evolutionary rate estimation with multilocus data. Syst. Biol. 2002;51:689–702. doi: 10.1080/10635150290102456. [DOI] [PubMed] [Google Scholar]
- Thuiller W, Lavergne S, Roquet C, Boulangeat I, Araujo MB. Consequences of climate change on the tree of life in Europe. Nature. 2011;470:531–534. doi: 10.1038/nature09705. [DOI] [PubMed] [Google Scholar]
- Thuiller W, Lavorel S, Midgley G, Lavergne S, Rebelo T. Relating plant traits and species distributions along bioclimatic gradients for 88 Leucadendron taxa. Ecology. 2004;85:1688–1699. [Google Scholar]
- Thuiller W, Richardson DM, Pysek P, Midgley GF, Hughes GO, Rouget M. Niche-based modelling as a tool for predicting the risk of alien plant invasions at a global scale. Glob. Change Biol. 2005;11:2234–2250. doi: 10.1111/j.1365-2486.2005.001018.x. [DOI] [PubMed] [Google Scholar]
- Tutin TG, Burges NA, Chater AO, Edmondson JR, Heywood VH, Moore DM, Valentine DH, Walters SM, Webb DA. Flora Europaea. Cambridge Univ. Press; Cambridge: 1964-1993. [Google Scholar]
- Verdú M. Tempo, mode and phylogenetic associations of relative embryo size evolution in angiosperms. J. Evol. Biol. 2006;19:625–634. doi: 10.1111/j.1420-9101.2005.00998.x. [DOI] [PubMed] [Google Scholar]
- Wang YJ, Li XJ, Hao G, Liu JQ. Molecular phylogeny and biogeography of Androsace (Primulaceae) and the convergent evolution of cushion morphology. Acta Phytotaxon. Sin. 2004;42:481–499. [Google Scholar]
- Wiens JJ, Brandley MC, Reeder TW. Why does a trait evolve multiple times within a clade? Repeated evolution of snakelike body form in squamate reptiles. Evolution. 2006;60:123–141. [PubMed] [Google Scholar]
- Wiens JJ. Commentary on Losos (2008): niche conservatism deja vu. Ecol. Lett. 2008;11:1004–1005. doi: 10.1111/j.1461-0248.2008.01238.x. [DOI] [PubMed] [Google Scholar]
- Wiens JJ, Ackerly DD, Allen AP, Anacker BL, Buckley LB, Cornell HV, Damschen EI, Davies TJ, Grytnes JA, Harrison SP, et al. Niche conservatism as an emerging principle in ecology and conservation biology. Ecol. Lett. 2010;13:1310–1324. doi: 10.1111/j.1461-0248.2010.01515.x. [DOI] [PubMed] [Google Scholar]
- Wikström N, Savolainen V, Chase MW. Evolution of the angiosperms: calibrating the family tree. Proc. R. Soc. Lond. B. 2001;268:2211–2220. doi: 10.1098/rspb.2001.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witter MS, Carr GD. Adaptive radiation and genetic differentiation in the Hawaiian silversword alliance (Compositae, Madiinae) Evolution. 1988;42:1278–1287. doi: 10.1111/j.1558-5646.1988.tb04187.x. [DOI] [PubMed] [Google Scholar]
- Woodward FI. The impact of low temperatures in controlling the geographical distribution of plants. Philos. Trans. R. Soc. Lond. B. 1990;326:585–593. [Google Scholar]
- Woodward FI. Predicting plant responses to global environmental change. New Phytol. 1992;122:239–251. doi: 10.1111/j.1469-8137.1992.tb04228.x. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Yesson C, Culham A. A phyloclimatic study of Cyclamen. BMC Evol. Biol. 2006a;6:72. doi: 10.1186/1471-2148-6-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yesson C, Culham A. Phyloclimatic modeling: combining phylogenetics and bioclimatic modeling. Syst. Biol. 2006b;55:785–802. doi: 10.1080/1063515060081570. [DOI] [PubMed] [Google Scholar]
- Yesson C, Toomey NH, Culham A. Cyclamen: time, sea and speciation biogeography using a temporally calibrated phylogeny. J. Biogeogr. 2009;36:1234–1252. [Google Scholar]
- Zachos JC, Dickens GR, Zeebe RE. An early Cenozoic perspective on greenhouse warming and carbon-cycle dynamics. Nature. 2008;451:279–283. doi: 10.1038/nature06588. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.