We used a combination of whole-genome sequencing and in vitro validation to show that mutations that activated at least two pro-growth/survival pathways mediated intrinsic resistance to BRAF inhibition in a melanoma patient. These data demonstrate how in-depth analysis can reveal intrinsic resistance to standard of care, providing an opportunity for alternative therapeutic strategies for patients who are likely to fail first-line treat-575 ment.
Keywords: BRAF, melanoma, mechanisms of resistance, intra-tumour heterogeneity, tumour evolution
Abstract
Background
BRAF is mutated in ∼42% of human melanomas (COSMIC. http://www.sanger.ac.uk/genetics/CGP/cosmic/) and pharmacological BRAF inhibitors such as vemurafenib and dabrafenib achieve dramatic responses in patients whose tumours harbour BRAFV600 mutations. Objective responses occur in ∼50% of patients and disease stabilisation in a further ∼30%, but ∼20% of patients present primary or innate resistance and do not respond. Here, we investigated the underlying cause of treatment failure in a patient with BRAF mutant melanoma who presented primary resistance.
Methods
We carried out whole-genome sequencing and single nucleotide polymorphism (SNP) array analysis of five metastatic tumours from the patient. We validated mechanisms of resistance in a cell line derived from the patient's tumour.
Results
We observed that the majority of the single-nucleotide variants identified were shared across all tumour sites, but also saw site-specific copy-number alterations in discrete cell populations at different sites. We found that two ubiquitous mutations mediated resistance to BRAF inhibition in these tumours. A mutation in GNAQ sustained mitogen-activated protein kinase (MAPK) signalling, whereas a mutation in PTEN activated the PI3 K/AKT pathway. Inhibition of both pathways synergised to block the growth of the cells.
Conclusions
Our analyses show that the five metastases arose from a common progenitor and acquired additional alterations after disease dissemination. We demonstrate that a distinct combination of mutations mediated primary resistance to BRAF inhibition in this patient. These mutations were present in all five tumours and in a tumour sample taken before BRAF inhibitor treatment was administered. Inhibition of both pathways was required to block tumour cell growth, suggesting that combined targeting of these pathways could have been a valid therapeutic approach for this patient.
introduction
Melanoma is a potentially deadly form of skin cancer. The metastatic form of the disease has a poor prognosis with a median survival of 6–9 months and an overall survival of 10%–15% [1]. BRAF, which encodes a protein kinase, is mutated in ∼42% of human melanomas. Drugs that inhibit BRAF, such as vemurafenib and dabrafenib, can achieve dramatic responses in patients whose tumours harbour V600 BRAF mutations, with objective responses in ∼50% of patients and disease stabilisation in a further ∼30% [2, 3]. Thus, BRAF is a validated therapeutic target in melanoma, but despite these impressive results, acquired (secondary) resistance is almost universal, even when the initial response leads to profound tumour regression. In most cases, acquired resistance to BRAF inhibitors is due to MAPK pathway reactivation and is driven by events that can occur upstream, downstream or even at the level of BRAF [4, 5]. In the remainder of cases, resistance is driven by mitogen-activated protein kinase /extracellular signal-regulated kinase (MEK/ERK)-independent mechanisms. Notably, inter- and intra-lesional molecular heterogeneity can also shape acquired resistance to BRAF inhibitors, resulting in different mechanisms of resistance in individual tumours from a single patient [6, 7] or the co-existence of sensitive and resistant clones within a single tumour [8].
In addition to treatment failures due to acquired resistance, ∼20% of patients present intrinsic (primary/innate) resistance and do not derive any benefit from BRAF inhibitors [2, 3].
However, while impressive advances have been made in our knowledge of mechanisms of acquired resistance, there is limited mechanistic insight into intrinsic resistance. RAC1P29S mutations have recently been reported in one patient with intrinsic resistance and two patients with transiently stable disease followed by disease progression after administration of a BRAF inhibitor [9] but the functional significance of these intriguing observations was not elucidated. We used whole-genome sequencing (WGS) to characterise five metastatic tumours from a BRAF mutant melanoma patient who presented intrinsic resistance. We observed evidence of inter-lesional heterogeneity that pointed to clonal complexity, but identified concomitant mutations in GNAQ and PTEN as the mechanisms of resistance in all five tumours. Thus, we describe complex but ubiquitous mechanisms of intrinsic resistance that explain treatment failure in this patient.
methods
tissue and blood collection
Following patient consent, tumour tissue and blood were obtained at surgery and in the outpatients department, respectively. All samples were anonymised and access to samples and clinical data was restricted in accordance with the Human Tissue Act and Multi-centre Research Ethics Committees (MREC) guidelines. All the studies were conducted in accordance with a study protocol CCR3097, approved by the Royal Marsden Hospital Research Ethics Committee on 21 October 2008.
DNA extraction
The tissue was disrupted using Precellys®24 tissue homogeniser (Precellys, Dublin, Ireland). Subsequently, the tissue was lysed in ATL buffer (Qiagen, Manchester, UK) and Proteinase K (Qiagen) for 24 h at 56°C. The DNA was purified using the DNAaesy Blood and Tissue spin-column Kit (Qiagen) according to manufacturer's instructions. DNA from buffy coat and cultured cells was isolated using the same kit. DNA was quantified using the PicoGreen dsDNA Quantification Reagent (Invitrogen) according to manufacturer's recommendations. The structural integrity of the tumour DNA was assessed by gel electrophoresis.
sanger sequencing
Regions of interest were amplified by PCR. The products were directly sequenced using dye-terminator chemistry. Samples were analysed on a 3100 Genetic Analyser (Applied Biosystems, Paisley, UK). Sequences were visualised using Sequencher software (Gene Codes Corporation, Ann Arbor, MI, USA).
WGS and somatic variant detection
Sequencing of the matched normal and tumour samples was carried out using unchained combinatorial probe anchor ligation chemistry on arrays of self-assembling DNA nanoballs [10]. The reads were aligned to the NCBI build 37 reference genome. The gross mapping yield, haploid coverage and percentage of fully called bases for each genome are reported in supplementary Table S1, available at Annals of Oncology online. Variations between the reference genome (NCBI build 37) and each of the samples were called and scored using a local de novo assembly algorithm [10].
Somatic single-nucleotide variants (SNVs) and indels between the normal and tumour samples uncovered using the calldiff function of cgatools (http://cgatools.sourceforge.net/), which assigns a somatic score to each SNV and additionally a somatic score to short insertions and deletions. The somatic score is a measure of the confidence that each SNV/indel is a true somatic variant. Somatic variants with a somatic score ≥−10 were considered high-confidence variants. Variants present in single nucleotide polymorphism database (dbSNP) were excluded from further analysis. SNVs with somatic scores <−10 in a genome were designated true somatic variants if present in one of the other genomes with a somatic score ≥−10. A somatic score threshold of ≥0 was used to identify high-confidence site-specific SNVs.
Somatic structural variations (SVs) in the primary and metastatic tumours were called using the junctiondiff function of cgatools. High-confidence SV junctions were those that had at least 10 mate-pairs in a cluster, in which de novo assembly of the junction was successful, had a high mapping diversity and for which there was an absence of specific repeat sequences on the left and right side of the junction.
copy number alterations
SNP array
Genome wide genotyping was conducted on the Affymetrix Genome-Wide Human SNP Array 6.0. Genotypes were called using the apt-probeset-genotype algorithm (birdseed-v2 method) of Affymetrix Power Tools. logR ratios (LRRs) and B-allele frequencies (BAFs) were estimated by PennCNV-Affy and allele-specfic copy-number alterations between the germline and tumour genomes were called by the ASCAT algorithm using default parameters [11].
whole-genome sequencing
Read depth was corrected for GC content and normalised for average haploid genome coverage. Log2 ratios of the tumour to normal average read depths for windows of 2 kb were calculated. These values were smoothed and segmented as implemented in the BioConductor package DNAcopy using default parameters [12]. Bootstrapped, unsupervised clustering of the WGS CNA data was conducted using the R package pvclust (http://www.is.titech.ac.jp/~shimo/prog/pvclust/).
estimation of tumour purity
We conducted an analysis of the purity of the tumours using the SNP array data with the ASCAT algorithm [11]. This algorithm reports tumour cell fractions of 69%, 62%, 59%, 80% and 66% for sites 1, 2, 3, 4 and 5, respectively.
development of a continuous cell line from patient's tumour
Tumour tissue was minced using sterile crossed blades under sterile conditions. The pieces were plated sparsely in 6-well uncoated or collagen coated plates. Enough culture media was added to cover each tissue piece and the plates incubated in a humidified atmosphere at 37°C and 5% CO2 and either 20% O2 or 3% O2. When cells emerging from tissue pieces became confluent, they were passaged at dilution of 1:2.
immunofluorescence of cultured cells
Cells were grown on cover slips in 24-well plates and fixed with 4% formaldehyde in phosphate buffer solution (PBS) for 15 min. The cells rinsed in PBS three times, for 5 min each. Blocking buffer (1% bovine serum albumin (BSA) in PBS) was applied for 60 min, following which the specimen was incubated with the primary antibody overnight at 4°C. The specimen was rinsed in PBS three times for 5 min each, and the secondary antibody applied for 2 h at room temperature. The secondary antibody was rinsed off with PBS once and DAPI once, followed by three PBS washes, 5 min each. Coverslips were mounted with Vectashield and slides sealed with clear nail varnish. The results were visualised in a confocal microscope.
reagents
For western blotting, the following antibodies were used: rabbit anti-ERK2 (C-14) (Santa Cruz Biotechnology, Heidelberg, Germany); mouse anti-Tubulin, and mouse anti-ppERK1/2 (Sigma, Dorset, UK); rabbit anti-PTEN and rabbit anti-ppAKT (Cell Signalling, Hitchin, UK). For immunofluorescence, the following antibodies were used: rabbit anti-S100 (Dako, Ely, UK) and mouse anti-HMB45 (Abcam, Cambridge, UK). MK2206 was obtained from Stratech Laboratories (Suffolk, UK). PD184352 and PLX4720 were synthesised in-house. All drugs were prepared in Dimethyl sulfoxide (DMSO). Fluorescence in situ hybridisation (FISH) probes for chromosome 3, 7 and 17 were obtained from UroVysion, Abbott Molecular.
cell culture techniques
A375P cells were cultured in DMEM supplemented with 10% foetal bovine serum. A total of 22 092 cells were cultured in MCDB153 supplemented with 20% Leibovitz's L-15, 2% foetal bovine serum, insulin (5 µg/ml) and CaCl2 (1.68 mM). A375P cells were incubated at 37°C, 10% CO2 and 20% O2. A total of 22 092 cells were incubated at 37°C, 5% CO2 and 3% O2. For inhibitor treatment, the drugs were dissolved in DMSO and added to the medium for 3 h, unless otherwise stated.
cell proliferation assays
96-well plate format
Cells were seeded at a density of 1000–5000 cells per well in 100 μl medium. Cells were treated with drug in triplicate for 3–10 days. For the longer incubations (>5 days), the media and the drugs were replenished every 3 days. Cell growth was measured using the CellTiter-Glo Luminescent Cell Viability Assay (Promega, Southampton, UK). The luminescence was recorded on a luminometer (SpectraMax M5, Molecular Devices, Wokingham, UK).
6-well plate format
Cells were seeded at a density of10 000–20 000cells per well in 1000 μl medium. Cells were treated with drug for 10 days. The media and drug were replenished every 3 days. The media were removed and cells rinsed with PBS. Two millilitres of a mixture of 6% glutaraldehyde and 0.5% crystal violet were added to each well and incubated for 30 min. Crystal violet staining was quantified using ImageJ software (www.rsb.info.nih.gov/ij).
spheroids assays
One hundred microlitre of 50% Matrigel (diluted with medium) was placed into each well of a 96-well plate. The plate was incubated at 37°C for 1 h to set the matrigel and cells (1000/well) were seeded on top of the matrigel plug. Drugs were added at the time of cell seeding and both media and drugs were replenished every 2 days. Ten to 14 days after seeding images were taken for quantification of spheroid size and branching. At least four representative images were taken of each well, and ImageJ software was used for the analysis.
small interfering RNA transfection
2.5 × 105 cells were seeded per well in a 6-well dish and incubated overnight. The following day the cells were transfected with Lipofectamine (Invitrogen, Paisley, UK) and 20 nM of the following small interfering RNA (siRNA) pools: sc-35429 (Santa-Cruz), s5886 (Life Technologies, Paisley, UK) and s5887 (Life Technologies), or scrambled control. Cells were incubated for 48 h before preparation of cell lysates, or re-plating for a proliferations assay.
expression constructs
A plasmid with the entire GNAQ coding region was purchased from Open Biosystems. The Q209P mutation was created by standard PCR-directed mutagenesis approaches. A SpeI/SphI restriction fragment was released and introduced into a similarly digested pMCEF− vector. pMCEF− was derived from pMC1neo [13].
DNA transfections
For transient transfection of A375P cells, 2.5 × 105 cells were seeded per well in a 6-well dish and incubated overnight. Lipofectamine (Invitrogen) was used according to manufacturer's instructions. Cell lysates were prepared 2 days following transfection. For generation of stable cell lines, A375P cells were transfected with the various vectors carrying a NeoR cassette using Lipofectamine and selected with G418 (Invitrogen). The cells were harvested for isolation of mRNA and positive clones selected by quantitative real-time PCR (qRT-PCR).
quantitative real-time PCR
Primers for each candidate gene were designed off 3′ regions of the cDNA sequences. GNAQ probe (Hs00387073_m1) and the GAPDH Endogenous Control probe were obtained from Applied Biosystems. For each reaction, 10 μl of Precision Mastermix (PrimerDesign), 1 μl of the probe and 25 ng of the cDNA template were added and made up with to 20 μl nuclease-free H2O. All samples were tested in triplicates and H2O was used as a negative control. qRT-PCR was carried out on an Applied Biosystems 7900HT Fast Real Time Machine (Applied Biosystems). Relative expression was calculated using the ΔΔCt method with GAPDH as an internal control.
data access
WGS and SNP array data will be available from the European Genome-phenome Archive under study accession number EGAS0000100058.
results
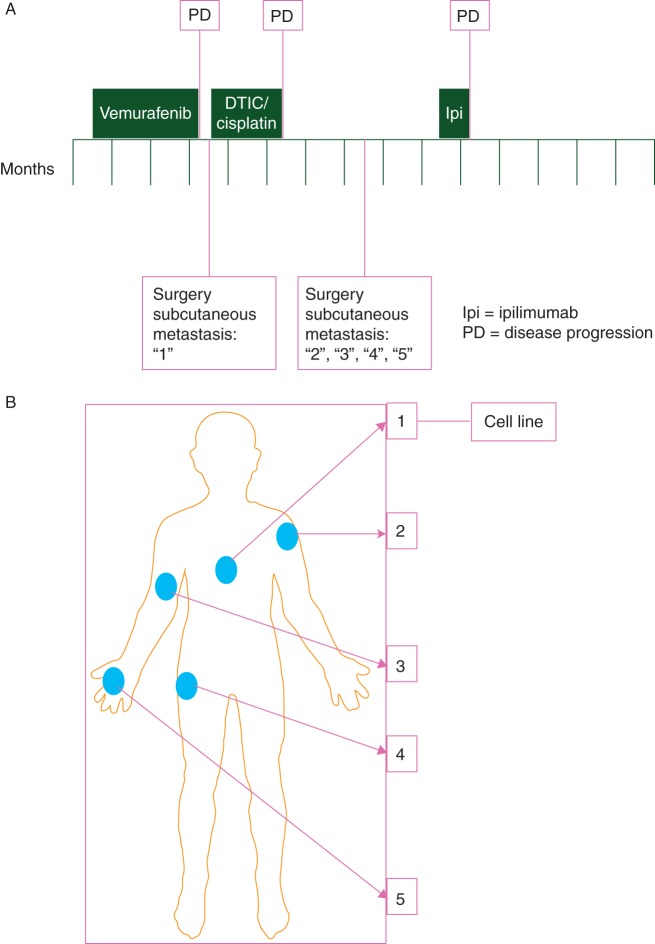
A 26-year-old male presented with a thick ulcerated cutaneous melanoma that, despite surgical resection, metastasised to the mediastinal lymph nodes, so systemic therapy was considered. Genotyping of archival tumour tissue revealed a T>A, p.V600E mutation in BRAF, and the patient was enrolled into a clinical trial with vemurafenib (BRIM-3) [14]. However, radiological assessment at 8 weeks revealed disease progression with a fivefold increase in the volume of the mediastinal mass and so vemurafenib was discontinued (Figure 1A). Five weeks later, a new subcutaneous tumour appeared and was removed (site 1; Figure 1A and B), and fresh tissue obtained for study. Two cycles of decarbazine and cisplatin were administered, but the patient derived no benefit. Seven weeks after completion of chemotherapy a further four subcutaneous tumours were removed (sites 2, 3, 4, 5; Figure 1A and B) and we obtained fresh tissue for study. A formalin-fixed subcutaneous metastasis removed 1 month before vemurafenib therapy was available as a baseline sample.
Figure 1.
Clinical case under study. (A) The clinical course of the patient is depicted over a period of 12 months, with time points and duration of different therapeutic interventions indicated. Metastases from sites ‘1’, ‘2’, ‘3’, ‘4’ and ‘5’ were obtained as fresh tissue. (B) Diagram showing the site of the subcutaneous sites from which metastatic tissue was removed. The tumour from site 1 was propagated as a continuous cell line.
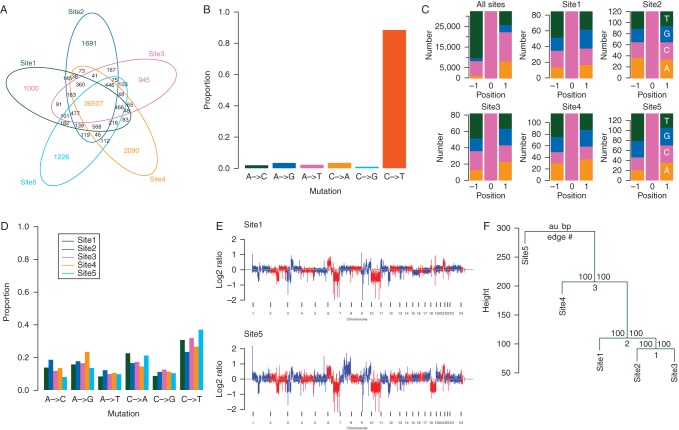
We confirmed that the T>A, p.V600E mutation in BRAF was present in all five tumours (supplementary Figure S1, available at Annals of Oncology online), so to investigate mechanisms of resistance to vemurafenib, we carried out WGS using the Complete Genomics platform [10] (supplementary Table S1, available at Annals of Oncology online). Compared with the patient's germline DNA, we observed ∼40 000 predicted somatic SNVs per tumour (supplementary Table S1, available at Annals of Oncology online), a level that is comparable with the mutation burden in other sun-exposed melanomas [15, 16]. The majority (36 507; 77%) of the SNVs were present in all tumours (Figure 2A) and decarbazine/cisplatin treatment did not significantly increase SNV load in tumours #2, #3, #4 and #5 (supplementary Table S1, available at Annals of Oncology online). Approximately 85% of the common somatic SNVs were C>T (G>A) transitions (Figure 2B; supplementary Table S1, available at Annals of Oncology online), of which over 90% were at the 3′ end of pyrimidine dinucleotides (Figure 2C, ‘All sites’; supplementary Table S1, available at Annals of Oncology online), consistent with a UV-induced DNA damage signature [17]. Notably, the high-confidence private SNVs from the individual tumours did not bear this signature (Figure 2C, sites 1–5; Figure 2D). Our data identified 283 predicted non-synonymous (ns) SNVs in one or more tumours, 219 of which were common to all five sites. Sanger sequencing revealed that the majority of the non-ubiquitous nsSNVs were false positives or miscalled germline variants (supplementary Table S2, available at Annals of Oncology online), and no nsSNVs were confirmed as exclusive. Genome wide, we predicted ∼424 somatic small deletions and ∼430 somatic small insertions, only 295 of which were present in all tumours. Sanger sequencing of these mutations confirmed only two coding region indels, both of which were present in all five tumours (supplementary Table S3, available at Annals of Oncology online).
Figure 2.
Genomic analysis of tumours. (A) Venn diagram showing numerical overlap in the high-confidence SNVs in tumour sites 1–5. (B) Graph showing mutation signature of the 36 507 common SNVs. (C) Stacked histograms showing the mutation context of the common C to T (G to A) transitions in all five tumours (All sites; top left panel), and for the high-confidence (somatic score ≥0) private C to T (G to A) transitions in each individual tumour (sites 1–5). The proportion of each nucleotide ±1 bp of the mutation is shown (A: orange, C: pink, G: blue, T: green). (D) Graph showing mutation signature of the high-confidence (somatic score ≥0) private (site-specific) SNVs for each individual tumour (sites 1–5). (E) Genome plots of somatic copy-number alterations in the site 1 (top panel) and site 5 (bottom panel) tumours as determined by normalised coverage across 2 kb windows. (F) Figure showing bootstrapped unsupervised clustering of the WGS copy-number alteration data. The approximately unbiased (au; left) P-value and bootstrap probability (bp; right) values are indicated.
We used WGS and SNP array analysis [11] to reveal somatic SVs and copy-number alterations (CNAs). The SV analysis predicted 89–129 high-confidence structural variants, 29 of which were common to all tumours (supplementary Figure S2 and Table S4, available at Annals of Oncology online). The CNA analysis revealed very similar landscapes for sites 1, 2 and 3, whereas sites 4 and 5 showed higher levels of aneuploidy (Figure 2E; supplementary Figure S3, available at Annals of Oncology online; supplementary Table S5, available at Annals of Oncology online). In particular, site 5 showed increased copy number of chromosome 7 (supplementary Figure S3, available at Annals of Oncology online), and this tumour presented higher levels of nuclear polymorphism (supplementary Figure S4, available at Annals of Oncology online, supplementary Table S6, available at Annals of Oncology online). Bootstrapped unsupervised clustering of the WGS CNA data revealed a phylogeny in which sites 1, 2 and 3 segregated from sites 4 and 5 (Figure 2F). We carried out FISH, and this revealed that the cells in sites 2 and 3 were homogeneous and had largely diploid nuclei, whereas those in sites 4 and 5 were aneuploid (supplementary Table S5, available at Annals of Oncology online). Specifically, site 4 presented three to five chromosomes per nucleus, and site 5 was comprised of a mixed cell population of small cells with predominantly diploid nuclei and large cells with up to 15 chromosomes per nucleus (supplementary Table S6, available at Annals of Oncology online). Thus, all tumours shared several common events, including being hemizygous for chromosome 10 (Figure 2E; supplementary Figure S3, available at Annals of Oncology online), but more recently, the heterogeneous cells in sites 4 and 5 appeared to have acquired additional chromosomal gains.
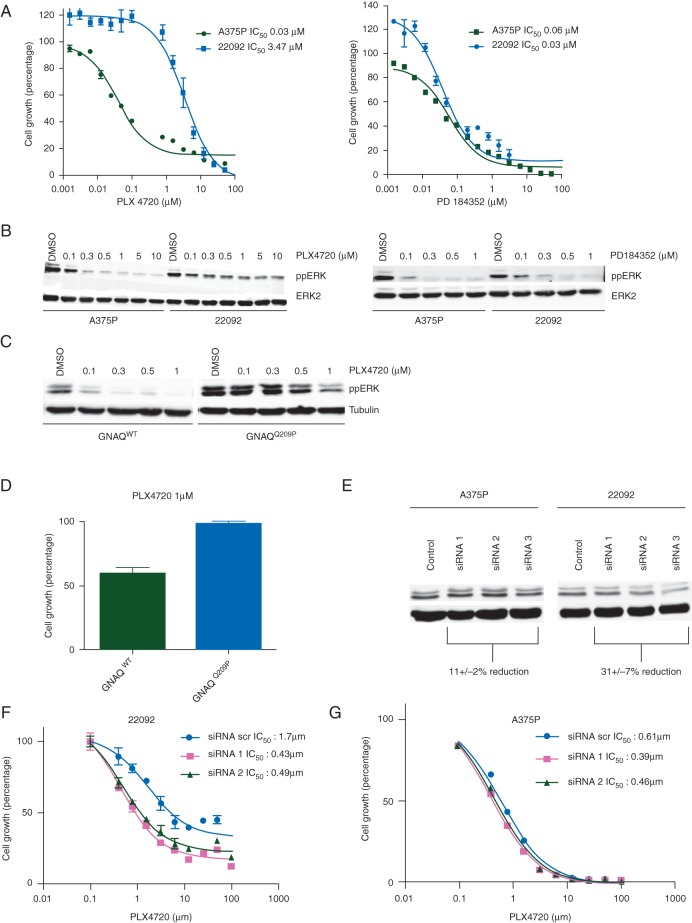
We established a continuous cell line (designated 22 092 cells) from tumour #1 and confirmed that the T>A, p.V600E mutation in BRAF was present in this line (supplementary Figure S5A, available at Annals of Oncology online). The cells stained positive for the melanoma markers HMB45 and S100 (supplementary Figure S5B, available at Annals of Oncology online), and the majority of the SNVs identified in tumour #1 persisted in this line (supplementary Figure S5C, available at Annals of Oncology online). Notably, compared with A375 melanoma cells, which also express BRAFV600E, the 22 092 cells were significantly less sensitive to the BRAF inhibitor PLX4720, but displayed equipotency to the MEK inhibitor PD184352 (Figure 3A). Accordingly, ERK phosphorylation was inhibited by PLX4720 in A375 cells, but persisted in 22 092 cells, whereas PD184352 inhibited ERK equally in both lines (Figure 3B). Thus, despite the BRAFV600E mutation, 22 092 cells were resistant to BRAF inhibitors.
Figure 3.
Oncogenic GNAQ contributes to BRAFi resistance. (A) Graph showing A375P and 22 092 cell growth in the presence of increasing concentrations of PLX4720 and PD184352. (B) Western blots for phospho-ERK (ppERK) and total ERK2 (loading control) in A375P and 22 092 cells treated with the indicated concentrations in micromolar of PLX4720 or PD184352 for 3 h. (C) Western blot for phospho-ERK (ppERK) and tubulin (loading control) in A375P cells expressing wild-type (GNAQWT) or mutant (GNAQQ209P) GNAQ and treated with the indicated concentrations of PLX4720 for 3 h. (D) Graph showing growth of A375P cells expressing wild-type (GNAQWT) or mutant (GNAQQ209P) GNAQ and treated with 1 μM PLX4720 for 10 days. Cell growth was quantified by crystal violet. (E) Western blot for phospho-ERK (ppERK) and tubulin (loading control) in A375P and 22 092 cells transfected with scrambled control (Control), or three different GNAQ siRNAs (siRNA 1, siRNA 2, siRNA 3) targeting GNAQ for 48 h. ppERK levels were quantified using ImageJ and the mean reduction (± SD) in levels is shown. (F) Graph showing growth of 22092 cells transfected with scrambled control (siRNA scr) or two GNAQ siRNAs (siRNA 1, siRNA 2) for 48 h and then treated with PLX4720 for 72 h. (G) Graph showing growth of A375P cells transfected with scrambled control (siRNA scr) or two GNAQ (siRNA 1, siRNA 2) siRNAs for 48 h and then treated with PLX4720 for 72 h.
No mutations were observed in RAS, MEK or ERK, but all five tumours harboured an A>C, p.Q209P mutation in GNAQ (supplementary Figure S6, available at Annals of Oncology online). Sanger sequencing of the pre-treatment lesion confirmed that this mutation was present before vemurafenib therapy (supplementary Figure S6, available at Annals of Oncology online). Activating mutations in GNAQ occur in ∼50% of uveal melanomas [18] but are rare in cutaneous melanoma [18, 19]. To date, three cases of concurrent mutations in GNAQ and BRAF have been reported in melanoma (http://www.cbioportal.org/), but the mutations in GNAQ (D333fs, G64A and P185S/A302G) were not at the common Q209 hotspot, so the functional significance of those alterations is unclear. However, oncogenic GNAQ activates ERK in uveal melanoma [18, 19, 20], so we investigated its effects in BRAF-mutant melanoma cells. When GNAQQ209P was expressed in A375 cells (supplementary Figure S7, available at Annals of Oncology online), it increased basal ERK phosphorylation and reduced ERK sensitivity to PLX4720 (Figure 3C) and, critically, it also reduced the sensitivity of A375 cells to PLX4720 (Figure 3D). Conversely, when GNAQQ209P was depleted in 22 092 cells using siRNA (supplementary Figure S8, available at Annals of Oncology online), ERK activity was suppressed, whereas depletion of wild-type GNAQ from A375 cells had less effect on ERK activity (Figure 3E). Notably, depletion of GNAQQ209P increased the sensitivity of 22 092 cells to PLX4720 (Figure 3F), whereas depletion of wild-type GNAQ did not increase A375 cell sensitivity to this drug (Figure 3G).
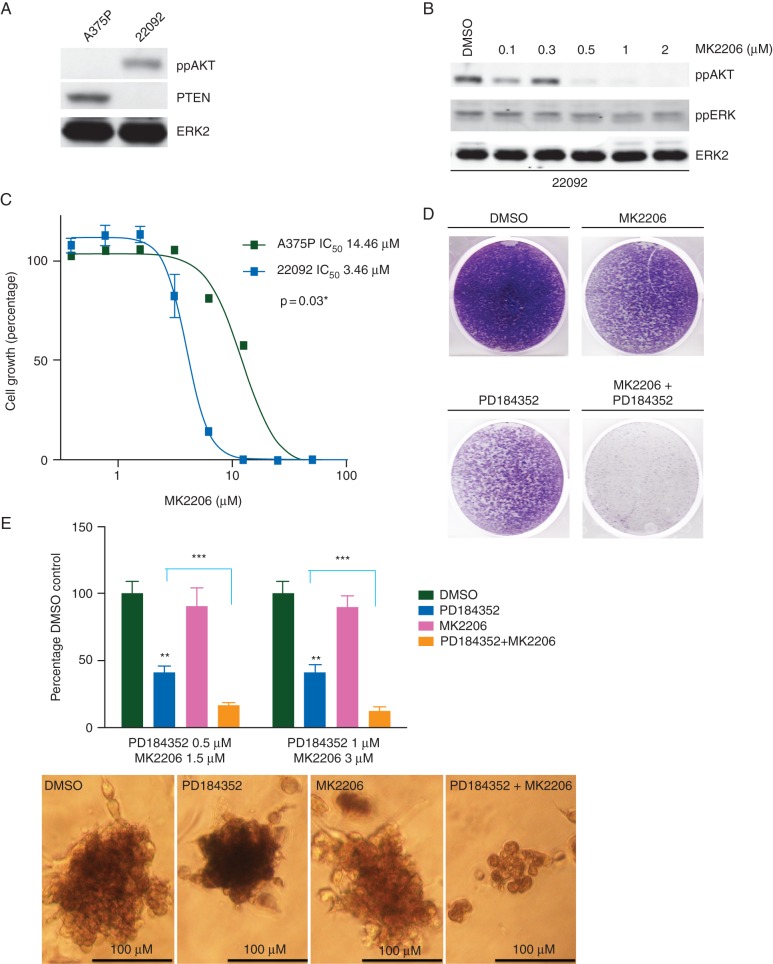
Intriguingly, one of the other confirmed mutations in all of the tumour samples, including the pre-treatment sample, was a 4-bp deletion in PTEN c.950_953delTACT (supplementary Figure S9, available at Annals of Oncology online) that introduces a frameshift at codon V317 and a stop at codon 319 within the C2 binding domain. Sanger sequencing revealed wild-type PTEN sequence traces in the tumours, but not the 22 092 cells (supplementary Figure S8, available at Annals of Oncology online), suggesting that the wild-type PTEN sequence in the tumour samples is the result of stromal contamination. Accordingly, we observed chromosome 10 loss in all tumours (Figure 2E; supplementary Figure S3, available at Annals of Oncology online) and observed that, in the 22 092 cells, PTEN was not expressed and AKT phosphorylation was elevated (Figure 4A). The allosteric AKT inhibitor MK2206 inhibited AKT phosphorylation in 22 092 cells (Figure 4B) and 22 092 cells were more sensitive to MK2206 than A375 cells (Figure 4C). Taken together, our data show that, in the tumours from this patient, ERK activity was elevated due to the GNAQ P209 mutation, and AKT activity was elevated by loss of PTEN expression. Accordingly, PD184352 and MK2206 synergised to inhibit the growth of 22 092 cells in long-term proliferation assays (Figure 4D), and when the cells were grown in suspension (Figure 4E).
Figure 4.
Loss of PTEN contributes to BRAFi resistance in patient cells (A) Western blot for phospho-AKT (ppAKT), PTEN and total ERK2 (loading control) in A375P and 22 092 cells. (B) Western blot for ppAKT, phospho-ERK (ppERK) and total ERK2 (loading control) in 22 092 cells treated with the indicated concentrations of MK2206 for 3 h. (C) Graph showing growth of A375P and 22 092 treated with increasing concentrations of MK2206 for 10 days. (D) Photograph showing crystal violet staining of 22 092 cells treated with DMSO, MK2206 (1 µM), PD184352 (0.1 µM), or both for 10 days. (E) Graph showing growth of 22 092 spheroids in collagen following treatment with the indicated concentrations of PD184352, MK2206 or both for 7 days. Representative images of the treated spheroids are shown below and were assessed by measuring individual spheroids with ImageJ software in triplicate for each condition. **P value ≤0.01 individual treatments compared with DMSO-treated control; ***P value ≤0.001 combination treatment compared with PD184352 alone.
discussion
Here, we analysed the genomes of five metastatic BRAFV600E melanomas from a patient who presented intrinsic resistance to vemurafenib. Previous studies have reported discordance in BRAF mutation status in different tumours from other patients [21] and this could drive differential response to BRAF inhibitors. However, we confirmed that the T>A, p.V600E mutation in BRAF persisted in all five sites and that it was present in the pre-treatment sample. Our WGS revealed an A>C, p.Q209P mutation in GNAQ that was also present in all five tumours. We demonstrated that GNAQQ209P sustained ERK activity in BRAF mutant melanoma cells in the presence of a BRAF inhibitor, allowing the cells to grow even when BRAFV600E was inhibited. Our sequencing also revealed a ubiquitous PTEN frame-shift deletion that was associated with AKT hyper-activation. Both GNAQ and PTEN mutations were detected in the pre-vemurafenib sample, observations that are consistent with features of intrinsic mechanism of resistance; however, since the first assessment was carried out at 8 weeks after starting treatment, we cannot exclude the possibility that the patient had a transient early response followed by rapid progression.
It has been reported that mutations in the RAS small G-proteins can mediate acquired resistance to BRAF inhibitors in BRAF mutant melanoma [6] and that GNAQ-mutant uveal melanomas do not respond to BRAF inhibitors [22]. However, mutations in GNAQ have not previously been reported to mediate resistance to BRAF inhibitors in BRAF-mutant melanoma. BRAF and GNAQ mutations are thought to be mutually exclusive and activating GNAQ mutations have not been reported to occur concomitant with BRAF mutations. It has been reported previously that PTEN mutations can mediate acquired resistance to BRAF inhibition in melanoma pre-clinical models [23, 24]. Although down-regulation of PTEN has an association with overall lower responses and shorter progression-free interval in patients [25, 26], the presence of PTEN alterations does not always preclude a response to BRAF inhibitors [9]. Critically, in our study, we show that both the MEK/ERK and AKT pathways needed to be targeted to mediate effective inhibition of proliferation of cells derived from this patient's tumour, suggesting that loss of PTEN did contribute to intrinsic resistance to BRAF inhibition in this patient.
An important consequence of the clonal nature of cancers is spatial and temporal variation in tumour composition [27]. Spatial heterogeneity has been demonstrated in several tumour types, including renal [28] and pancreatic [29] cancers. We observed a high degree of homogeneity at the single-nucleotide level in the tumours from this patient, but nevertheless observed more CNAs in the tumours at sites 4 and 5. Furthermore, at site 5 we observed a mixture of cells with distinct genomic and phenotypic features. This suggests that, while the tumours all evolved from a common metastatic progenitor, the cancer cells at sites 4 and 5 acquired additional genomic alterations after disease dissemination. We note that all of the tumours had lost a copy of chromosome 18 and that loss of 18q is linked to chromosomal instability [30]. Furthermore, although the DNA damaging agents cisplatin and decarbazine did not increase SNV burden or alter the mutation signature in sites 2, 3, 4 and 5 compared with site 1, we cannot exclude the possibility that these agents caused the additional chromosomal alterations we observed at sites 4 and 5. Critically, these findings of intra- and inter-lesional heterogeneity caution against the sampling bias that can occur when genomic data from single-tumour biopsies are analysed.
Our data reveal that resistance in this patient was not mediated by acquired mutations or sub-clonal evolution under selection pressure exerted by treatment, but by a combination of pre-existing mutations in two genes that activate at least two pro-growth/pro-survival pathways. These mutations were present in all tumours and predated vemurafenib administration, explaining why the patient presented intrinsic resistance. Two recent studies have also reported multiple mechanisms of acquired resistance to BRAF inhibitors within the same tumour biopsy [9, 31]. Together with the findings reported here, these observations highlight the genetic complexity that can drive drug resistance in melanoma patients.
We demonstrated that AKT and MEK inhibitors synergised to block the growth of the tumour cells, suggesting that combined inhibition of these pathways may have been an effective approach for this patient. Our findings show how comprehensive tumour profiling combined with knowledge of signalling biology can identify biomarkers that may predict treatment resistance and may also reveal effective drug combinations. Clearly, this will help formulate more effective therapeutic strategies for individual patients.
funding
This study was supported by Cancer Research UK (ref: C107/A10433, C5759/A12328), The Harry J Lloyd Charitable Trust and the Complete Genomics Cancer Grant Programme (ref: ICR_RM083111FA1).
disclosure
The authors have declared no conflicts of interest.
Supplementary Material
acknowledgements
We thank Kim Edmonds, Anna Carlisle and Jade Griffiths for assistance with tissue collection; Mahrokh Nohadani for tissue processing; and Saima Awan for assistance with Sanger sequencing. FISH studies were carried out by Advanced Diagnostics Lab, UCL, London.
references
- 1.Thirlwell C, Nathan P. Melanoma—part 2: management. BMJ. 2008;00:a2488. doi: 10.1136/bmj.a2488. [DOI] [PubMed] [Google Scholar]
- 2.Sosman JA, Kim KB, Schuchter L, et al. Survival in BRAF V600-mutant advanced melanoma treated with vemurafenib. N Engl J Med. 2012;366:707–714. doi: 10.1056/NEJMoa1112302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hauschild A, Grob JJ, Demidov LV, et al. Dabrafenib in BRAF-mutated metastatic melanoma: a multicentre, open-label, phase 3 randomised controlled trial. Lancet. 2012;380:358–365. doi: 10.1016/S0140-6736(12)60868-X. [DOI] [PubMed] [Google Scholar]
- 4.Jang S, Atkins MB. Which drug, and when, for patients with BRAF-mutant melanoma? Lancet Oncol. 2013;14:e60–e69. doi: 10.1016/S1470-2045(12)70539-9. [DOI] [PubMed] [Google Scholar]
- 5.Sullivan RJ, Flaherty KT. Resistance to BRAF-targeted therapy in melanoma. Eur J Cancer. 2013;49:1297–1304. doi: 10.1016/j.ejca.2012.11.019. [DOI] [PubMed] [Google Scholar]
- 6.Nazarian R, Shi H, Wang Q, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–977. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Romano E, Pradervand S, Paillusson A, et al. Identification of multiple mechanisms of resistance to vemurafenib in a patient with BRAFV600E-mutated cutaneous melanoma successfully rechallenged after progression. Clin Cancer Res. 2013;19(20):5749–57. doi: 10.1158/1078-0432.CCR-13-0661. [DOI] [PubMed] [Google Scholar]
- 8.Wilmott JS, Tembe V, Howle JR, et al. Intratumoral molecular heterogeneity in a BRAF-mutant, BRAF inhibitor-resistant melanoma: a case illustrating the challenges for personalized medicine. Mol Cancer Ther. 2012;11:2704–2708. doi: 10.1158/1535-7163.MCT-12-0530. [DOI] [PubMed] [Google Scholar]
- 9.Van Allen EM, Wagle N, Sucker A, et al. The genetic landscape of clinical resistance to RAF inhibition in metastatic melanoma. Cancer Discov. 2014;4:94–109. doi: 10.1158/2159-8290.CD-13-0617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drmanac R, Sparks AB, Callow MJ, et al. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science. 2010;327:78–81. doi: 10.1126/science.1181498. [DOI] [PubMed] [Google Scholar]
- 11.Van Loo P, Nordgard SH, Lingjaerde OC, et al. Allele-specific copy number analysis of tumors. Proc Natl Acad Sci USA. 2010;107:16910–16915. doi: 10.1073/pnas.1009843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Venkatraman ES, Olshen AB. A faster circular binary segmentation algorithm for the analysis of array CGH data. Bioinformatics. 2007;23:657–663. doi: 10.1093/bioinformatics/btl646. [DOI] [PubMed] [Google Scholar]
- 13.Marais R, Spooner RA, Light Y, et al. Gene-directed enzyme prodrug therapy with a mustard prodrug/carboxypeptidase G2 combination. Cancer Res. 1996;56:4735–4742. [PubMed] [Google Scholar]
- 14.Chapman PB, Hauschild A, Robert C, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berger MF, Hodis E, Heffernan TP, et al. Melanoma genome sequencing reveals frequent PREX2 mutations. Nature. 2012;485:502–506. doi: 10.1038/nature11071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pleasance ED, Cheetham RK, Stephens PJ, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463:191–196. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pfeifer GP, You YH, Besaratinia A. Mutations induced by ultraviolet light. Mutat Res. 2005;571:19–31. doi: 10.1016/j.mrfmmm.2004.06.057. [DOI] [PubMed] [Google Scholar]
- 18.Van Raamsdonk CD, Bezrookove V, Green G, et al. Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature. 2009;457:599–602. doi: 10.1038/nature07586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Raamsdonk CD, Griewank KG, Crosby MB, et al. Mutations in GNA11 in uveal melanoma. N Engl J Med. 2010;363:2191–2199. doi: 10.1056/NEJMoa1000584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khalili JS, Yu X, Wang J, et al. Combination small molecule MEK and PI3K inhibition enhances uveal melanoma cell death in a mutant GNAQ- and GNA11-dependent manner. Clin Cancer Res. 2012;18:4345–4355. doi: 10.1158/1078-0432.CCR-11-3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yancovitz M, Litterman A, Yoon J, et al. Intra- and inter-tumor heterogeneity of BRAF(V600E))mutations in primary and metastatic melanoma. PLoS One. 2012;7:e29336. doi: 10.1371/journal.pone.0029336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mitsiades N, Chew SA, He B, et al. Genotype-dependent sensitivity of uveal melanoma cell lines to inhibition of B-Raf, MEK, and Akt kinases: rationale for personalized therapy. Invest Ophthalmol Vis Sci. 2011;52:7248–7255. doi: 10.1167/iovs.11-7398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paraiso KH, Xiang Y, Rebecca VW, et al. PTEN loss confers BRAF inhibitor resistance to melanoma cells through the suppression of BIM expression. Cancer Res. 2011;71:2750–2760. doi: 10.1158/0008-5472.CAN-10-2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Villanueva J, Vultur A, Lee JT, et al. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer Cell. 2010;18:683–695. doi: 10.1016/j.ccr.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nathanson KL, Martin AM, Wubbenhorst B, et al. Tumor genetic analyses of patients with metastatic melanoma treated with the BRAF inhibitor dabrafenib (GSK2118436) Clin Cancer Res. 2013;19:4868–4878. doi: 10.1158/1078-0432.CCR-13-0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trunzer K, Pavlick AC, Schuchter L, et al. Pharmacodynamic effects and mechanisms of resistance to vemurafenib in patients with metastatic melanoma. J Clin Oncol. 2013;31:1767–1774. doi: 10.1200/JCO.2012.44.7888. [DOI] [PubMed] [Google Scholar]
- 27.Aparicio S, Caldas C. The implications of clonal genome evolution for cancer medicine. N Engl J Med. 2013;368:842–851. doi: 10.1056/NEJMra1204892. [DOI] [PubMed] [Google Scholar]
- 28.Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Campbell PJ, Yachida S, Mudie LJ, et al. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature. 2010;467:1109–1113. doi: 10.1038/nature09460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burrell RA, McClelland SE, Endesfelder D, et al. Replication stress links structural and numerical cancer chromosomal instability. Nature. 2013;494:492–496. doi: 10.1038/nature11935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi H, Hugo W, Kong X, et al. Acquired resistance and clonal evolution in melanoma during BRAF inhibitor therapy. Cancer Discov. 2014;4:80–93. doi: 10.1158/2159-8290.CD-13-0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.