Abstract
Background
Human bone marrow derived mesenchymal stem cells (hMSCs) are capable of differentiation into multiple cell lineages and demonstrate a wide variety of use in various therapeutic applications. Only recently has research begun to understand the gene expression profiles of hMSCs and their differentiated counterparts in vivo and ex vivo.
Purpose
The research presented here aimed at gaining a better understanding of gene expression patterns present during hMSC invasion through a basement membrane.
Methods
Changes in gene expression were evaluated between invasive and non-invasive cells using Agilent’s gene expression arrays and Matrigel invasion chambers. The cells were specifically attracted to a defined stem cell media called SCM.
Results
A total 435 genes were up-regulated by 2- fold or more in the invasive population of cells and classified into developmental programs and immunological/inflammatory signaling pathways determined by Ingenuity Pathway Analysis (IPA). This list included a variety of regulators of growth and differentiation including NANOG, STAT3 and STAT5A and members of the polycomb repressive complex-2 (PCRC2) EZH2 and SUZ12. The known regulator of inflammation and hypoxia HIF-1α was also increased suggesting that regulation of the microenvironment is important during this process. Finally, the invasion process could be reversed using the STAT3 inhibitor Static.
Conclusions
Overall these data will increase the understanding of the genetic pathways functioning during hMSC invasion and aid in the development of their therapeutic applications.
Keywords: hMSCs, Invasion, Gene expression, STAT3
Introduction
Adult human bone marrow derived mesenchymal stromal cells (hMSCs), also called mesenchymal stem cells, were first identified as highly proliferative/radiation resistant cells isolated from human bone marrow transplants [1]. These cells are capable of undergoing self-renewal and are multipotent, meaning that under the appropriate stimuli are capable of differentiating into multiple cell lineages including osteoblasts, chondrocytes, adipocytes and myoblasts [2–4]. With their diverse potential, these cells demonstrate a wide variety of use in many therapeutic applications such as osteogenesis imperfecta [5], myocardial infarction [6], muscular dystrophy [7], spinal cord injury [8,9], graft versus host disease [10,11] and have even recently been examined for their role in the treatment of cancer [12–14]. Previous research has characterized that injured or wounded tissues produce inflammatory cytokines which seem to attract hMSCs expressing CD29, CD44, CD51, CD73, CD105, CD166 and Stro-1 (bone morphogenic protein-3: BMP3) [15,16]. The expression of certain markers, however, does seem to be specific to the microenvironment. Additionally, the process of hypoxia, which is a reduction in the amount of available oxygen, is exhibited by a variety of tumors.
This process is regulated by hypoxia-inducible factor-1 α (HIF1α) [17] and injured tissues or tumors which are present in a hypoxic microenvironment produce these same angiogenic and inflammatory cytokines, further perpetuating the homing of hMSCs. Stem cell niches are often located in anatomical regions characterized by hypoxic conditions as they require low levels of oxygen to minimize damage caused by DNA oxidation. The relationship between oxygen and MSCs is under intense investigation as MSCs reside in locations close to the vascular structures, yet the tissues where MSCs are found exhibit low oxygen levels [18]. The exact mechanism(s) by which oxygen regulates MSCs is unknown, but it is clear that oxygen is a critical regulator of MSC fate.
The research presented here is aimed at gaining a better understanding of what these additional pathways regulating hMSCs might be. An in depth investigation of gene expression patterns present during hMSC invasion through a basement membrane was performed using a 44,000 probe-based gene expression array. These data have provided a unique gene expression signature of invasion toward a stem cell niche.
The research presented here is aimed at gaining a better understanding of what these additional pathways regulating hMSCs might be. An in depth investigation of gene expression patterns present during hMSC invasion through a basement membrane was performed using a 44,000 probe-based gene expression array. These data have provided a unique gene expression signature of invasion toward a stem cell niche.
Methods
Cell lines and reagents
Early passage (P3) human bone marrow derived mesenchymal stem cells (hMSCs) were obtained from Lonza (Gaithersburg, MD) and maintained using their recommended conditions with addition of 500 U/mL penicillin/500 μg/mL streptomycin and 0.250 μg/mL amphotericin-B all from Gibco (Invitrogen, Carlsbad, CA). The cultures were maintained in 5% CO2 air at 37°C. The STAT3 inhibitor Stattic (Calbiochem, Gibbstown, NJ) was dissolved in molecular grade ethanol.
Matrigel invasion assay
Matrigel-coated 24-well inserts (8 μM pore size) and non-coated control inserts purchased from BD Biosciences Clontech (Palo Alto, CA) were used according to manufacturer’s instructions. Between 60,000 to 100,000 cells were seeded for the invasion in serum-free RPMI and migrated toward media specific for stem cells (SCM) containing DMEM/F12 with human supplementation of 10 ng/mL bFGF, 20 ng/mL EGF and 5 μg/mL insulin along with 0.4% BSA (each from Sigma, St. Louis, MO). Routine invasion assays were performed for 24 hours and then stained with the Diffi-Quick Staining kit (Dade Behring, Deerfield, IL). The direct STAT-3 inhibitor Static was used at 1 μM. Three to five microscopic fields (20X) were photographed and counted for each sample. The experiment was repeated three independent times.
Microarray Analysis
RNA was isolated and labelled as previously described from invasive and non-invasive cells [19], with the following modifications: Reverse transcriptase was heat inactivated at 65°C for 10 minutes and again 100 ng of RNA was amplified using the Message Amp a RNA Amplification Kit. A total of 2 μg of Universal Reference RNA (Stratagene, La Jolla, CA) was labelled with Cy3-dUTP and experimental samples were labelled with Cy5-dUTP. Samples were hybridized to an Agilent whole genome gene expression array following manufacturer’s directions. Arrays were scanned using a GenePix 4000B scanner and extracted using the mAdb portal from the National Cancer Institute. The data was normalized using the Loess method and bad/or not found spots were excluded for extraction. The array data was further analyzed using Cluster and Tree view offered by Michael B. Eisen as freeware (http://rana.lbl.gov/EisenSoftware.htm) to produce the heat map. The complete list of genes whose expression changed ≥ 2- or ≤ 2- fold is available in Supplementary Table S1.
Proliferation Assays
Cells were seeded overnight in a 96 well plate in 100 μL of regular media at a density of 2000 cells per well. Cells were treated with either DSMO or 1 μM Stattic. Cell proliferation was measured using the CellTiter-Glo assay from Promega (Madison, WI, USA) after 24 hours using 100 μL of reagent and an incubation time of 20 minutes. The relative luciferase units (RLU) were quantified using a Tecan Infinite 200 plate reader. Samples were performed using an N of 8.
Quantitative Real Time Polymerase Chain Reaction (QRT-PCR)
Total RNA was isolated using TRIzol (Invitrogen Corporation, Carlsbad, CA). RNA from ‘top’ cells was isolated using a cell pellet acquired from trypsinizing cells from one membrane after bottom cell were removed with a cotton swab. Conversely, RNA from the bottom cells was isolated by combining three membranes where the top cells were removed using a cotton swab. The membranes were pooled and placed in TRIzol for 10 minutes at room termperature, and the conventional procedure for isolation of RNA was then followed. To increase the yield of RNA, 5 μg of linear arcylamide (Ambion, Austin, TX) was added prior to precipitation of RNA with isopropanol. Additional to increase overall yield, 100 ng of RNA was amplified using the Message Amp aRNA Amplification Kit (Ambion, Austin, TX). cDNA was prepared using the SuperScript®III First-Strand Synthesis System (Invitrogen Corporation, Carlsbad, CA). Quantitative real time polymerase chain reaction (QRT-PCR) analysis was performed using a StepOne Real-time PCR machine (Applied Biosystems, Foster City, CA) with TaqMan Gene Expression Assay reagents and probes (Applied Biosystems). A total of 4 μL of cDNA was used in a 20 μL reaction resulting in a 1:5 dilution. The following SYBR based human probes were used: ER-β (Hs01112040_m1), EZH2 (Hs00544830_ m1), HIF1α (Hs00153153_m1), NANOG (Hs02387400_g1), STAT3 (Hs01047580_m1), STAT5A (Hs00559643_m1), SUZ12 (Hs00248742_ m1) and 18S rRNA (Hs99999901_s1). Relative fold induction of mRNA was compared between top and bottom cells using the Delta-Delta CT method of quantitation, and 18S rRNA was used as a loading control.
Ingenuity and Genomatrix Software
Agilent databases (excel 2003 format) were uploaded for analysis using software available at http://www.ingenuity.com/ or http://www.genomatix.de/
Statistical Analysis
Using Graph Pad Prisim (Version 4) a Two-way ANOVA with a Bonferroni post test was performed to compare groups and * represents a p-value of <0.05 comparing invasive to non-invasive cells.
Results
Microarray analysis of invasive hMSCs
Initially it was determined that a small percentage (~20%) of hMSC were able to invade though the Matrigel toward a highly defined media called stem cell media (SCM) previously shown to attract invasive prostate cancer stem cells [19–21] (Figure 1A). A very small number of cells were able to migrate across the control membrane toward SCM, demonstrating the importance of Matrigel (or a basement membrane) in hMSC homing. To determine which genes are differentially regulated within this population of invaded cells compared to the non-invasive cells, a gene expression microarray was performed using Agilent whole genome expression array (1×44K). Arrays were extracted using the mAdb Gateway from the National Cancer Institute (NCI) and after background subtraction and normalization to universal RNA, a total of 435 genes were found to be up-regulated by 2-fold or more in the invasive population of cells (Supplemental Table 1).
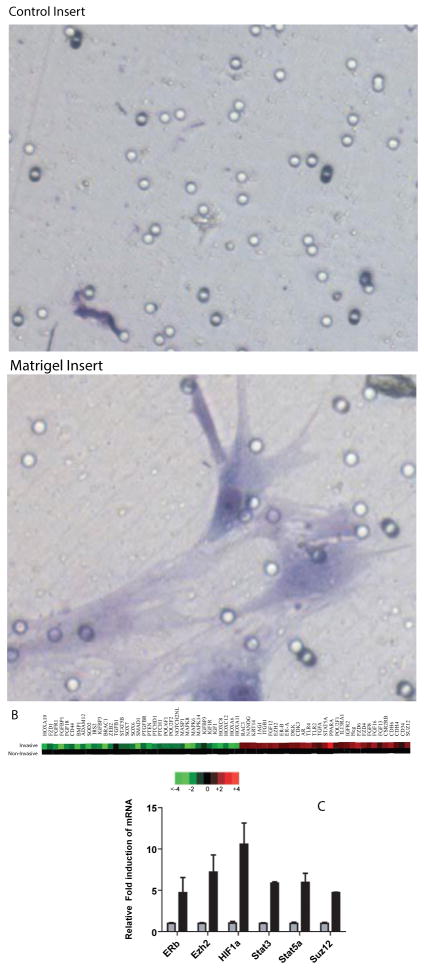
Figure 1. Gene expression changes in invasive hMSCs.
A) Microscopic images (20X) of cells invaded toward SCM after 24 hours using either control inserts or inserts containing Matrigel. Cells were staining with the Diffi-Quick Staining kit.
B) Heat Maps demonstrating increased (red) or decreased (green) expression of a select number genes from the invasive compared to non-invasive cells. Samples were hybridized to an Agilent whole genome gene expression array following manufacturer’s directions. Arrays were scanned using a GenePix 4000B scanner (Molecular Devices, Sunnyvale, CA) and analyzed using Cluster and Treeview offered by Michael B. Eisen as freeware (http://rana.lbl.gov/EisenSoftware.htm). The complete list of genes whose expression changed ≥1.8- or ≤1.8-fold is available in Supplemental Table S1.
C)A select number of up-regulated genes were verified using qRT-PCR for increased expression in the invasive cells. Gray bars represent the non-invasive cell expression normalized to 1 and black bars are relative fold-induction of mRNA from invasive cells. Fold induction was calculated using the Delta-Delta CT method where the non-invasive cells were set at 1.0 as the control, and 18S rRNA was used as a loading control. Data is shown as transformed log2. A Twoway ANOVA with a Bonferroni post-test was performed to compare groups and * represents a p-value of < 0.05 comparing invasive to non-invasive cells.
Heat maps generated by using TreeView demonstrated the variety of genes related to ‘stemness’ and migration/invasion/metastasis (Figure 1B and 1C) which increased in the invasive cells. When carefully analyzing the specific genes in the array that are increased after invasion (Table 1), a variety of receptors were found to up-regulated including androgen receptor (AR), estrogen receptor-β (ER-β or ESR2) and a number of immune based receptors increased including interleukin-13 receptor-α1 (IL13RA1) and toll-like receptor-2 and 4 (TLR2 and TRL4). Growth and differentiation genes such as fibroblast growth factor-12 (FGF12) and insulin-like growth factor-2 receptor (IGF2) were also increased. The stem cell regulator NANOG and additional genes implicated in regulation of stem cells including cytokeratin-14 (KRT14), intergrin- β1 (ITGβ1), the Ras protein ras-related C3 botulinum toxin substrate-3 (RAC3), the notch-1 receptor ligand jagged-1 (JAG1) and finally the signal transducer and activator of transcription-5A (STAT5A) were also increased. Two members of the polycomb repressive complex-2 (PCRC2) enhancer of zeste-2 (EZH2) and suppressor of zeste-12 (SUZ12) were increased as well.
Table 1. Genes up-regulated in invasive hMSC cells determined by microarray analysis.
Significant changes in selected genes demonstrating a 1.6-fold or higher increase. The Agilent ID corresponds to the probe ID from the array, values for non-invasive and invasive hMSCs as well as their fold change (Invasive-Non-Invasive) are provided.
| Agilent ID | hMSC Non- Invasive | hMSC_Invasive | Fold Change | UNIQID | Gene Name |
|---|---|---|---|---|---|
| A_24_P919916 | −1.7055 | 5.8604 | 7.5659 | WID:6488014 | PTF1A |
| A_23_P42065 | −2.8123 | 0.9734 | 3.7857 | WID:6484664 | TNFRSF21 |
| A_23_P30736 | −2.4033 | 0.8032 | 3.2065 | WID:6496619 | HLA-DOB |
| A_23_P60306 | −1.43 | 1.7307 | 3.1607 | WID:6502707 | TLR4 |
| A_23_P113111 | −1.7102 | 1.2511 | 2.9613 | WID:6488464 | AR |
| A_23_P77223 | −1.0517 | 1.9013 | 2.953 | WID:6513234 | MESP1 |
| A_23_P210763 | −2.7955 | 0.1168 | 2.9123 | WID:6486716 | JAG1 |
| A_24_P38387 | −3.0463 | −0.1468 | 2.8995 | WID:6504089 | NDRG1 |
| A_32_P122579 | 0.6126 | 3.5082 | 2.8956 | WID:6480399 | EZH2 |
| A_23_P207367 | −2.2092 | 0.5955 | 2.8047 | WID:6481655 | STAT5A |
| A_23_P84320 | −1.8188 | 0.8944 | 2.7132 | WID:6495148 | HMX1 |
| A_24_P265346 | −2.8338 | −0.1382 | 2.6956 | WID:6504873 | KRT14 |
| A_23_P77440 | −2.7784 | −0.1771 | 2.6013 | WID:6502102 | NFATC3 |
| A_32_P160537 | 2.0805 | 4.6637 | 2.5832 | WID:6492904 | FGF12 |
| A_23_P206441 | −2.1401 | 0.3806 | 2.5207 | WID:6586963 | FANCA |
| A_24_P598836 | 1.5009 | 3.9523 | 2.4514 | WID:6517296 | ITGB1 |
| A_23_P214011 | −1.9233 | 0.4855 | 2.4088 | WID:6494488 | CDH6 |
| A_23_P23947 | −1.2674 | 1.096 | 2.3634 | WID:6489062 | MAP3K8 |
| A_23_P156953 | −1.7175 | 0.6025 | 2.32 | WID:6517216 | IGF2R |
| A_23_P125001 | −0.4863 | 1.8332 | 2.3195 | WID:6482994 | RAC3 |
| A_23_P204640 | −4.0766 | −1.7784 | 2.2982 | WID:6487751 | NANOG |
| A_23_P92499 | 0.3371 | 2.5755 | 2.2384 | WID:6486692 | TLR2 |
| A_24_P889103 | −2.3142 | 0.6211 | 2.9353 | WID:6516766 | SUZ12P |
| A-23_P54100 | −1.5936 | 0.4465 | 2.0401 | WID:6506930 | ESR2 |
| A_23_P137196 | −1.3207 | 0.6947 | 2.0154 | WID:6488748 | IL13RA1 |
| A_23_P46964 | 0.0039 | 1.6847 | 1.6808 | WID:6478939 | HIF1AN |
| A_23_P100795 | −0.1833 | 1.42 | 1.6083 | WID:6480615 | STAT3 |
qRT-PCR confirmation of microarray data
Small panels of these genes were analyzed for confirmation of their expression patterns using qRT-PCR (Fig. 1C). The genes analyzed included a 4.7-fold increase in ER-β, as well as a 7.2- fold increase in the PCRC2 members EZH2 and 4.7-fold increase in SUZ12. Finally, the growth regulators STAT3 and STAT5A demonstrated a 5.8-fold and 5.9- fold increase, respectively, and although STAT3 was only up-regulated by 1.6-fold in the array we further analyzed the gene due its growing importance in stem cell maintenance. The stem cell gene NANOG was found to increased 2.29-fold in the array (Table 1), yet only 1.4-fold by qRT-PCR (data not shown).
Ingenuity pathway analysis
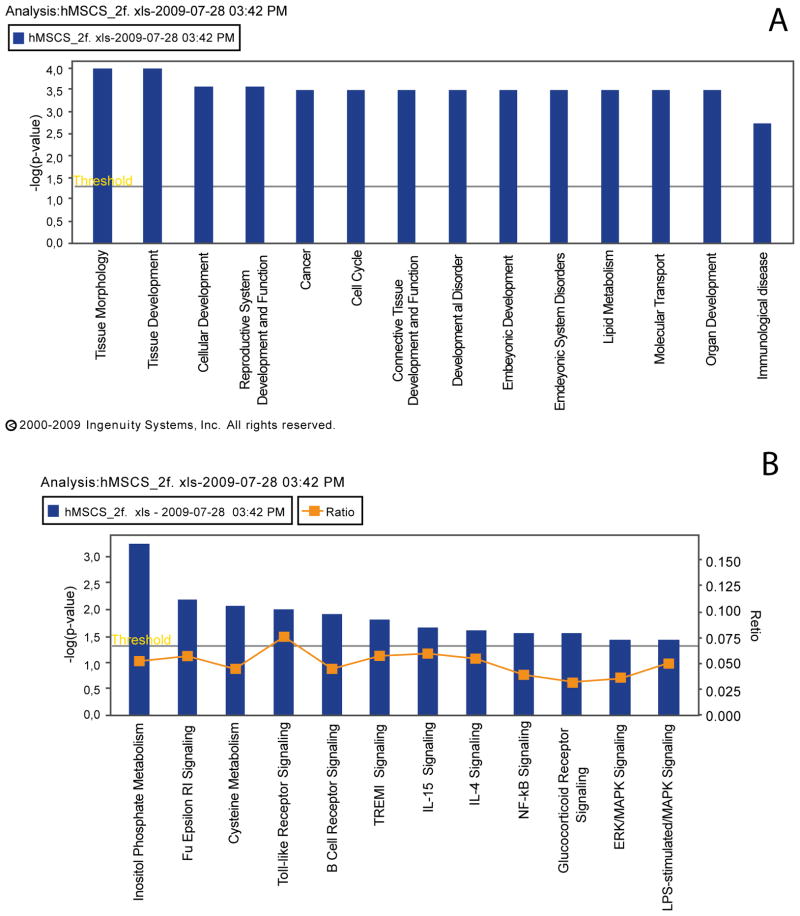
The majority of genes demonstrating an increase in expression classified into developmental programs including tissue development, cellular development, reproductive system development, connective tissue development, embryonic development and organ development (Figure 2A). A number of genes also belong to the immunological disease category (Figure 2A) and the canonical immunological/inflammatory signalling pathways including Fc Epsilon RI signalling, Toll-like receptor signalling, B-Cell receptor signalling, TREM1 signalling, IL-15 and IL-4 signalling and finally the NFκB pathway (Figure 2B).
Figure 2. Ingenuity pathway analysis demonstrating significant changes.
(A) genes in developmental pathways
(B) immunological based pathways in invasive hMSC cells.
Any pathway above the yellow threshold bar demonstrates significant changes in gene expression
STAT3 regulates hMSC invasion
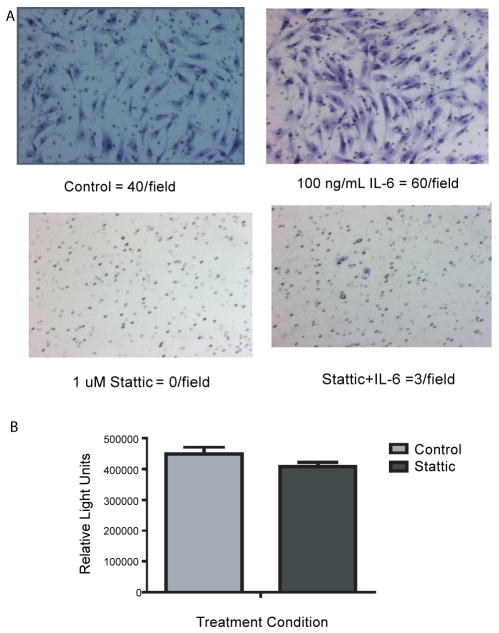
Recent data suggests a number of these pathways that are increasing with invasion of hMSCs intersect using the STAT3 pathway [22–25]. Using this same invasion assay, our lab has recently demonstrated that the invasion of prostate cancer cells toward SCM can be blocked using the STAT3 inhibitor Stattic [26]. This prompted us to test if Stattic could also block invasion of hMSCs toward SCM. In the presence of Stattic, invasion of the hMSCs was completely abolished without affecting normal cell proliferation, suggesting a strong role for the STAT proteins in mediating this process (Figure 3A and 3B).
Figure 3. Inhibition of invasive hMSCs toward SCM using the STAT3 inhibitor Stattic.
A) Matrigel invasion assays were seeded with 60,000 cells per well in RPMI and invaded toward SCM with or without 100 ng/mL of IL-6, and with or without 1 μM Stattic. After 24 hours the cells were stained using the Diffi-Quick Staining kit and 4 independent fields were counted and averaged. The experiment was repeated twice and each time no cells were able to invade toward SCM in the presence of Stattic.
B) Cell-titer glow experiment for cell proliferation of hMSCs treated with or without 1 μM Stattic for 24 hours
Discussion
Human bone marrow derived mesenchymal stem cells (hMSCs) have the potential to revolutionize medicine with their donor specific treatment of diseases. hMSCs represent an attractive model for a number of reasons, including the lack of ethical controversy regarding their isolation since the cells themselves can be isolated from the patient’s own blood/tissues [1], as well as their lack of significant immunogenicity allowing for allogenic transplantation without the use of potentially harmful immunosuppressive drugs [27]. Albeit as provocative as this system may seem there is still much unknown about the behaviour of these patient derived hMSCs. In order to begin to determine which genes might be differentially regulated during the process of hMSCs invasion we analyzed changes in gene expression patterns of the cells invading through a basement membrane of Matrigel. Using the defined media SCM we were able to identity a specific population of invasive hMSCs moving across the Matrigel membrane. This demonstrates that only a portion of the MSCs have the ability to be attracted by the SCM. Clearly, it can be assumed that the invasive cells express receptors for the components of the SCM, yet there is a significant amount of cross-talk and activation that occurs in response to signalling by each EGF, bFGF and insulin independently and synergistically when used in combination. From the microarray, we found that 435 genes were up-regulated by 2-fold or more after normalization and background subtraction. An increase in a number of classical chemokine receptors such as IL13RA1and TLR2 and TLR4 was observed, in addition to an increase in AR and ER-β. Traditionally these sex hormone receptors were not thought to be expressed on stem cells, but in a recent review by Ray et al., the authors summarize their roles in the differentiation of hMSCs and embryonic stem (ES) cells [28]. For example, in four independent mouse ES cell line there is functional expression of AR which can interact with transfected androgen response elements [29]. In addition, treatment of hMSCs with 17β-estradiaol is able to help induce osteogenic differentiation [30]. Furthermore, AR expression has been validated on CD34+ cells [31] and with regards to more tissue specific stem cells, AR expression has been seen in putative stem cell population from prostate tumor lines [32]. The increased expression of these sex steroid hormone receptors within the invasive population of hMSCs could be utilized for further differentiation of the cells depending on the environment and/or niche to which they are homed to. The increased expression of TLRs are very interesting in the context of this study. Toll-like receptors (TLRs) involved in mediating stress responses of hMSCs occur through the activation of many pathways, including NFκB, AKT and MAPK [33]. Additionally, activation of TLRs with their various ligands, including lipopolysaccharide (LPS)-induced expression of a variety of chemokines and cytokines including C-X-C motif chemokine 10 (CXCL10), interferon-1β (INF1β) and interleukin-6 (IL-6) resulted in an enhanced migratory ability [33]. Furthermore, the importance and role of TLRs, as well as other growth hormones, chemokine/cytokines receptors and adhesion molecules expressed by hMSCs is summarized nicely by Spaeth et al. [16] in a review regarding ‘the migratory itinerary of mesenchymal stem cells.’ Observing an increased expression of TLR2 and TLR4 in the invasive cells was not surprising based on this evidence. Evolutionarily conserved co-expression of transcription factors in human ES cells have been investigated for a number of years, and of interest to our research is the overlap in expression of the such genes as NANOG and STATs [34]. Our data demonstrates an increase in each of these gene families within the invasive population of hMSCs. As previously mentioned, we observed a significant increase in STAT5A within the invasive cells, and a slight increase in STAT3, both of which were validated by qRT-PCR. The role of STATs in the regulation of stem cells has been investigated for a number of years, but only recently has their role in the regulation of hMSCs, tissue specific stem cells and invasion surfaced within the literature [24,25,35–41]. To further determine the importance of STATs/IL-6 since this has been a longstanding interest in our lab, we placed the STAT3 specific inhibitor Stattic in the chemoattractive media and we observed a complete block in invasion (Figure 3). Although addition of exogenous IL-6 did slightly increase invasion, the presence of this inhibitor was again able to block this invasion (Figure 3). The observation that inflammatory regulator HIF-1α was also increased in the invasive cells was of interest and in direct relation to the STAT data. Recently, it has been shown that HIF-1α regulates IL-6 in low oxygen environments, and IL- 6 increases when HIF-1α is active [42]. A recent review relates the relationships of HIFs with such pathways governing stem cell regulation such as octamer binding transcription factor 4 (OCT4), bone morphogenic proteins (BMPs), NOTCH, sonic hedgehog (SHH) and β-catenin/WNT [43]. Thus, taken together with our data, we believe that IL-6/STAT plays a key role in hMSC invasion and homing and should be taken into consideration when developing therapeutic uses of hMSCs. Additionally, selective withdrawal of IL-6 and induction with tissue specific factors could ensure proper timing and specificity of hMSCs differentiation. Additional changes we observed that we believe are important in the regulation of hMSC homing and invasion based on previous research by our lab and others include increases in gene members of the Glinsky Signature (SUZ12 and EZH2), which determine likelihood of death from metastatic disease [44–46]. EZH2 is involved in histone methylation and deacetylation in stem cells and a recent paper shows that retroviral overexpression of EZH2 in mouse embryonic fibroblasts (MEFs) resulted in bypassing of the senescence program [47]. With regards to normal HSCs, which were rapidly exhausted after serial transplantations, overexpression of EZH2 completely conserved long-term repopulating potential [47]. Finally, recent data suggests that EZH2 regulated by STAT3 is correlated to the pathological stage and progression of prostate cancer [48].
Conclusion
Overall, we have determined that a number of genes involved in immunological maintenance, developmental processes and the regulation of ‘stemness’ are increased in invasive hMSCs. By understanding the pathways functioning during hMSC invasion we believe this will aid in the development of their therapeutic applications.
Supplementary Material
Acknowledgments
This work was supported by the Division of Preclinical Innovation, National Center for Advancing Translational Sciences; the Molecular Libraries Initiative of the National Institutes of Health Roadmap for Medical Research; the Intramural Research Program of the National Human Genome Research Institute and the Frederick National Laboratory for Cancer Research, National Institutes of Health including contract HHSN261200800001E and grant # U54CA143930. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This Research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. The authors would also like to thank Dr. Stephanie Cabarcas from the lab for extensive editing of this manuscript.
Abbreviations
- hMSCs
Human Bone Marrow Derived Mesenchymal Stem Cells
- SCM
Stem Cell Media
- qRT-PCR
Quantitative Real Time Polymerase Chain Reaction
- IPA
Ingenuity Pathway Analysis
- STAT3
Signal Transducer And Activator Of Transcription 3
- STAT5A
Signal Transducer And Activator Of Transcription 5A
- PCRC2
Polycomb Repressive Complex-2
- EZH2
Enhancer of Zeste-2
- SUZ12
Suppressor of Zeste-12
- HIF-1α
Hypoxia- Inducible Factor-1 Alpha
- EGF
Epidermal Growth Factor
- FGF
Fibroblast Growth Factor
- ERβ
Estrogen Receptor Beta
- NCI
National Cancer Institute
- AR
Androgen Receptor
- IL13RA1
Interleukin-13 Receptor Alpha-1
- TLR2/4
Toll-Like Receptor-2/4
- FGF12/7
Fibroblast Growth Factor-12/716
- IGF2
Insulin-Like Growth Factor-2
- KRT14
Cytokeratin-14
- ITGβ1/4
Intergrin Beta-1/4
- RAC3
Ras Related C3 Botulinum Toxin Substrate-3
- JAG1
Jagged-1
- TREM1
Triggering Receptor Expressed On Myeloid Cells-1
- IL
Interleukin
- NFκB
Nuclear Factor Kappa B
- ES
Embryonic Stem
- AKT
Protein Kinase B
- MAPK
Mitogen Activated Protein Kinase
- INF1β
Interferon 1 Beta
- CXCR4/10
C-X-C Chemokine Receptor Type-4/10
- OCT4
Octamer-Binding Transcription Factor 4
- BMPs
Bone Morphogenic Proteins
- SHH
Sonic Hedgehog
- WNT
Wingless-Type MMTV Integration Site Family
- EPCs
Endothelial Precursor Cells
- VCAM-1
Vascular Cellular Adhesion Molecule-1
- MEF
Mouse Embryonic Fibroblast
References
- 1.Golde DW, Hocking WG, Quan SG, Sparkes RS, Gale RP. Origin of human bone marrow fibroblasts. Br J Haematol. 1980;44:183–187. doi: 10.1111/j.1365-2141.1980.tb01200.x. [DOI] [PubMed] [Google Scholar]
- 2.Friedenstein AJ. Stromal mechanisms of bone marrow: cloning in vitro and retransplantation in vivo. Haematol Blood Transfus. 1980;25:19–29. doi: 10.1007/978-3-642-67319-1_3. [DOI] [PubMed] [Google Scholar]
- 3.Owen M. Lineage of osteogenic cells and their relationship to the stromal system. Journal of Bone and Mineral Research. 1985:1–25. [Google Scholar]
- 4.Dexter TM. Stromal cell associated haemopoiesis. J Cell Physiol. 1982:87–94. doi: 10.1002/jcp.1041130414. [DOI] [PubMed] [Google Scholar]
- 5.Caplan AI. Osteogenesis imperfecta, rehabilitation medicine, fundamental research and mesenchymal stem cells. Connect Tissue Res. 1995;31:S9–14. doi: 10.3109/03008209509116826. [DOI] [PubMed] [Google Scholar]
- 6.Pittenger MF, Martin BJ. Mesenchymal stem cells and their potential as cardiac therapeutics. Circ Res. 2004;95:9–20. doi: 10.1161/01.RES.0000135902.99383.6f. [DOI] [PubMed] [Google Scholar]
- 7.Gonçalves MA, de Vries AA, Holkers M, van de Watering MJ, van der Velde I, et al. Human mesenchymal stem cells ectopically expressing full-length dystrophin can complement Duchenne muscular dystrophy myotubes by cell fusion. Hum Mol Genet. 2006;15:213–221. doi: 10.1093/hmg/ddi438. [DOI] [PubMed] [Google Scholar]
- 8.Zhao LR. Human bone marrow stem cells exhibit neural phenotypes and ameliorate neurological deficits after grafting into the ischemic brain of rats. Exp Neurol. 2000;174:11–20. doi: 10.1006/exnr.2001.7853. [DOI] [PubMed] [Google Scholar]
- 9.Sanchez-Ramos J, Song S, Cardozo-Pelaez F, Hazzi C, Stedeford T, et al. Adult bone marrow stromal cells differentiate into neural cells in vitro. Exp Neurol. 2000;164:247–256. doi: 10.1006/exnr.2000.7389. [DOI] [PubMed] [Google Scholar]
- 10.Ikehara S. A novel strategy for allogeneic stem cell transplantation: perfusion method plus intra-bone marrow injection of stem cells. Exp Hematol. 2003;31:1142–1146. doi: 10.1016/j.exphem.2003.08.020. [DOI] [PubMed] [Google Scholar]
- 11.van Laar JM, Tyndall A. Adult stem cells in the treatment of autoimmune diseases. Rheumatology (Oxford) 2006;45:1187–1193. doi: 10.1093/rheumatology/kel158. [DOI] [PubMed] [Google Scholar]
- 12.Nakamizo A, Marini F, Amano T, Khan A, Studeny M, et al. Human bone marrow-derived mesenchymal stem cells in the treatment of gliomas. Cancer Res. 2005;65:3307–3318. doi: 10.1158/0008-5472.CAN-04-1874. [DOI] [PubMed] [Google Scholar]
- 13.Stoff-Khalili MA, Rivera AA, Mathis JM, Banerjee NS, Moon AS, et al. Mesenchymal stem cells as a vehicle for targeted delivery of CRAds to lung metastases of breast carcinoma. Breast Cancer Res Treat. 2007;105:157–167. doi: 10.1007/s10549-006-9449-8. [DOI] [PubMed] [Google Scholar]
- 14.Qiao L, Xu ZL, Zhao TJ, Ye LH, Zhang XD. Dkk-1 secreted by mesenchymal stem cells inhibits growth of breast cancer cells via depression of Wnt signalling. Cancer Lett. 2008;269:67–77. doi: 10.1016/j.canlet.2008.04.032. [DOI] [PubMed] [Google Scholar]
- 15.Ries C, Egea V, Karow M, Kolb H, Jochum M, et al. MMP-2, MT1-MMP, and TIMP-2 are essential for the invasive capacity of human mesenchymal stem cells: differential regulation by inflammatory cytokines. Blood. 2007;109:4055–4063. doi: 10.1182/blood-2006-10-051060. [DOI] [PubMed] [Google Scholar]
- 16.Spaeth E, Klopp A, Dembinski J, Andreeff M, Marini F. Inflammation and tumor microenvironments: defining the migratory itinerary of mesenchymal stem cells. Gene Ther. 2008;15:730–738. doi: 10.1038/gt.2008.39. [DOI] [PubMed] [Google Scholar]
- 17.Dvorak HF. Tumors: wounds that do not heal. Similarities between tumor stroma generation and wound healing. N Engl J Med. 1986;315:1650–1659. doi: 10.1056/NEJM198612253152606. [DOI] [PubMed] [Google Scholar]
- 18.Mohyeldin A, Garzón-Muvdi T, Quiñones-Hinojosa A. Oxygen in stem cell biology: a critical component of the stem cell niche. Cell Stem Cell. 2010;7:150–161. doi: 10.1016/j.stem.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 19.Hurt EM, Kawasaki BT, Klarmann GJ, Thomas SB, Farrar WL. CD44+ CD24(−) prostate cells are early cancer progenitor/stem cells that provide a model for patients with poor prognosis. Br J Cancer. 2008;98:756–765. doi: 10.1038/sj.bjc.6604242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mathews LA, Hurt EM, Zhang X, Farrar WL. Epigenetic regulation of CpG promoter methylation in invasive prostate cancer cells. Mol Cancer. 2010;9:267. doi: 10.1186/1476-4598-9-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klarmann GJ, Hurt EM, Mathews LA, Zhang X, Duhagon MA, et al. Invasive prostate cancer cells are tumor initiating cells that have a stem cell-like genomic signature. Clin Exp Metastasis. 2009;26:433–446. doi: 10.1007/s10585-009-9242-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim HL, Cassone M, Otvos L, Jr, Vogiatzi P. HIF-1alpha and STAT3 client proteins interacting with the cancer chaperone Hsp90: therapeutic considerations. Cancer Biol Ther. 2008;7:10–14. doi: 10.4161/cbt.7.1.5458. [DOI] [PubMed] [Google Scholar]
- 23.Xu Q, Briggs J, Park S, Niu G, Kortylewski M, et al. Targeting Stat3 blocks both HIF-1 and VEGF expression induced by multiple oncogenic growth signaling pathways. Oncogene. 2005;24:5552–5560. doi: 10.1038/sj.onc.1208719. [DOI] [PubMed] [Google Scholar]
- 24.Rattigan Y, Hsu JM, Mishra PJ, Glod J, Banerjee D. Interleukin 6 mediated recruitment of mesenchymal stem cells to the hypoxic tumor milieu. Exp Cell Res. 2010;316:3417–3424. doi: 10.1016/j.yexcr.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 25.Nilsson CL, Dillon R, Devakumar A, Shi SD, Greig M, et al. Quantitative phosphoproteomic analysis of the STAT3/IL-6/HIF1alpha signaling network: an initial study in GSC11 glioblastoma stem cells. J Proteome Res. 2010;9:430–443. doi: 10.1021/pr9007927. [DOI] [PubMed] [Google Scholar]
- 26.Mathews LA, Hurt EM, Zhang X, Farrar WL. Epigenetic regulation of CpG promoter methylation in invasive prostate cancer cells. Mol Cancer. 2010;9:267. doi: 10.1186/1476-4598-9-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pansky A, Roitzheim B, Tobiasch E. Differentiation potential of adult human mesenchymal stem cells. Clin Lab. 2007;53:81–84. [PubMed] [Google Scholar]
- 28.Ray R, Novotny NM, Crisostomo PR, Lahm T, Abarbanell A, et al. Sex steroids and stem cell function. Mol Med. 2008;14:493–501. doi: 10.2119/2008-00004.Ray. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goldman-Johnson DR, de Kretser DM, Morrison JR. Evidence that androgens regulate early developmental events, prior to sexual differentiation. Endocrinology. 2008;149:5–14. doi: 10.1210/en.2007-1123. [DOI] [PubMed] [Google Scholar]
- 30.Wang Q, Yu JH, Zhai HH, Zhao QT, Chen JW, et al. Temporal expression of estrogen receptor alpha in rat bone marrow mesenchymal stem cells. Biochem Biophys Res Commun. 2006;347:117–123. doi: 10.1016/j.bbrc.2006.06.070. [DOI] [PubMed] [Google Scholar]
- 31.Sinha-Hikim I, Taylor WE, Gonzalez-Cadavid NF, Zheng W, Bhasin S. Androgen receptor in human skeletal muscle and cultured muscle satellite cells: up-regulation by androgen treatment. J Clin Endocrinol Metab. 2004;89:5245–5255. doi: 10.1210/jc.2004-0084. [DOI] [PubMed] [Google Scholar]
- 32.Sharifi N, Hurt EM, Farrar WL. Androgen receptor expression in prostate cancer stem cells: is there a conundrum? Cancer Chemother Pharmacol. 2008;62:921–923. doi: 10.1007/s00280-007-0659-5. [DOI] [PubMed] [Google Scholar]
- 33.Tomchuck SL, Zwezdaryk KJ, Coffelt SB, Waterman RS, Danka ES, et al. Toll-like receptors on human mesenchymal stem cells drive their migration and immunomodulating responses. Stem Cells. 2008;26:99–107. doi: 10.1634/stemcells.2007-0563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun Y, Li H, Liu Y, Mattson MP, Rao MS, et al. Evolutionarily conserved transcriptional co-expression guiding embryonic stem cell differentiation. PLoS One. 2008;3:e3406. doi: 10.1371/journal.pone.0003406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dahéron L, Opitz SL, Zaehres H, Lensch MW, Andrews PW, et al. LIF/STAT3 signaling fails to maintain self-renewal of human embryonic stem cells. Stem Cells. 2004;22:770–778. doi: 10.1634/stemcells.22-5-770. [DOI] [PubMed] [Google Scholar]
- 36.Taga T, Fukuda S. Role of IL-6 in the neural stem cell differentiation. Clin Rev Allergy Immunol. 2005;28:249–256. doi: 10.1385/CRIAI:28:3:249. [DOI] [PubMed] [Google Scholar]
- 37.Katoh M, Katoh M. STAT3-induced WNT5A signaling loop in embryonic stem cells, adult normal tissues, chronic persistent inflammation, rheumatoid arthritis and cancer (Review) Int J Mol Med. 2007;19:273–278. [PubMed] [Google Scholar]
- 38.Liang J, Wan M, Zhang Y, Gu P, Xin H, et al. Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nat Cell Biol. 2008;10:731–739. doi: 10.1038/ncb1736. [DOI] [PubMed] [Google Scholar]
- 39.Pricola KL, Kuhn NZ, Haleem-Smith H, Song Y, Tuan RS. Interleukin-6 maintains bone marrow-derived mesenchymal stem cell stemness by an ERK1/2-dependent mechanism. J Cell Biochem. 2009;108:577–588. doi: 10.1002/jcb.22289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sherry MM, Reeves A, Wu JK, Cochran BH. STAT3 is required for proliferation and maintenance of multipotency in glioblastoma stem cells. Stem Cells. 2009;27:2383–2392. doi: 10.1002/stem.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang M, Atkinson RL, Rosen JM. Selective targeting of radiation-resistant tumor-initiating cells. Proc Natl Acad Sci U S A. 2010;107:3522–3527. doi: 10.1073/pnas.0910179107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Das R, Jahr H, van Osch GJ, Farrell E. The role of hypoxia in bone marrow-derived mesenchymal stem cells: considerations for regenerative medicine approaches. Tissue Eng Part B Rev. 2010;16:159–168. doi: 10.1089/ten.TEB.2009.0296. [DOI] [PubMed] [Google Scholar]
- 43.Keith B, Simon MC. Hypoxia-inducible factors, stem cells, and cancer. Cell. 2007;129:465–472. doi: 10.1016/j.cell.2007.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Glinsky GV. “Stemness” genomics law governs clinical behavior of human cancer: implications for decision making in disease management. J Clin Oncol. 2008;26:2846–2853. doi: 10.1200/JCO.2008.17.0266. [DOI] [PubMed] [Google Scholar]
- 45.Glinsky GV. Stem cell origin of death-from-cancer phenotypes of human prostate and breast cancers. Stem Cell Rev. 2007;3:79–93. doi: 10.1007/s12015-007-0011-9. [DOI] [PubMed] [Google Scholar]
- 46.Gebauer G, Krones-Herzig A, Glinskii AB, Glinsky GV. Identification of genes associated with tumor progression using microarrays. Anticancer Res. 2005;25:1477–1482. [PubMed] [Google Scholar]
- 47.Kamminga LM, Bystrykh LV, de Boer A, Houwer S, Douma J, et al. The Polycomb group gene Ezh2 prevents hematopoietic stem cell exhaustion. Blood. 2006;107:2170–2179. doi: 10.1182/blood-2005-09-3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yeh HY, Cheng SW, Lin YC, Yeh CY, Lin SF, et al. Identifying significant genetic regulatory networks in the prostate cancer from microarray data based on transcription factor analysis and conditional independency. BMC Med Genomics. 2009;2:70. doi: 10.1186/1755-8794-2-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.